Microbial Type IA Topoisomerase C-Terminal Domain Sequence Motifs, Distribution and Combination
Abstract
:1. Introduction
2. Results
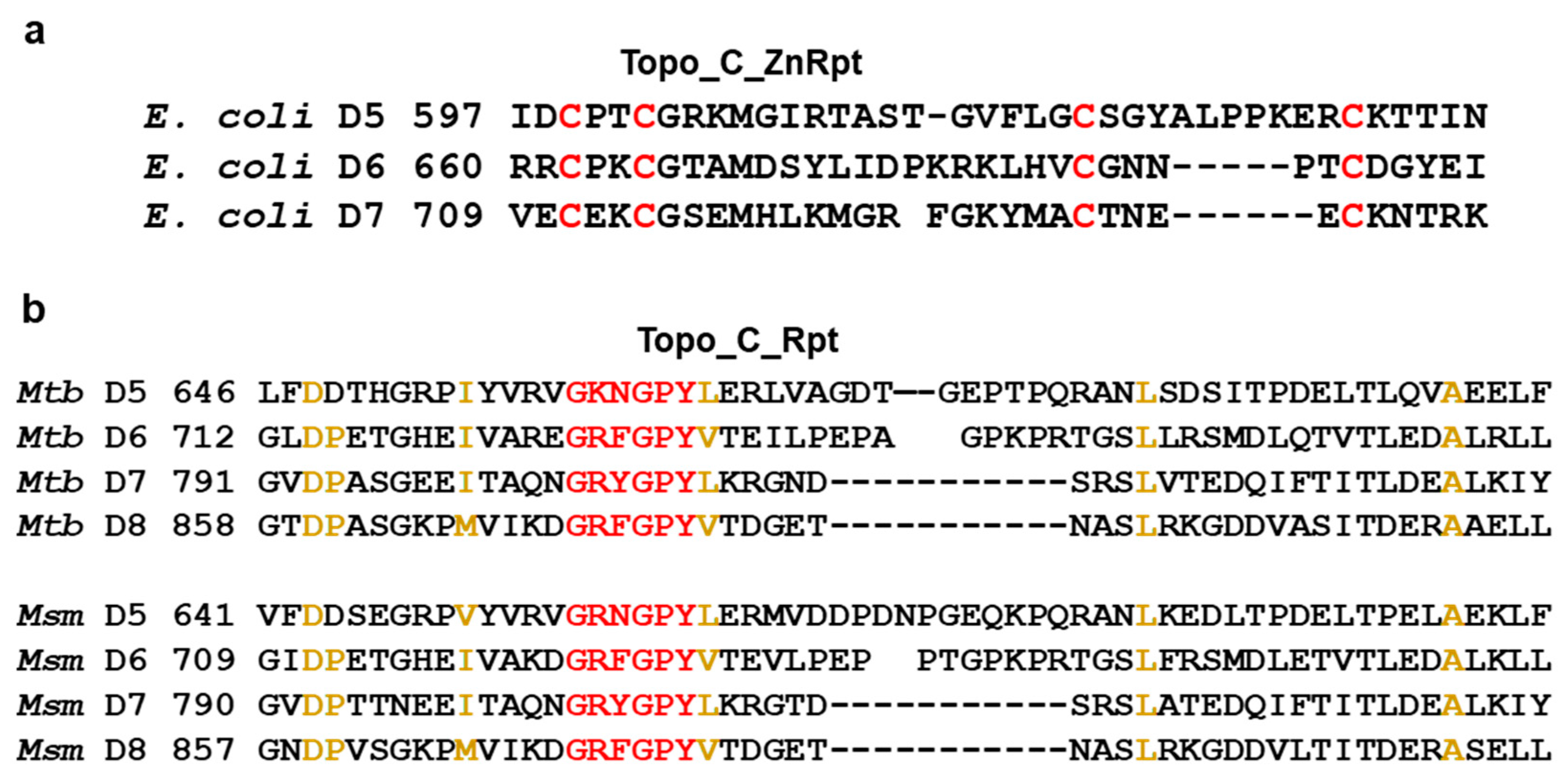
2.1. Topo_C_ZnRpt and Topo_C_Rpt in Bacterial Topoisomerase I C-terminal Domains
2.1.1. Distribution
2.1.2. Consensus Sequence for Topo_C_ZnRpt and Topo_C_Rpt
2.1.3. Combinations of Topo_C_ZnRpt and Topo_C_Rpt Repeats in Individual Bacterial Topoisomerase I Sequences
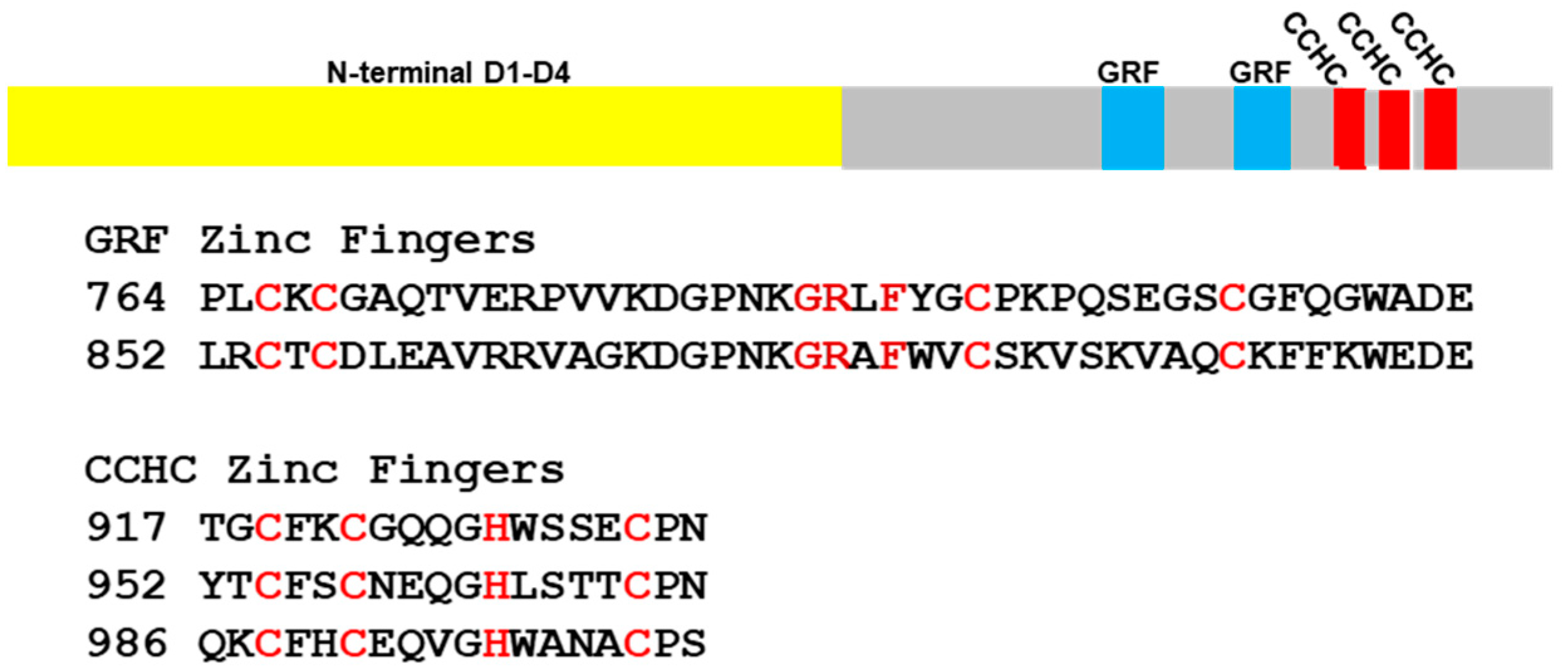
2.2. Observation of New Types of C-Terminal Repeats in Fungal Topoisomerase III
2.2.1. Distribution of Top3 C-Terminal Repeats in Fungal Phyla
2.2.2. Combinations of zf-GRF and zf-CCHC Zinc Fingers in Fungal Species
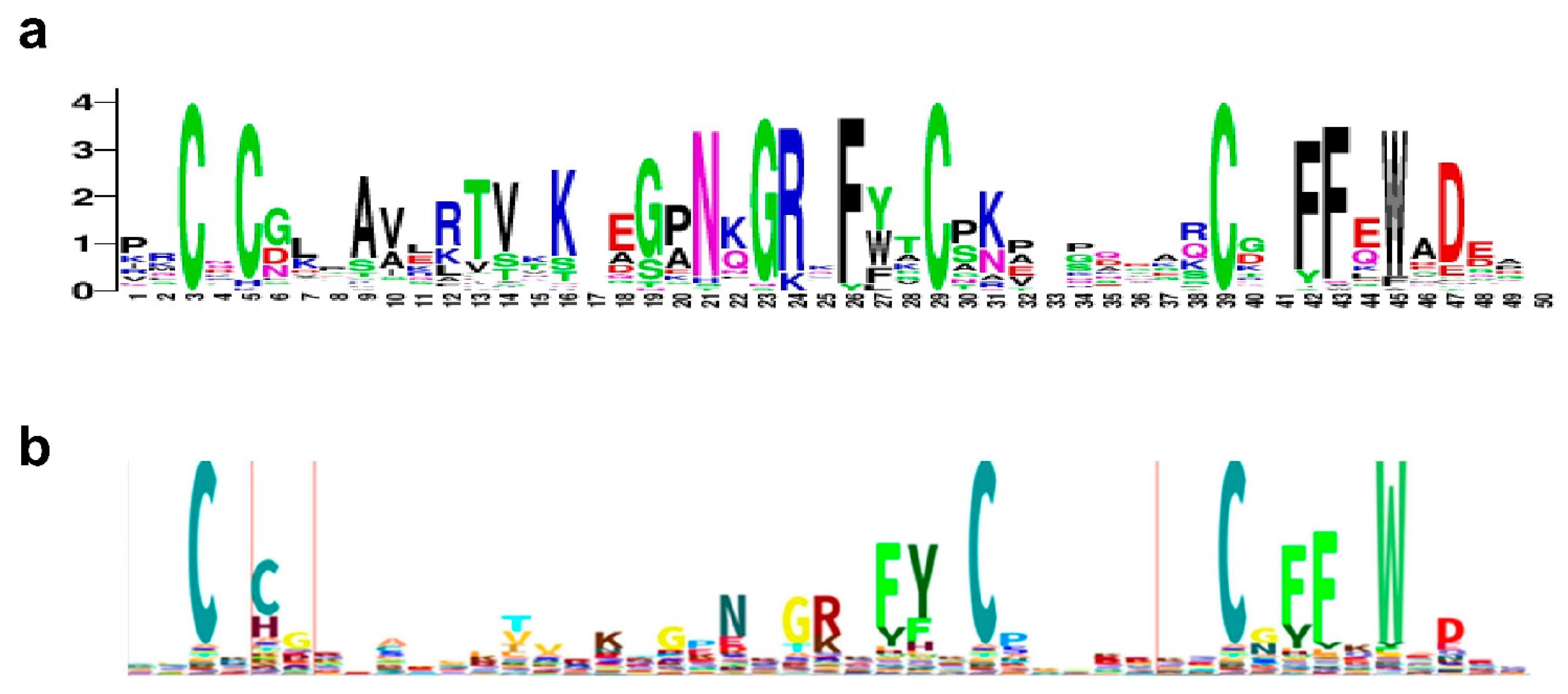
2.2.3. Consensus Sequence for zf-GRF and zf-CCHC Zinc Fingers Found in Fungal Topoisomerase III
2.2.4. Predicted Structures and Nucleic Acid Interactions for zf-GRF Domains of Puccinia graminis f. sp. tritici Topoisomerase III
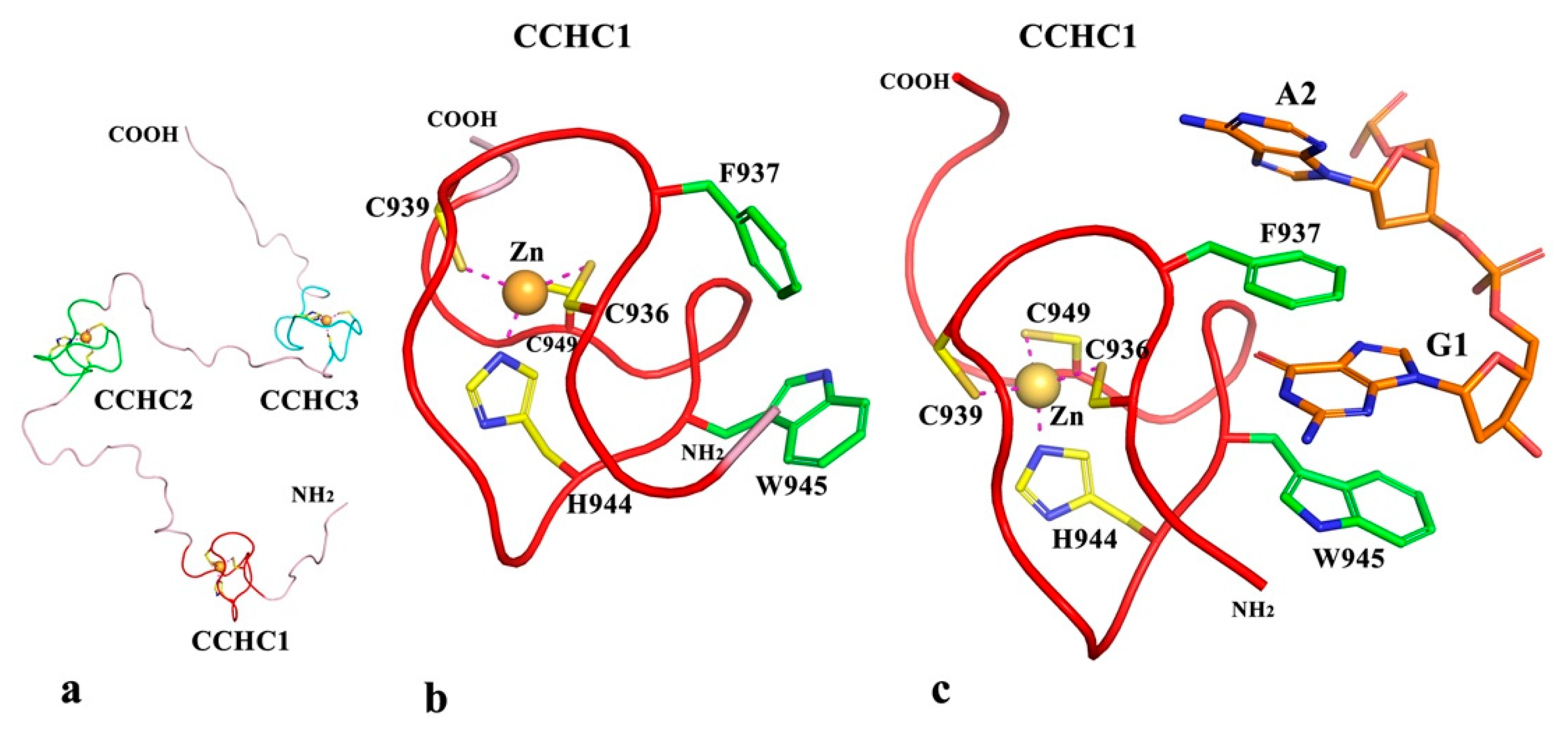
2.2.5. Predicted Structures and Nucleic Acid Interactions for zf-CCHC Domains of Puccinia graminis f. sp. Tritici Topoisomerase III
3. Discussion
4. Materials and Methods
4.1. Sequence Database Search
4.2. Generation of Taxonomy Common Tree
4.3. Sequence Alignment
4.4. Structure Prediction and Model Building
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bizard, A.H.; Hickson, I.D. The many lives of type IA topoisomerases. J. Biol. Chem. 2020, 295, 7138–7153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brochu, J.; Breton, E.V.; Drolet, M. Supercoiling, R-loops, replication and the functions of bacterial type 1A topoisomerases. Genes 2020, 11, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garnier, F.; Debat, H.; Nadal, M. Type IA DNA topoisomerases: A universal core and multiple activities. Methods Mol. Biol. 2018, 1703, 1–20. [Google Scholar] [CrossRef]
- Ahmad, M.; Xu, D.; Wang, W. Type IA topoisomerases can be “magicians” for both DNA and RNA in all domains of life. RNA Biol. 2017, 14, 854–864. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.M.; Rajan, R.; Mondragón, A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009, 37, 693–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Cheng, B.; Tse-Dinh, Y.C. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 2011, 108, 6939–6944. [Google Scholar] [CrossRef] [Green Version]
- Bocquet, N.; Bizard, A.H.; Abdulrahman, W.; Larsen, N.B.; Faty, M.; Cavadini, S.; Bunker, R.D.; Kowalczykowski, S.C.; Cejka, P.; Hickson, I.D.; et al. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIalpha and RMI1. Nat. Struct. Mol. Biol. 2014, 21, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.; Cao, N.; Cheng, B.; Joachimiak, A.; Tse-Dinh, Y.C. Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold. J. Mol. Biol. 2016, 428, 182–193. [Google Scholar] [CrossRef] [Green Version]
- Goto-Ito, S.; Yamagata, A.; Takahashi, T.S.; Sato, Y.; Fukai, S. Structural basis of the interaction between Topoisomerase IIIbeta and the TDRD3 auxiliary factor. Sci. Rep. 2017, 7, 42123. [Google Scholar] [CrossRef] [Green Version]
- Cao, N.; Tan, K.; Annamalai, T.; Joachimiak, A.; Tse-Dinh, Y.C. Investigating mycobacterial topoisomerase I mechanism from the analysis of metal and DNA substrate interactions at the active site. Nucleic Acids Res. 2018, 46, 7296–7308. [Google Scholar] [CrossRef]
- Hansen, G.; Harrenga, A.; Wieland, B.; Schomburg, D.; Reinemer, P. Crystal structure of full length topoisomerase I from Thermotoga maritima. J. Mol. Biol. 2006, 358, 1328–1340. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Hevener, K.E. Crystal structure of the 65-kilodalton amino-terminal fragment of DNA topoisomerase I from the gram-positive model organism Streptococcus mutans. Biochem. Biophys. Res. Commun. 2019, 516, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, T.; Ferdous, S.; Tse-Dinh, Y.C. Mechanism of type IA topoisomerases. Molecules 2020, 25, 4769. [Google Scholar] [CrossRef] [PubMed]
- Tse-Dinh, Y.C.; Beran-Steed, R.K. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J. Biol. Chem. 1988, 263, 15857–15859. [Google Scholar] [CrossRef]
- Ahumada, A.; Tse-Dinh, Y.C. The Zn(II) binding motifs of E. coli DNA topoisomerase I is part of a high-affinity DNA binding domain. Biochem. Biophys. Res. Commun. 1998, 251, 509–514. [Google Scholar] [CrossRef]
- Grishin, N.V. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J. Mol. Biol. 2000, 299, 1165–1177. [Google Scholar] [CrossRef]
- Tan, K.; Zhou, Q.; Cheng, B.; Zhang, Z.; Joachimiak, A.; Tse-Dinh, Y.C. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res. 2015, 43, 11031–11046. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Bhat, A.G.; Leelaram, M.N.; Menon, S.; Nagaraja, V. Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage. Nucleic Acids Res. 2013, 41, 7462–7471. [Google Scholar] [CrossRef] [Green Version]
- Strzalka, A.; Szafran, M.J.; Strick, T.; Jakimowicz, D. C-terminal lysine repeats in Streptomyces topoisomerase I stabilize the enzyme-DNA complex and confer high enzyme processivity. Nucleic Acids Res. 2017, 45, 11908–11924. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.L.; Malpure, S.; Li, Z.; Hiasa, H.; DiGate, R.J. The role of the carboxyl-terminal amino acid residues in Escherichia coli DNA topoisomerase III-mediated catalysis. J. Biol. Chem. 1996, 271, 9039–9045. [Google Scholar] [CrossRef] [Green Version]
- Cao, N.; Tan, K.; Zuo, X.; Annamalai, T.; Tse-Dinh, Y.C. Mechanistic insights from structure of Mycobacterium smegmatis topoisomerase I with ssDNA bound to both N- and C-terminal domains. Nucleic Acids Res. 2020, 48, 4448–4462. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Zhu, C.X.; Ji, C.; Ahumada, A.; Tse-Dinh, Y.C. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J. Biol. Chem. 2003, 278, 30705–30710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banda, S.; Cao, N.; Tse-Dinh, Y.C. Distinct mechanism evolved for mycobacterial RNA polymerase and topoisomerase I protein-protein interaction. J. Mol. Biol. 2017, 429, 2931–2942. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, T.; Bagui, T.K.; Sikder, D.; Nagaraja, V. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J. Biol. Chem. 1998, 273, 13925–13932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster-Böckler, B.; Schultz, J.; Rahmann, S. HMM Logos for visualization of protein families. BMC Bioinform. 2004, 5, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Zhu, C.X.; Tse-Dinh, Y.C.; Fesik, S.W. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry 1995, 34, 7622–7628. [Google Scholar] [CrossRef] [PubMed]
- Terekhova, K.; Marko, J.F.; Mondragon, A. Single-molecule analysis uncovers the difference between the kinetics of DNA decatenation by bacterial topoisomerases I and III. Nucleic Acids Res. 2014, 42, 11657–11667. [Google Scholar] [CrossRef] [Green Version]
- DiGate, R.J.; Marians, K.J. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988, 263, 13366–13373. [Google Scholar] [CrossRef]
- Liu, D.; Shao, Y.; Chen, G.; Tse-Dinh, Y.C.; Piccirilli, J.A.; Weizmann, Y. Synthesizing topological structures containing RNA. Nat. Commun. 2017, 8, 14936. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.; Xue, Y.; Lee, S.K.; Martindale, J.L.; Shen, W.; Li, W.; Zou, S.; Ciaramella, M.; Debat, H.; Nadal, M.; et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016, 44, 6335–6349. [Google Scholar] [CrossRef] [Green Version]
- Wallis, J.W.; Chrebet, G.; Brodsky, G.; Rolfe, M.; Rothstein, R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 1989, 58, 409–419. [Google Scholar] [CrossRef]
- Goodwin, A.; Wang, S.-W.; Toda, T.; Norbury, C.; Hickson, I.D. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999, 27, 4050–4058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spatafora, J.W.; Aime, M.C.; Grigoriev, I.V.; Martin, F.; Stajich, J.E.; Blackwell, M. The fungal tree of life: From molecular systematics to genome-scale phylogenies. Microbiol. Spectr. 2017, 5, 1–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5. [Google Scholar] [CrossRef]
- Wallace, B.D.; Berman, Z.; Mueller, G.A.; Lin, Y.; Chang, T.; Andres, S.N.; Wojtaszek, J.L.; DeRose, E.F.; Appel, C.D.; London, R.E.; et al. APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. USA 2017, 114, 304–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, W.; Lu, J.; Huang, M.; Gao, L.; Li, D.; Wang, G.G.; Song, J. Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 2019, 10, 5042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, A.A.; Wojtaszek, J.L.; Greer, B.H.; Haldar, T.; Gates, K.S.; Williams, R.S.; Eichman, B.F. An autoinhibitory role for the GRF zinc finger domain of DNA glycosylase NEIL3. J. Biol. Chem. 2020, 295, 15566–15575. [Google Scholar] [CrossRef] [PubMed]
- Summers, M.F.; South, T.L.; Kim, B.; Hare, D.R. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry 1990, 29, 329–340. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Pang, Y.; Yu, H.; Zhang, W.; Zhao, X.; Yu, J. The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol. 2021, 18, 2107–2126. [Google Scholar] [CrossRef]
- Weisbrich, A.; Honnappa, S.; Jaussi, R.; Okhrimenko, O.; Frey, D.; Jelesarov, I.; Akhmanova, A.; Steinmetz, M.O. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 2007, 14, 959–967. [Google Scholar] [CrossRef] [PubMed]
- South, T.L.; Summers, M.F. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Sci. 1993, 2, 3–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Hamill, S.; Wolin, S.L.; Reinisch, K.M. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl. Acad. Sci. USA 2010, 107, 15045–15050. [Google Scholar] [CrossRef] [Green Version]
- Amodeo, P.; Castiglione Morelli, M.A.; Ostuni, A.; Battistuzzi, G.; Bavoso, A. Structural features in EIAV NCp11: A lentivirus nucleocapsid protein with a short linker. Biochemistry 2006, 45, 5517–5526. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, H.; Zhang, C.; Su, S.; Chen, Y.; Chen, X.; Li, Y.; Shao, Z.; Zhang, Y.; Shao, Q.; et al. Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei. Nucleic Acids Res. 2021, 49, 568–583. [Google Scholar] [CrossRef]
- Wallen, R.M.; Perlin, M.H. An overview of the function and maintenance of sexual reproduction in dikaryotic fungi. Front. Microbiol. 2018, 9, 503. [Google Scholar] [CrossRef] [Green Version]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Séguéla-Arnaud, M.; Choinard, S.; Larchevêque, C.; Girard, C.; Froger, N.; Crismani, W.; Mercier, R. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 2017, 45, 1860–1871. [Google Scholar] [CrossRef] [Green Version]
- Dello Stritto Maria, R.; Vojtassakova, N.; Velkova, M.; Hamminger, P.; Ulm, P.; Jantsch, V. The topoisomerase 3 zinc finger domain cooperates with the RMI1 scaffold to promote stable association of the BTR complex to recombination intermediates in the Caenorhabditis elegans germline. Nucleic Acids Res. 2022, 50, 5652–5671. [Google Scholar] [CrossRef]
- How to Generate a Common Tree for a Set of Taxa. Available online: https://www.ncbi.nlm.nih.gov/guide/howto/gen-com-tree/ (accessed on 1 August 2022).
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. D Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Cryst. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef] [Green Version]



| Phylum 1 | Topo_C_ZnRpt (PF01396) | Topo_C_Rpt (PF13368) |
|---|---|---|
| Actinobacteria | 55 | 1003 |
| Bacteroidetes | 32 | 607 |
| Firmicutes | 1067 | 0 |
| Proteobacteria | 1679 | 1011 |
| Species 1 | Phylum | UniProt ID | Topo_C_ZnRpt | Topo_C_Rpt |
|---|---|---|---|---|
| Acidobacterium capsulatum | Acidobacteria | C1F6V0 | 5 | 0 |
| Mycobacterium avium | Actinobacteria | X8B6F9 | 0 | 1 |
| Streptomyces inhibens | Actinobacteria | A0A371PYR4 | 0 | 4 |
| Flavobacterium fontis | Bacteroidetes | A0A1M5B513 | 0 | 2 |
| Caldilinea aerophila | Chloroflexi | I0I048 | 0 | 3 |
| Lactobacillus plantarum | Firmicutes | A0A0G9F8X8 | 2 | 0 |
| Staphylococcus aureus | Firmicutes | Q2FZ32 | 3 | 0 |
| Caulobacter crescentus | Proteobacteria | Q9A5J6 | 1 | 3 |
| Helicobacter pylori | Proteobacteria | P55991 | 4 | 0 |
| Methylocapsa palsarum | Proteobacteria | A0A1I4AKP7 | 1 | 4 |
| Rickettsia bellii | Proteobacteria | Q1RIM1 | 1 | 2 |
| Thermotoga maritima | Thermotoga | P46799 | 1 | 0 |
| Species 1 | Phylum | UniProt ID | Zf-GRF | Zf-CCHC |
|---|---|---|---|---|
| Candida auris | Ascomycota | A0A0L0P6P7 | 0 | 0 |
| Wallemia ichthyophaga | Basidiomycota | R9AS06 | 1 | 1 |
| Grifola frondosa | Basidiomycota | A0A1C7M1I3 | 1 | 2 |
| Steccherinum ochraceum | Basidiomycota | A0A4R0RRI7 | 1 | 3 |
| Ustilago maydis | Basidiomycota | A0A0D1C790 | 2 | 2 |
| Puccinia graminis f. sp. tritici | Basidiomycota | A0A5B0PD53 | 2 | 3 |
| Spizellomyces punctatus | Chytridiomycota | A0A0L0HVJ1 | 1 | 0 |
| Rozella allomycis | Cryptomycota | A0A075AT24 | 3 | 1 2 |
| Rhizopus azygosporus | Mucoromycota | A0A367JWR4 | 2 | 0 |
| Coemansia reversa | Zoopagomycota | A0A2G5B3Y2 | 2 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz, B.; Mederos, C.; Tan, K.; Tse-Dinh, Y.-C. Microbial Type IA Topoisomerase C-Terminal Domain Sequence Motifs, Distribution and Combination. Int. J. Mol. Sci. 2022, 23, 8709. https://doi.org/10.3390/ijms23158709
Diaz B, Mederos C, Tan K, Tse-Dinh Y-C. Microbial Type IA Topoisomerase C-Terminal Domain Sequence Motifs, Distribution and Combination. International Journal of Molecular Sciences. 2022; 23(15):8709. https://doi.org/10.3390/ijms23158709
Chicago/Turabian StyleDiaz, Brenda, Christopher Mederos, Kemin Tan, and Yuk-Ching Tse-Dinh. 2022. "Microbial Type IA Topoisomerase C-Terminal Domain Sequence Motifs, Distribution and Combination" International Journal of Molecular Sciences 23, no. 15: 8709. https://doi.org/10.3390/ijms23158709
APA StyleDiaz, B., Mederos, C., Tan, K., & Tse-Dinh, Y.-C. (2022). Microbial Type IA Topoisomerase C-Terminal Domain Sequence Motifs, Distribution and Combination. International Journal of Molecular Sciences, 23(15), 8709. https://doi.org/10.3390/ijms23158709






