The Roles of Histone Post-Translational Modifications in the Formation and Function of a Mitotic Chromosome
Abstract
1. Introduction
2. Chromosome Condensation
3. Histone Modifications, CENP-A Incorporation, and Kinetochore Function
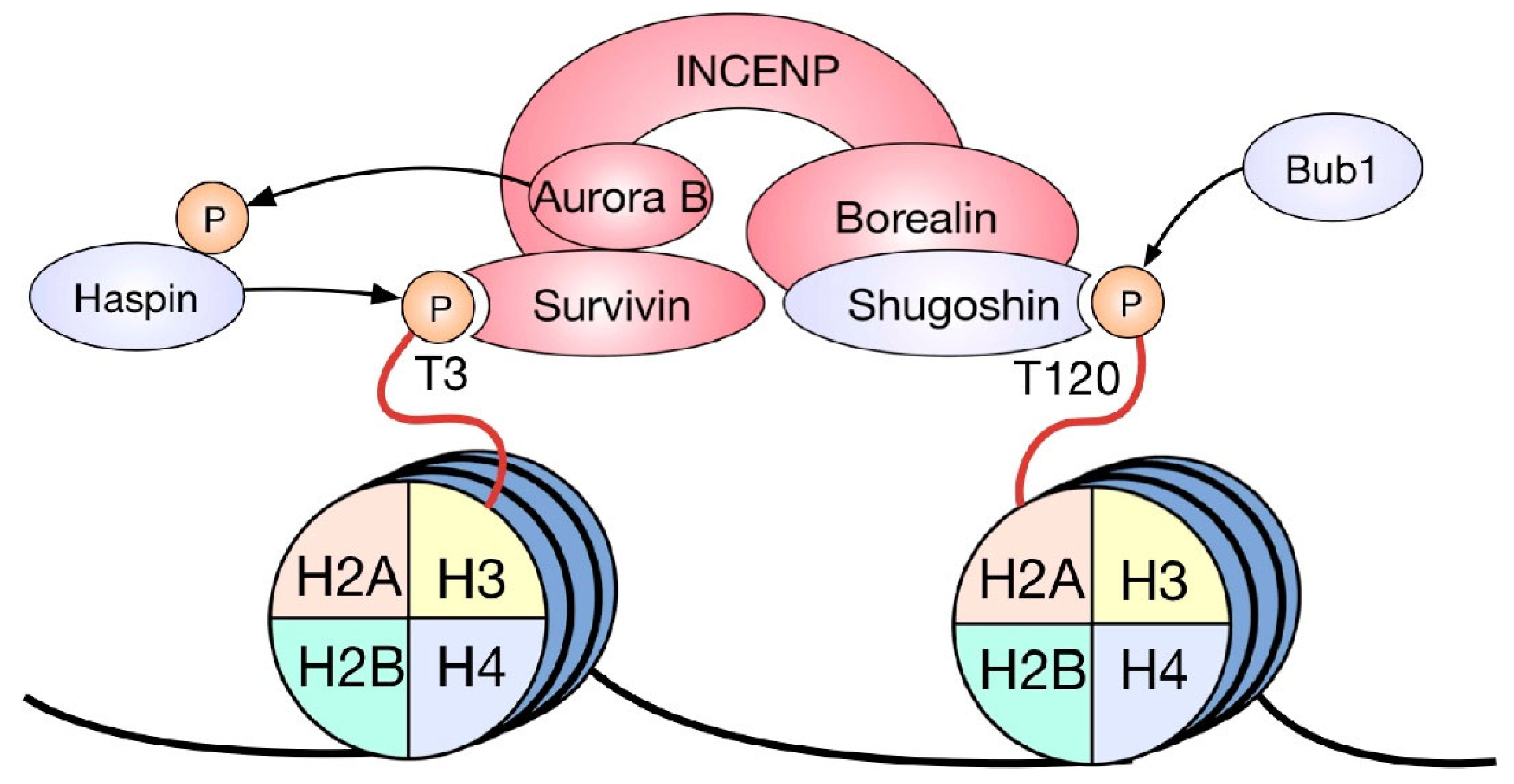
3.1. HPTMs and the Chromosomal Passenger Complex
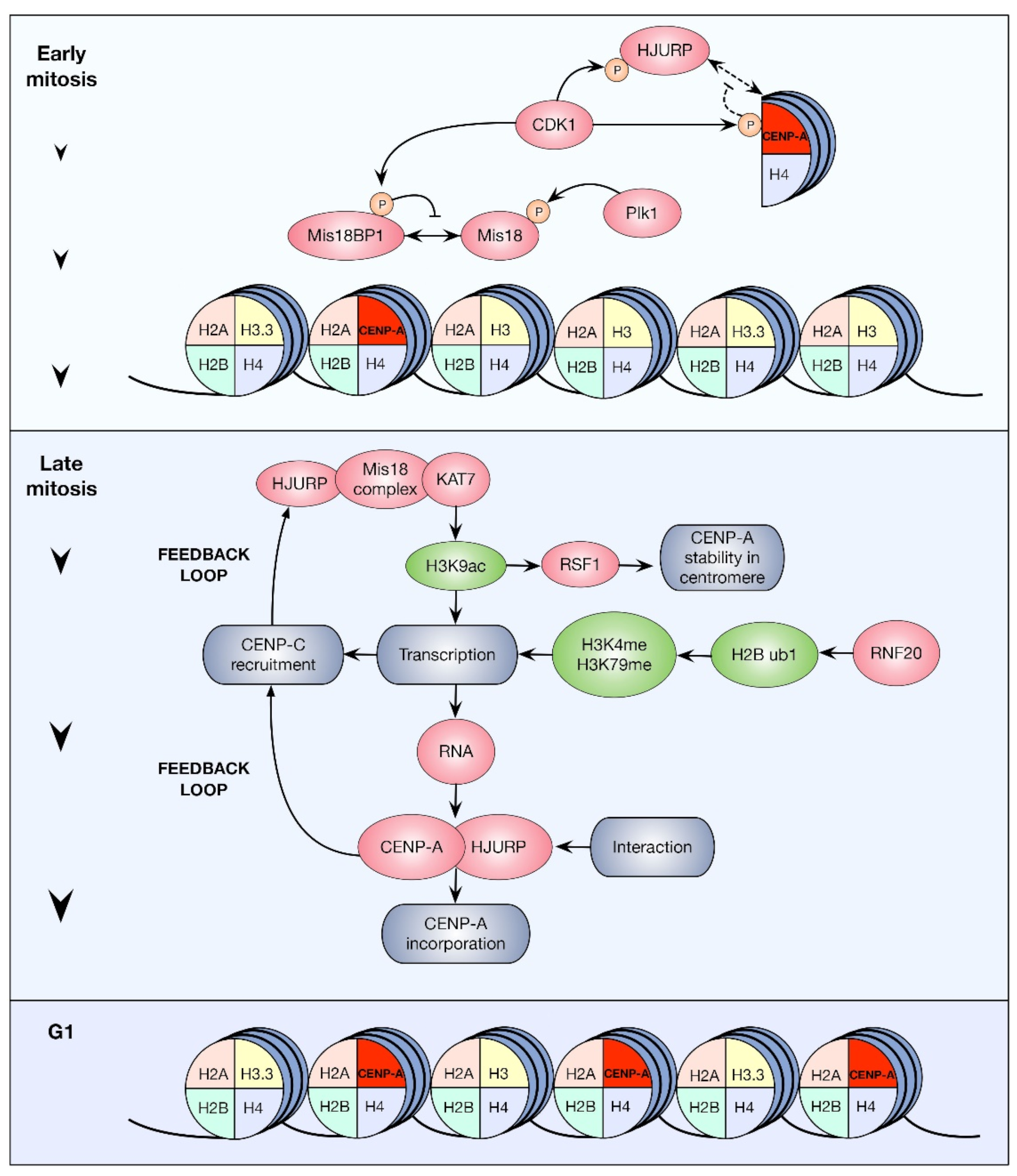
3.2. HPTMs and CENP-A Centromeric Incorporation
4. HPTMs on SAC Activity
5. Environmental Factors, Histone Post-Translational Modifications, and Genome Maintenance
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cutter, A.R.; Hayes, J.J. A Brief Review of Nucleosome Structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef]
- DesJarlais, R.; Tummino, P.J. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry 2016, 55, 1584–1599. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.P.; Zhao, Y.T.; Zhao, T.C. Histone Deacetylases and Mechanisms of Regulation of Gene Expression. Crit. Rev. Oncog. 2015, 20, 35–47. [Google Scholar] [CrossRef]
- Verdin, E.; Ott, M. 50 Years of Protein Acetylation: From Gene Regulation to Epigenetics, Metabolism and Beyond. Nat. Rev. Mol. Cell Biol. 2015, 16, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The Interplay of Histone Modifications—Writers That Read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, K.J.; Scelfo, A.; Jammula, S.; Cuomo, A.; Barozzi, I.; Stützer, A.; Fischle, W.; Bonaldi, T.; Pasini, D. Polycomb-Dependent H3K27me1 and H3K27me2 Regulate Active Transcription and Enhancer Fidelity. Mol. Cell 2014, 53, 49–62. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone Phosphorylation: A Chromatin Modification Involved in Diverse Nuclear Events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Du, H.-N. Transcription, DNA Damage and beyond: The Roles of Histone Ubiquitination and Deubiquitination. Curr. Protein Pept. Sci. 2012, 13, 447–466. [Google Scholar] [CrossRef]
- Man, T.; Witt, H.; Peterman, E.J.G.; Wuite, G.J.L. The Mechanics of Mitotic Chromosomes. Q. Rev. Biophys. 2021, 54, e10. [Google Scholar] [CrossRef]
- Stellfox, M.E.; Bailey, A.O.; Foltz, D.R. Putting CENP-A in Its Place. Cell. Mol. Life Sci. 2013, 70, 387–406. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, D.W.; Mao, Y.; Sullivan, K.F. Centromeres and Kinetochores: From Epigenetics to Mitotic Checkpoint Signaling. Cell 2003, 112, 407–421. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The Chromosomal Passenger Complex (CPC): From Easy Rider to the Godfather of Mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef]
- Zheng, Y.; Sanchez-Guerra, M.; Zhang, Z.; Joyce, B.T.; Zhong, J.; Kresovich, J.K.; Liu, L.; Zhang, W.; Gao, T.; Chang, D.; et al. Traffic-Derived Particulate Matter Exposure and Histone H3 Modification: A Repeated Measures Study. Environ. Res. 2017, 153, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Shintomi, K.; Takahashi, T.S.; Hirano, T. Reconstitution of Mitotic Chromatids with a Minimum Set of Purified Factors. Nat. Cell Biol. 2015, 17, 1014–1023. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; Van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-Specific Phosphorylation of Histone H3 Initiates Primarily within Pericentromeric Heterochromatin during G2 and Spreads in an Ordered Fashion Coincident with Mitotic Chromosome Condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef]
- Wei, Y.; Yu, L.; Bowen, J.; Gorovsky, M.A.; Allis, C.D. Phosphorylation of Histone H3 Is Required for Proper Chromosome Condensation and Segregation. Cell 1999, 97, 99–109. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Sun, Z.W.; Li, X.; Reuben, M.; Tatchell, K.; Bishop, D.K.; Grushcow, J.M.; Brame, C.J.; Caldwell, J.A.; Hunt, D.F.; et al. Mitotic Phosphorylation of Histone H3 Is Governed by Ipl1/Aurora Kinase and Glc7/PP1 Phosphatase in Budding Yeast and Nematodes. Cell 2000, 102, 279–291. [Google Scholar] [CrossRef]
- Adams, R.R.; Maiato, H.; Earnshaw, W.C.; Carmena, M. Essential Roles of Drosophila Inner Centromere Protein (INCENP) and Aurora B in Histone H3 Phosphorylation, Metaphase Chromosome Alignment, Kinetochore Disjunction, and Chromosome Segregation. J. Cell Biol. 2001, 153, 865–880. [Google Scholar] [CrossRef]
- Ditchfield, C.; Johnson, V.L.; Tighe, A.; Ellston, R.; Haworth, C.; Johnson, T.; Mortlock, A.; Keen, N.; Taylor, S.S. Aurora B Couples Chromosome Alignment with Anaphase by Targeting BubR1, Mad2, and Cenp-E to Kinetochores. J. Cell Biol. 2003, 161, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Van Hooser, A.; Goodrich, D.W.; Allis, C.D.; Brinkley, B.R.; Mancini, M.A. Histone H3 Phosphorylation Is Required for the Initiation, but Not Maintenance, of Mammalian Chromosome Condensation. J. Cell Sci. 1998, 111 Pt 23, 3497–3506. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, B.J.; Rall, N.A.; Ostwal, Y.; Kruitwagen, T.; Hiragami-Hamada, K.; Winkler, M.; Barral, Y.; Fischle, W.; Neumann, H. A Cascade of Histone Modifications Induces Chromatin Condensation in Mitosis. Science 2014, 343, 77–80. [Google Scholar] [CrossRef]
- Kruitwagen, T.; Denoth-Lippuner, A.; Wilkins, B.J.; Neumann, H.; Barral, Y. Axial Contraction and Short-Range Compaction of Chromatin Synergistically Promote Mitotic Chromosome Condensation. Elife 2015, 4, e10396. [Google Scholar] [CrossRef]
- Mora-Bermúdez, F.; Gerlich, D.; Ellenberg, J. Maximal Chromosome Compaction Occurs by Axial Shortening in Anaphase and Depends on Aurora Kinase. Nat. Cell Biol. 2007, 9, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Petrova, B.; Dehler, S.; Kruitwagen, T.; Hériché, J.-K.; Miura, K.; Haering, C.H. Quantitative Analysis of Chromosome Condensation in Fission Yeast. Mol. Cell Biol. 2013, 33, 984–998. [Google Scholar] [CrossRef]
- Vagnarelli, P.; Hudson, D.F.; Ribeiro, S.A.; Trinkle-Mulcahy, L.; Spence, J.M.; Lai, F.; Farr, C.J.; Lamond, A.I.; Earnshaw, W.C. Condensin and Repo-Man-PP1 Co-Operate in the Regulation of Chromosome Architecture during Mitosis. Nat. Cell Biol. 2006, 8, 1133–1142. [Google Scholar] [CrossRef]
- Qian, J.; Lesage, B.; Beullens, M.; Van Eynde, A.; Bollen, M. PP1/Repo-Man Dephosphorylates Mitotic Histone H3 at T3 and Regulates Chromosomal Aurora B Targeting. Curr. Biol. 2011, 21, 766–773. [Google Scholar] [CrossRef]
- Xin, G.; Fu, J.; Luo, J.; Deng, Z.; Jiang, Q.; Zhang, C. Aurora B Regulates PP1γ-Repo-Man Interactions to Maintain the Chromosome Condensation State. J. Biol. Chem. 2020, 295, 14780–14788. [Google Scholar] [CrossRef]
- Wike, C.L.; Graves, H.K.; Hawkins, R.; Gibson, M.D.; Ferdinand, M.B.; Zhang, T.; Chen, Z.; Hudson, D.F.; Ottesen, J.J.; Poirier, M.G.; et al. Aurora-A Mediated Histone H3 Phosphorylation of Threonine 118 Controls Condensin I and Cohesin Occupancy in Mitosis. Elife 2016, 5, e11402. [Google Scholar] [CrossRef]
- De la Barre, A.E.; Angelov, D.; Molla, A.; Dimitrov, S. The N-Terminus of Histone H2B, but Not That of Histone H3 or Its Phosphorylation, Is Essential for Chromosome Condensation. EMBO J. 2001, 20, 6383–6393. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Lu, S.; Shi, Y.; Pan, Y.; Limsakul, P.; Chernov, A.V.; Qiu, J.; Chai, X.; Shi, Y.; Wang, P.; et al. Coordinated Histone Modifications and Chromatin Reorganization in a Single Cell Revealed by FRET Biosensors. Proc. Natl. Acad. Sci. USA 2018, 115, E11681–E11690. [Google Scholar] [CrossRef] [PubMed]
- Legartová, S.; Lochmanová, G.; Bártová, E. The Highest Density of Phosphorylated Histone H1 Appeared in Prophase and Prometaphase in Parallel with Reduced H3K9me3, and HDAC1 Depletion Increased H1.2/H1.3 and H1.4 Serine 38 Phosphorylation. Life 2022, 12, 798. [Google Scholar] [CrossRef]
- McManus, K.J.; Biron, V.L.; Heit, R.; Underhill, D.A.; Hendzel, M.J. Dynamic Changes in Histone H3 Lysine 9 Methylations: Identification of a Mitosis-Specific Function for Dynamic Methylation in Chromosome Congression and Segregation. J. Biol. Chem. 2006, 281, 8888–8897. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.O.; Panchenko, T.; Shabanowitz, J.; Lehman, S.M.; Bai, D.L.; Hunt, D.F.; Black, B.E.; Foltz, D.R. Identification of the Post-Translational Modifications Present in Centromeric Chromatin. Mol. Cell. Proteom. 2016, 15, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.A.; Karpen, G.H. Centromeric Chromatin Exhibits a Histone Modification Pattern That Is Distinct from Both Euchromatin and Heterochromatin. Nat. Struct. Mol. Biol. 2004, 11, 1076–1083. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Weir, J.R.; Musacchio, A. Progress in the Structural and Functional Characterization of Kinetochores. Curr. Opin. Struct. Biol. 2016, 37, 152–163. [Google Scholar] [CrossRef]
- Ruchaud, S.; Carmena, M.; Earnshaw, W.C. Chromosomal Passengers: Conducting Cell Division. Nat. Rev. Mol. Cell Biol. 2007, 8, 798–812. [Google Scholar] [CrossRef]
- Van der Horst, A.; Lens, S.M.A. Cell Division: Control of the Chromosomal Passenger Complex in Time and Space. Chromosoma 2014, 123, 25–42. [Google Scholar] [CrossRef]
- Kim, S.; Kim, N.H.; Park, J.E.; Hwang, J.W.; Myung, N.; Hwang, K.-T.; Kim, Y.A.; Jang, C.-Y.; Kim, Y.K. PRMT6-Mediated H3R2me2a Guides Aurora B to Chromosome Arms for Proper Chromosome Segregation. Nat. Commun. 2020, 11, 612. [Google Scholar] [CrossRef]
- Lampson, M.A.; Cheeseman, I.M. Sensing Centromere Tension: Aurora B and the Regulation of Kinetochore Function. Trends Cell Biol. 2011, 21, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, Y.; Honda, T.; Tanno, Y.; Watanabe, Y. Two Histone Marks Establish the Inner Centromere and Chromosome Bi-Orientation. Science 2010, 330, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Dai, J.; Daum, J.R.; Niedzialkowska, E.; Banerjee, B.; Stukenberg, P.T.; Gorbsky, G.J.; Higgins, J.M.G. Histone H3 Thr-3 Phosphorylation by Haspin Positions Aurora B at Centromeres in Mitosis. Science 2010, 330, 231–235. [Google Scholar] [CrossRef]
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin Reads Phosphorylated Histone H3 Threonine 3 to Activate the Mitotic Kinase Aurora B. Science 2010, 330, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Earnshaw, W.C. The Centromere: Chromatin Foundation for the Kinetochore Machinery. Dev. Cell 2014, 30, 496–508. [Google Scholar] [CrossRef]
- McAinsh, A.D.; Meraldi, P. The CCAN Complex: Linking Centromere Specification to Control of Kinetochore-Microtubule Dynamics. Semin. Cell Dev. Biol. 2011, 22, 946–952. [Google Scholar] [CrossRef]
- Srivastava, S.; Zasadzińska, E.; Foltz, D.R. Posttranslational Mechanisms Controlling Centromere Function and Assembly. Curr. Opin. Cell Biol. 2018, 52, 126–135. [Google Scholar] [CrossRef]
- Takada, M.; Zhang, W.; Suzuki, A.; Kuroda, T.S.; Yu, Z.; Inuzuka, H.; Gao, D.; Wan, L.; Zhuang, M.; Hu, L.; et al. FBW7 Loss Promotes Chromosomal Instability and Tumorigenesis via Cyclin E1/CDK2-Mediated Phosphorylation of CENP-A. Cancer Res. 2017, 77, 4881–4893. [Google Scholar] [CrossRef]
- Yu, Z.; Zhou, X.; Wang, W.; Deng, W.; Fang, J.; Hu, H.; Wang, Z.; Li, S.; Cui, L.; Shen, J.; et al. Dynamic Phosphorylation of CENP-A at Ser68 Orchestrates Its Cell-Cycle-Dependent Deposition at Centromeres. Dev. Cell 2015, 32, 68–81. [Google Scholar] [CrossRef]
- Black, B.E.; Cleveland, D.W. Epigenetic Centromere Propagation and the Nature of CENP-a Nucleosomes. Cell 2011, 144, 471–479. [Google Scholar] [CrossRef]
- Chen, C.-C.; Mellone, B.G. Chromatin Assembly: Journey to the CENter of the Chromosome. J. Cell Biol. 2016, 214, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lacoste, N.; Woolfe, A.; Tachiwana, H.; Garea, A.V.; Barth, T.; Cantaloube, S.; Kurumizaka, H.; Imhof, A.; Almouzni, G. Mislocalization of the Centromeric Histone Variant CenH3/CENP-A in Human Cells Depends on the Chaperone DAXX. Mol. Cell 2014, 53, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Nechemia-Arbely, Y.; Fachinetti, D.; Miga, K.H.; Sekulic, N.; Soni, G.V.; Kim, D.H.; Wong, A.K.; Lee, A.Y.; Nguyen, K.; Dekker, C.; et al. Human Centromeric CENP-A Chromatin Is a Homotypic, Octameric Nucleosome at All Cell Cycle Points. J. Cell Biol. 2017, 216, 607–621. [Google Scholar] [CrossRef]
- Ohzeki, J.; Bergmann, J.H.; Kouprina, N.; Noskov, V.N.; Nakano, M.; Kimura, H.; Earnshaw, W.C.; Larionov, V.; Masumoto, H. Breaking the HAC Barrier: Histone H3K9 Acetyl/Methyl Balance Regulates CENP-A Assembly. EMBO J. 2012, 31, 2391–2402. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Cardinale, S.; Noskov, V.N.; Gassmann, R.; Vagnarelli, P.; Kandels-Lewis, S.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. Inactivation of a Human Kinetochore by Specific Targeting of Chromatin Modifiers. Dev. Cell 2008, 14, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, S.; Bergmann, J.H.; Kelly, D.; Nakano, M.; Valdivia, M.M.; Kimura, H.; Masumoto, H.; Larionov, V.; Earnshaw, W.C. Hierarchical Inactivation of a Synthetic Human Kinetochore by a Chromatin Modifier. Mol. Biol. Cell 2009, 20, 4194–4204. [Google Scholar] [CrossRef]
- Bergmann, J.H.; Rodríguez, M.G.; Martins, N.M.C.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.T.; Earnshaw, W.C. Epigenetic Engineering Shows H3K4me2 Is Required for HJURP Targeting and CENP-A Assembly on a Synthetic Human Kinetochore. EMBO J. 2011, 30, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Foltz, D.R. Posttranslational Modifications of CENP-A: Marks of Distinction. Chromosoma 2018, 127, 279–290. [Google Scholar] [CrossRef]
- Ohzeki, J.-I.; Shono, N.; Otake, K.; Martins, N.M.C.; Kugou, K.; Kimura, H.; Nagase, T.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation. Dev. Cell 2016, 37, 413–427. [Google Scholar] [CrossRef]
- Fujita, Y.; Hayashi, T.; Kiyomitsu, T.; Toyoda, Y.; Kokubu, A.; Obuse, C.; Yanagida, M. Priming of Centromere for CENP-A Recruitment by Human HMis18alpha, HMis18beta, and M18BP1. Dev. Cell 2007, 12, 17–30. [Google Scholar] [CrossRef]
- Quénet, D.; Dalal, Y. A Long Non-Coding RNA Is Required for Targeting Centromeric Protein A to the Human Centromere. Elife 2014, 3, e03254. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, L.; Siggens, L.; Svensson, J.P.; Ekwall, K. Centromeric Histone H2B Monoubiquitination Promotes Noncoding Transcription and Chromatin Integrity. Nat. Struct. Mol. Biol. 2014, 21, 236–243. [Google Scholar] [CrossRef]
- Kim, J.; Guermah, M.; McGinty, R.K.; Lee, J.-S.; Tang, Z.; Milne, T.A.; Shilatifard, A.; Muir, T.W.; Roeder, R.G. RAD6-Mediated Transcription-Coupled H2B Ubiquitylation Directly Stimulates H3K4 Methylation in Human Cells. Cell 2009, 137, 459–471. [Google Scholar] [CrossRef]
- McGinty, R.K.; Kim, J.; Chatterjee, C.; Roeder, R.G.; Muir, T.W. Chemically Ubiquitylated Histone H2B Stimulates HDot1L-Mediated Intranucleosomal Methylation. Nature 2008, 453, 812–816. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Shang, W.-H.; Toyoda, A.; Misu, S.; Monma, N.; Ikeo, K.; Molina, O.; Vargiu, G.; Fujiyama, A.; Kimura, H.; et al. Histone H4 Lys 20 Monomethylation of the CENP-A Nucleosome Is Essential for Kinetochore Assembly. Dev. Cell 2014, 29, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.B.; Oda, H.; Shen, S.S.; Reinberg, D. PR-Set7 and H4K20me1: At the Crossroads of Genome Integrity, Cell Cycle, Chromosome Condensation, and Transcription. Genes Dev. 2012, 26, 325–337. [Google Scholar] [CrossRef]
- ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Barra, V.; Logsdon, G.A.; Scelfo, A.; Hoffmann, S.; Hervé, S.; Aslanian, A.; Nechemia-Arbely, Y.; Cleveland, D.W.; Black, B.E.; Fachinetti, D. Phosphorylation of CENP-A on Serine 7 Does Not Control Centromere Function. Nat. Commun. 2019, 10, 175. [Google Scholar] [CrossRef]
- Shang, W.-H.; Hori, T.; Westhorpe, F.G.; Godek, K.M.; Toyoda, A.; Misu, S.; Monma, N.; Ikeo, K.; Carroll, C.W.; Takami, Y.; et al. Acetylation of Histone H4 Lysine 5 and 12 Is Required for CENP-A Deposition into Centromeres. Nat. Commun. 2016, 7, 13465. [Google Scholar] [CrossRef]
- Goutte-Gattat, D.; Shuaib, M.; Ouararhni, K.; Gautier, T.; Skoufias, D.A.; Hamiche, A.; Dimitrov, S. Phosphorylation of the CENP-A Amino-Terminus in Mitotic Centromeric Chromatin Is Required for Kinetochore Function. Proc. Natl. Acad. Sci. USA 2013, 110, 8579–8584. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, S.G.; Shelby, R.D.; Sullivan, K.F. CENP-A Is Phosphorylated by Aurora B Kinase and Plays an Unexpected Role in Completion of Cytokinesis. J. Cell Biol. 2001, 155, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Higgins, J.M.G. Histone Modifications and Mitosis: Countermarks, Landmarks, and Bookmarks. Trends Cell Biol. 2013, 23, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.O.; Panchenko, T.; Sathyan, K.M.; Petkowski, J.J.; Pai, P.-J.; Bai, D.L.; Russell, D.H.; Macara, I.G.; Shabanowitz, J.; Hunt, D.F.; et al. Posttranslational Modification of CENP-A Influences the Conformation of Centromeric Chromatin. Proc. Natl. Acad. Sci. USA 2013, 110, 11827–11832. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Liu, Y.; Wang, M.; Fang, J.; Huang, H.; Yang, N.; Li, Y.; Wang, J.; Yao, X.; Shi, Y.; et al. Structure of a CENP-A-Histone H4 Heterodimer in Complex with Chaperone HJURP. Genes Dev. 2011, 25, 901–906. [Google Scholar] [CrossRef]
- Fachinetti, D.; Logsdon, G.A.; Abdullah, A.; Selzer, E.B.; Cleveland, D.W.; Black, B.E. CENP-A Modifications on Ser68 and Lys124 Are Dispensable for Establishment, Maintenance, and Long-Term Function of Human Centromeres. Dev. Cell 2017, 40, 104–113. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, K.; Huang, L.; Zhao, J.; Chen, X.; Wu, Q.; Yu, Z.; Li, G. Ser68 Phosphoregulation Is Essential for CENP-A Deposition, Centromere Function and Viability in Mice. Sci. China Life Sci. 2022. [Google Scholar] [CrossRef]
- Hori, T.; Cao, J.; Nishimura, K.; Ariyoshi, M.; Arimura, Y.; Kurumizaka, H.; Fukagawa, T. Essentiality of CENP-A Depends on Its Binding Mode to HJURP. Cell Rep. 2020, 33, 108388. [Google Scholar] [CrossRef]
- Wang, K.; Yu, Z.; Liu, Y.; Li, G. Ser68 Phosphorylation Ensures Accurate Cell-Cycle-Dependent CENP-A Deposition at Centromeres. Dev. Cell 2017, 40, 5–6. [Google Scholar] [CrossRef]
- Niikura, Y.; Kitagawa, R.; Ogi, H.; Abdulle, R.; Pagala, V.; Kitagawa, K. CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev. Cell 2015, 32, 589–603. [Google Scholar] [CrossRef]
- Niikura, Y.; Kitagawa, R.; Fang, L.; Kitagawa, K. CENP-A Ubiquitylation Is Indispensable to Cell Viability. Dev. Cell 2019, 50, 683–689.e6. [Google Scholar] [CrossRef]
- Salinas-Luypaert, C.; Allu, P.K.; Logsdon, G.A.; Dawicki-McKenna, J.M.; Gambogi, C.W.; Fachinetti, D.; Black, B.E. Gene Replacement Strategies Validate the Use of Functional Tags on Centromeric Chromatin and Invalidate an Essential Role for CENP-AK124ub. Cell Rep. 2021, 37, 109924. [Google Scholar] [CrossRef]
- Bassett, E.A.; DeNizio, J.; Barnhart-Dailey, M.C.; Panchenko, T.; Sekulic, N.; Rogers, D.J.; Foltz, D.R.; Black, B.E. HJURP Uses Distinct CENP-A Surfaces to Recognize and to Stabilize CENP-A/Histone H4 for Centromere Assembly. Dev. Cell 2012, 22, 749–762. [Google Scholar] [CrossRef]
- α-Amino Trimethylation of CENP-A by NRMT Is Required for Full Recruitment of the Centromere—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/28266506/ (accessed on 3 July 2022).
- Ryu, H.-Y.; Hochstrasser, M. Histone Sumoylation and Chromatin Dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef] [PubMed]
- Ohkuni, K.; Takahashi, Y.; Fulp, A.; Lawrimore, J.; Au, W.-C.; Pasupala, N.; Levy-Myers, R.; Warren, J.; Strunnikov, A.; Baker, R.E.; et al. SUMO-Targeted Ubiquitin Ligase (STUbL) Slx5 Regulates Proteolysis of Centromeric Histone H3 Variant Cse4 and Prevents Its Mislocalization to Euchromatin. Mol. Biol. Cell 2016, 27, 1500–1510. [Google Scholar] [CrossRef] [PubMed]
- Ohkuni, K.; Suva, E.; Au, W.-C.; Walker, R.L.; Levy-Myers, R.; Meltzer, P.S.; Baker, R.E.; Basrai, M.A. Deposition of Centromeric Histone H3 Variant CENP-A/Cse4 into Chromatin Is Facilitated by Its C-Terminal Sumoylation. Genetics 2020, 214, 839–854. [Google Scholar] [CrossRef] [PubMed]
- Ohkuni, K.; Levy-Myers, R.; Warren, J.; Au, W.-C.; Takahashi, Y.; Baker, R.E.; Basrai, M.A. N-Terminal Sumoylation of Centromeric Histone H3 Variant Cse4 Regulates Its Proteolysis to Prevent Mislocalization to Non-Centromeric Chromatin. G3 Genes Genomes Genet. 2018, 8, 1215–1223. [Google Scholar] [CrossRef]
- Van de Pasch, L.A.L.; Miles, A.J.; Nijenhuis, W.; Brabers, N.A.C.H.; van Leenen, D.; Lijnzaad, P.; Brown, M.K.; Ouellet, J.; Barral, Y.; Kops, G.J.P.L.; et al. Centromere Binding and a Conserved Role in Chromosome Stability for SUMO-Dependent Ubiquitin Ligases. PLoS ONE 2013, 8, e65628. [Google Scholar] [CrossRef]
- Musacchio, A.; Salmon, E.D. The Spindle-Assembly Checkpoint in Space and Time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef]
- Schibler, A.; Koutelou, E.; Tomida, J.; Wilson-Pham, M.; Wang, L.; Lu, Y.; Cabrera, A.P.; Chosed, R.J.; Li, W.; Li, B.; et al. Histone H3K4 Methylation Regulates Deactivation of the Spindle Assembly Checkpoint through Direct Binding of Mad2. Genes Dev. 2016, 30, 1187–1197. [Google Scholar] [CrossRef]
- Beilharz, T.H.; Harrison, P.F.; Miles, D.M.; See, M.M.; Le, U.M.M.; Kalanon, M.; Curtis, M.J.; Hasan, Q.; Saksouk, J.; Margaritis, T.; et al. Coordination of Cell Cycle Progression and Mitotic Spindle Assembly Involves Histone H3 Lysine 4 Methylation by Set1/COMPASS. Genetics 2017, 205, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xu, X.; Hall, H.; Hyland, E.M.; Boeke, J.D.; Hazbun, T.; Kuo, M.-H. Histone H3 Exerts a Key Function in Mitotic Checkpoint Control. Mol. Cell. Biol. 2010, 30, 537–549. [Google Scholar] [CrossRef]
- Herrera, L.A.; Prada, D.; Andonegui, M.A.; Dueñas-González, A. The Epigenetic Origin of Aneuploidy. Curr. Genom. 2008, 9, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, S.; Liu, J.; Xiong, R.; Yu, T.; Makriyannis, A.; Chen, C. Chronic Low Dose Arsenic Exposure Preferentially Perturbs Mitotic Phase of the Cell Cycle. Genes Cancer 2019, 10, 39–51. [Google Scholar] [CrossRef] [PubMed]
- States, J.C. Disruption of Mitotic Progression by Arsenic. Biol. Trace Elem. Res. 2015, 166, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Hirano, S. Biotransformation of Arsenic and Toxicological Implication of Arsenic Metabolites. Arch. Toxicol. 2020, 94, 2587–2601. [Google Scholar] [CrossRef]
- Zhou, Q.; Xi, S. A Review on Arsenic Carcinogenesis: Epidemiology, Metabolism, Genotoxicity and Epigenetic Changes. Regul. Toxicol. Pharmacol. 2018, 99, 78–88. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Banerjee, M.; Giri, A.K. Role of Genomic Instability in Arsenic-Induced Carcinogenicity. A Review. Environ. Int. 2013, 53, 29–40. [Google Scholar] [CrossRef]
- Vega, L.; Gonsebatt, M.E.; Ostrosky-Wegman, P. Aneugenic Effect of Sodium Arsenite on Human Lymphocytes in Vitro: An Individual Susceptibility Effect Detected. Mutat. Res. 1995, 334, 365–373. [Google Scholar] [CrossRef]
- Yih, L.H.; Ho, I.C.; Lee, T.C. Sodium Arsenite Disturbs Mitosis and Induces Chromosome Loss in Human Fibroblasts. Cancer Res. 1997, 57, 5051–5059. [Google Scholar]
- Kitchin, K.T.; Wallace, K. The Role of Protein Binding of Trivalent Arsenicals in Arsenic Carcinogenesis and Toxicity. J. Inorg. Biochem. 2008, 102, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Kligerman, A.D.; Doerr, C.L.; Tennant, A.H. Oxidation and Methylation Status Determine the Effects of Arsenic on the Mitotic Apparatus. Mol. Cell. Biochem. 2005, 279, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miyazaki, K.; Kita, K.; Ochi, T. Trivalent Dimethylarsenic Compound Induces Histone H3 Phosphorylation and Abnormal Localization of Aurora B Kinase in HepG2 Cells. Toxicol. Appl. Pharmacol. 2009, 241, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, A.P. Acetylation and Methylation Patterns of Core Histones Are Modified after Heat or Arsenite Treatment of Drosophila Tissue Culture Cells. Nucleic Acids Res. 1983, 11, 1389–1404. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.; Ren, X.; Chasse, A.; Hickman, T.; Zhang, L.; Yuh, J.; Smith, M.T.; Burlingame, A.L. Quantitative Mass Spectrometry Reveals the Epigenome as a Target of Arsenic. Chem.-Biol. Interact. 2011, 192, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Howe, C.G.; Gamble, M.V. Influence of Arsenic on Global Levels of Histone Posttranslational Modifications: A Review of the Literature and Challenges in the Field. Curr. Environ. Health Rep. 2016, 3, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wu, D.; Zhao, L.; Yang, Y.; Ding, J.; Dong, L.; Hu, L.; Wang, F.; Zhao, X.; Cai, Y.; et al. Arsenic Trioxide Reduces Global Histone H4 Acetylation at Lysine 16 through Direct Binding to Histone Acetyltransferase HMOF in Human Cells. PLoS ONE 2015, 10, e0141014. [Google Scholar] [CrossRef]
- Yu, C.-W.; Liao, V.H.-C. Transgenerational Reproductive Effects of Arsenite Are Associated with H3K4 Dimethylation and SPR-5 Downregulation in Caenorhabditis Elegans. Environ. Sci. Technol. 2016, 50, 10673–10681. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Arita, A.; Sun, H.; Costa, M. Effects of Nickel, Chromate, and Arsenite on Histone 3 Lysine Methylation. Toxicol. Appl. Pharmacol. 2009, 236, 78–84. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, H.; Ellen, T.P.; Chen, H.; Costa, M. Arsenite Alters Global Histone H3 Methylation. Carcinogenesis 2008, 29, 1831–1836. [Google Scholar] [CrossRef]
- Tu, W.; Liu, Y.; Xie, C.; Zhou, X. Arsenite Downregulates H3K4 Trimethylation and H3K9 Dimethylation during Transformation of Human Bronchial Epithelial Cells. J. Appl. Toxicol. 2018, 38, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Paramasivam, M.; Cai, Q.; Dai, X.; Wang, P.; Lin, K.; Song, J.; Seidman, M.M.; Wang, Y. Arsenite Binds to the RING Finger Domains of RNF20-RNF40 Histone E3 Ubiquitin Ligase and Inhibits DNA Double-Strand Break Repair. J. Am. Chem. Soc. 2014, 136, 12884–12887. [Google Scholar] [CrossRef] [PubMed]
- Chervona, Y.; Arita, A.; Costa, M. Carcinogenic Metals and the Epigenome: Understanding the Effect of Nickel, Arsenic, and Chromium. Metallomics 2012, 4, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.S.; Buchner, V.; Tchounwou, P.B. Exploring the Molecular Mechanisms of Nickel-Induced Genotoxicity and Carcinogenicity: A Literature Review. Rev. Environ. Health 2011, 26, 81–92. [Google Scholar] [CrossRef]
- Ohshima, S. Induction of Aneuploidy by Nickel Sulfate in V79 Chinese Hamster Cells. Mutat. Res. 2001, 492, 39–50. [Google Scholar] [CrossRef]
- Seoane, A.I.; Dulout, F.N. Genotoxic Ability of Cadmium, Chromium and Nickel Salts Studied by Kinetochore Staining in the Cytokinesis-Blocked Micronucleus Assay. Mutat. Res. 2001, 490, 99–106. [Google Scholar] [CrossRef]
- Ohshima, S. Induction of Genetic Instability and Chromosomal Instability by Nickel Sulfate in V79 Chinese Hamster Cells. Mutagenesis 2003, 18, 133–137. [Google Scholar] [CrossRef][Green Version]
- Ellen, T.P.; Kluz, T.; Harder, M.E.; Xiong, J.; Costa, M. Heterochromatinization as a Potential Mechanism of Nickel-Induced Carcinogenesis. Biochemistry 2009, 48, 4626–4632. [Google Scholar] [CrossRef]
- Lee, Y.W.; Klein, C.B.; Kargacin, B.; Salnikow, K.; Kitahara, J.; Dowjat, K.; Zhitkovich, A.; Christie, N.T.; Costa, M. Carcinogenic Nickel Silences Gene Expression by Chromatin Condensation and DNA Methylation: A New Model for Epigenetic Carcinogens. Mol. Cell. Biol. 1995, 15, 2547–2557. [Google Scholar] [CrossRef]
- Wu, C.-H.; Tang, S.-C.; Wang, P.-H.; Lee, H.; Ko, J.-L. Nickel-Induced Epithelial-Mesenchymal Transition by Reactive Oxygen Species Generation and E-Cadherin Promoter Hypermethylation. J. Biol. Chem. 2012, 287, 25292–25302. [Google Scholar] [CrossRef]
- Ke, Q.; Li, Q.; Ellen, T.P.; Sun, H.; Costa, M. Nickel Compounds Induce Phosphorylation of Histone H3 at Serine 10 by Activating JNK-MAPK Pathway. Carcinogenesis 2008, 29, 1276–1281. [Google Scholar] [CrossRef] [PubMed]
- Broday, L.; Peng, W.; Kuo, M.H.; Salnikow, K.; Zoroddu, M.; Costa, M. Nickel Compounds Are Novel Inhibitors of Histone H4 Acetylation. Cancer Res. 2000, 60, 238–241. [Google Scholar] [PubMed]
- Sutherland, J.E.; Peng, W.; Zhang, Q.; Costa, M. The Histone Deacetylase Inhibitor Trichostatin A Reduces Nickel-Induced Gene Silencing in Yeast and Mammalian Cells. Mutat. Res. 2001, 479, 225–233. [Google Scholar] [CrossRef]
- Kang, J.; Zhang, Y.; Chen, J.; Chen, H.; Lin, C.; Wang, Q.; Ou, Y. Nickel-Induced Histone Hypoacetylation: The Role of Reactive Oxygen Species. Toxicol. Sci. 2003, 74, 279–286. [Google Scholar] [CrossRef]
- Zoroddu, M.A.; Peana, M.; Medici, S.; Casella, L.; Monzani, E.; Costa, M. Nickel Binding to Histone H4. Dalton Trans. 2010, 39, 787–793. [Google Scholar] [CrossRef]
- Chen, H.; Ke, Q.; Kluz, T.; Yan, Y.; Costa, M. Nickel Ions Increase Histone H3 Lysine 9 Dimethylation and Induce Transgene Silencing. Mol. Cell. Biol. 2006, 26, 3728–3737. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Kluz, T.; Zhang, P.; Chen, H.; Costa, M. Analysis of Specific Lysine Histone H3 and H4 Acetylation and Methylation Status in Clones of Cells with a Gene Silenced by Nickel Exposure. Toxicol. Appl. Pharmacol. 2003, 190, 272–277. [Google Scholar] [CrossRef]
- Ke, Q.; Ellen, T.P.; Costa, M. Nickel Compounds Induce Histone Ubiquitination by Inhibiting Histone Deubiquitinating Enzyme Activity. Toxicol. Appl. Pharmacol. 2008, 228, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Wise, S.S.; Wise, J.P. Aneuploidy as an Early Mechanistic Event in Metal Carcinogenesis. Biochem. Soc. Trans. 2010, 38, 1650–1654. [Google Scholar] [CrossRef]
- Xiao, C.; Liu, Y.; Xie, C.; Tu, W.; Xia, Y.; Costa, M.; Zhou, X. Cadmium Induces Histone H3 Lysine Methylation by Inhibiting Histone Demethylase Activity. Toxicol. Sci. 2015, 145, 80–89. [Google Scholar] [CrossRef]
- Miousse, I.R.; Chalbot, M.-C.G.; Pathak, R.; Lu, X.; Nzabarushimana, E.; Krager, K.; Aykin-Burns, N.; Hauer-Jensen, M.; Demokritou, P.; Kavouras, I.G.; et al. In Vitro Toxicity and Epigenotoxicity of Different Types of Ambient Particulate Matter. Toxicol. Sci. 2015, 148, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, W.X.; Bai, C.; Song, Y. Particulate Matter-Induced Epigenetic Changes and Lung Cancer. Clin. Respir. J. 2017, 11, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-Andrade, M.; Sánchez-Pérez, Y.; Chirino, Y.I.; Morales-Bárcenas, R.; Herrera, L.A.; García-Cuellar, C.M. Airborne Particulate Matter Induces Mitotic Slippage and Chromosomal Missegregation through Disruption of the Spindle Assembly Checkpoint (SAC). Chemosphere 2019, 235, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Arita, A.; Shamy, M.Y.; Chervona, Y.; Clancy, H.A.; Sun, H.; Hall, M.N.; Qu, Q.; Gamble, M.V.; Costa, M. The Effect of Exposure to Carcinogenic Metals on Histone Tail Modifications and Gene Expression in Human Subjects. J. Trace Elem. Med. Biol. 2012, 26, 174–178. [Google Scholar] [CrossRef]
- Goodman, S.; Chappell, G.; Guyton, K.Z.; Pogribny, I.P.; Rusyn, I. Epigenetic Alterations Induced by Genotoxic Occupational and Environmental Human Chemical Carcinogens: An Update of a Systematic Literature Review. Mutat. Res. Rev. Mutat. Res. 2022, 789, 108408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andonegui-Elguera, M.A.; Cáceres-Gutiérrez, R.E.; López-Saavedra, A.; Cisneros-Soberanis, F.; Justo-Garrido, M.; Díaz-Chávez, J.; Herrera, L.A. The Roles of Histone Post-Translational Modifications in the Formation and Function of a Mitotic Chromosome. Int. J. Mol. Sci. 2022, 23, 8704. https://doi.org/10.3390/ijms23158704
Andonegui-Elguera MA, Cáceres-Gutiérrez RE, López-Saavedra A, Cisneros-Soberanis F, Justo-Garrido M, Díaz-Chávez J, Herrera LA. The Roles of Histone Post-Translational Modifications in the Formation and Function of a Mitotic Chromosome. International Journal of Molecular Sciences. 2022; 23(15):8704. https://doi.org/10.3390/ijms23158704
Chicago/Turabian StyleAndonegui-Elguera, Marco A., Rodrigo E. Cáceres-Gutiérrez, Alejandro López-Saavedra, Fernanda Cisneros-Soberanis, Montserrat Justo-Garrido, José Díaz-Chávez, and Luis A. Herrera. 2022. "The Roles of Histone Post-Translational Modifications in the Formation and Function of a Mitotic Chromosome" International Journal of Molecular Sciences 23, no. 15: 8704. https://doi.org/10.3390/ijms23158704
APA StyleAndonegui-Elguera, M. A., Cáceres-Gutiérrez, R. E., López-Saavedra, A., Cisneros-Soberanis, F., Justo-Garrido, M., Díaz-Chávez, J., & Herrera, L. A. (2022). The Roles of Histone Post-Translational Modifications in the Formation and Function of a Mitotic Chromosome. International Journal of Molecular Sciences, 23(15), 8704. https://doi.org/10.3390/ijms23158704







