Protective Effects of Recombinant Human Angiogenin in Keratinocytes: New Insights on Oxidative Stress Response Mediated by RNases
Abstract
:1. Introduction
2. Results
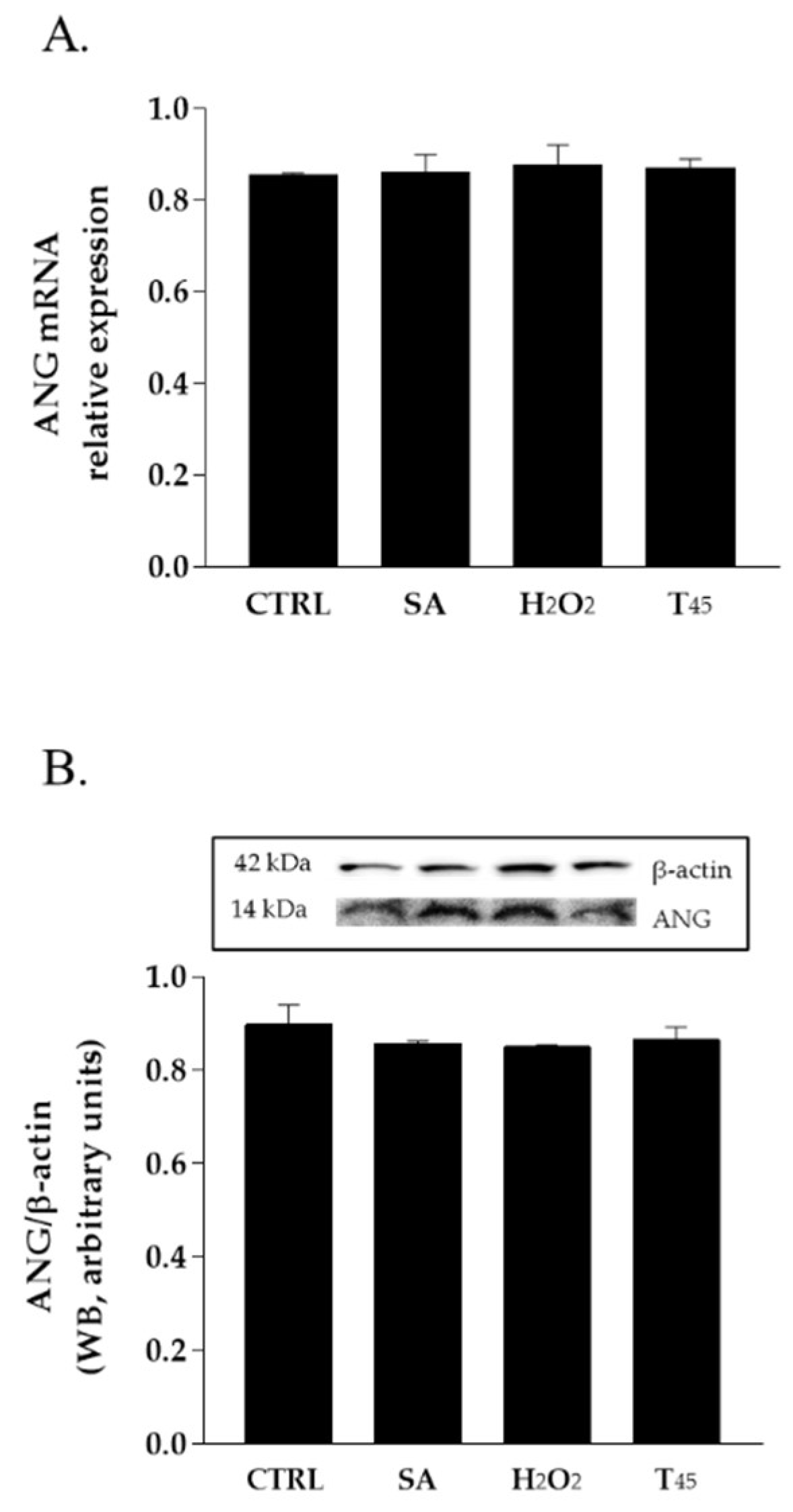
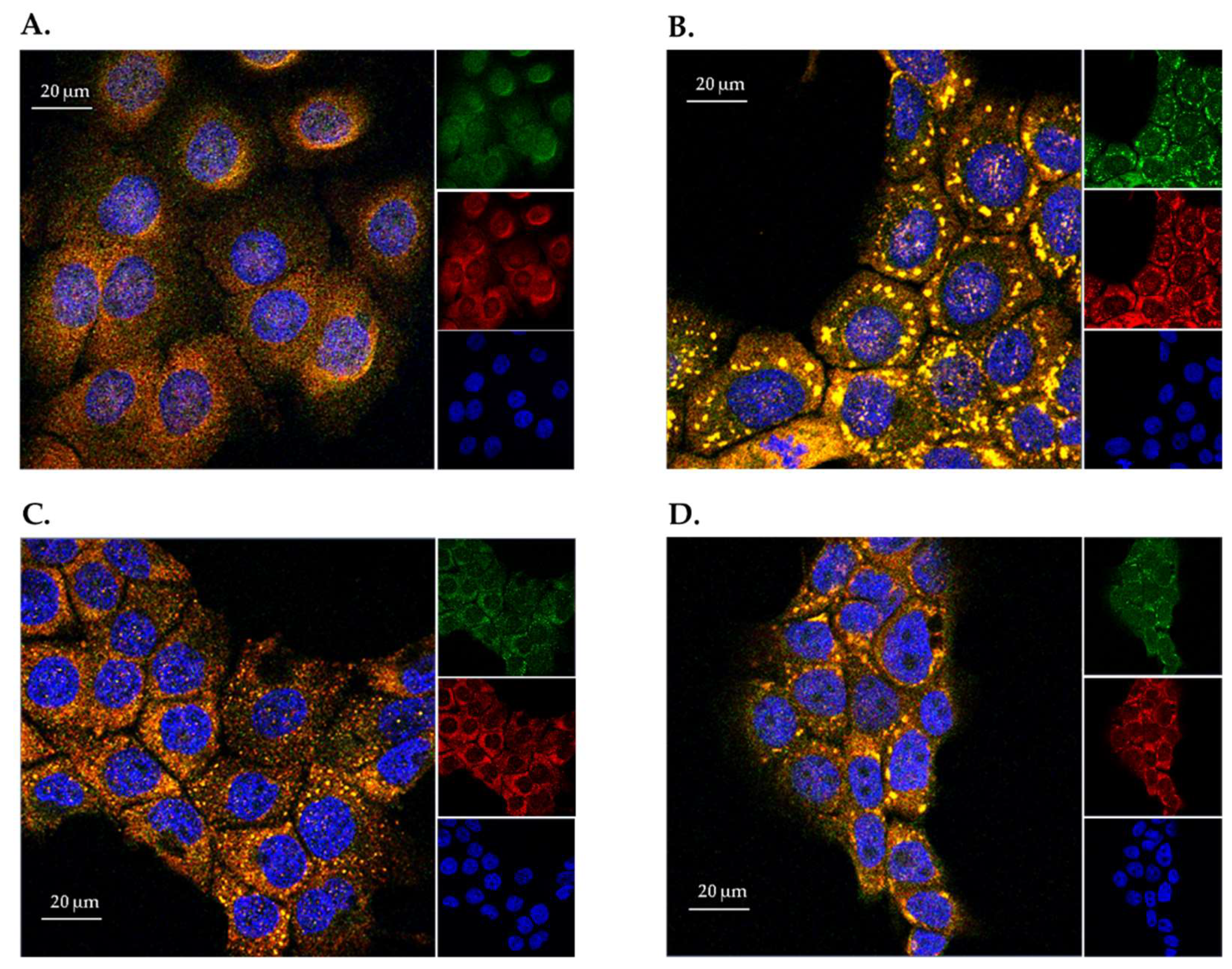
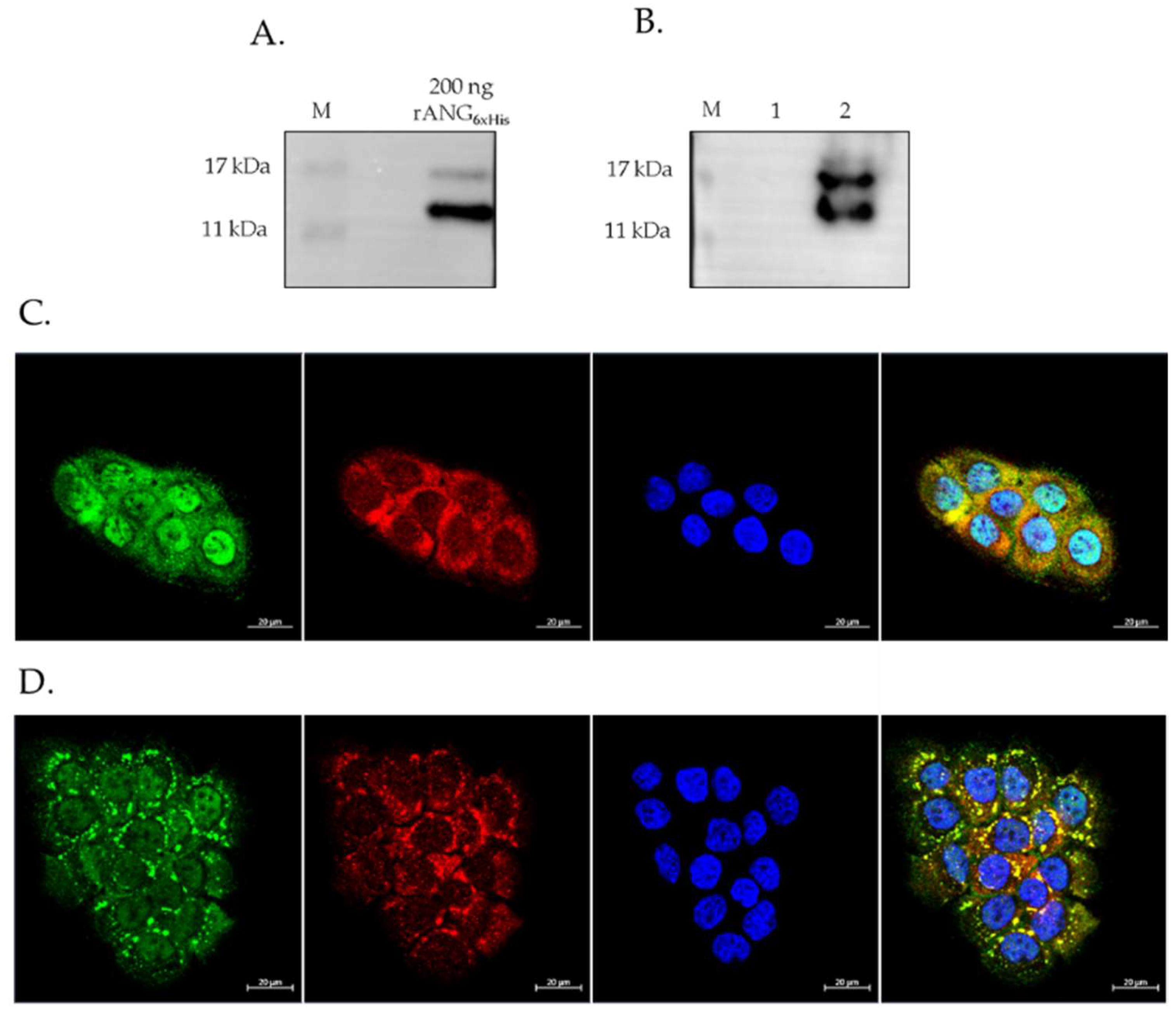
2.1. Expression and Localization of Endogenous ANG in HaCaT Cells
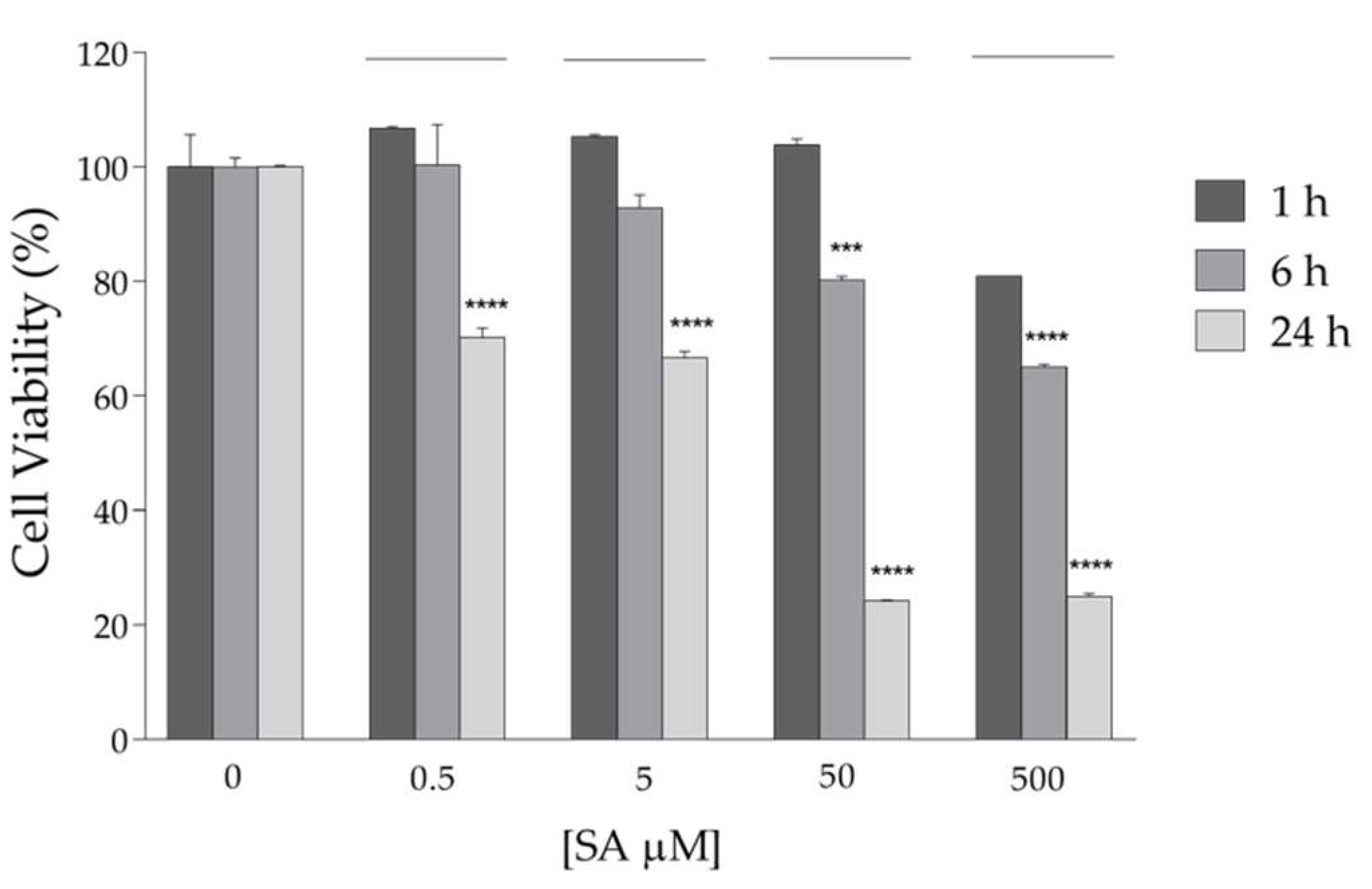
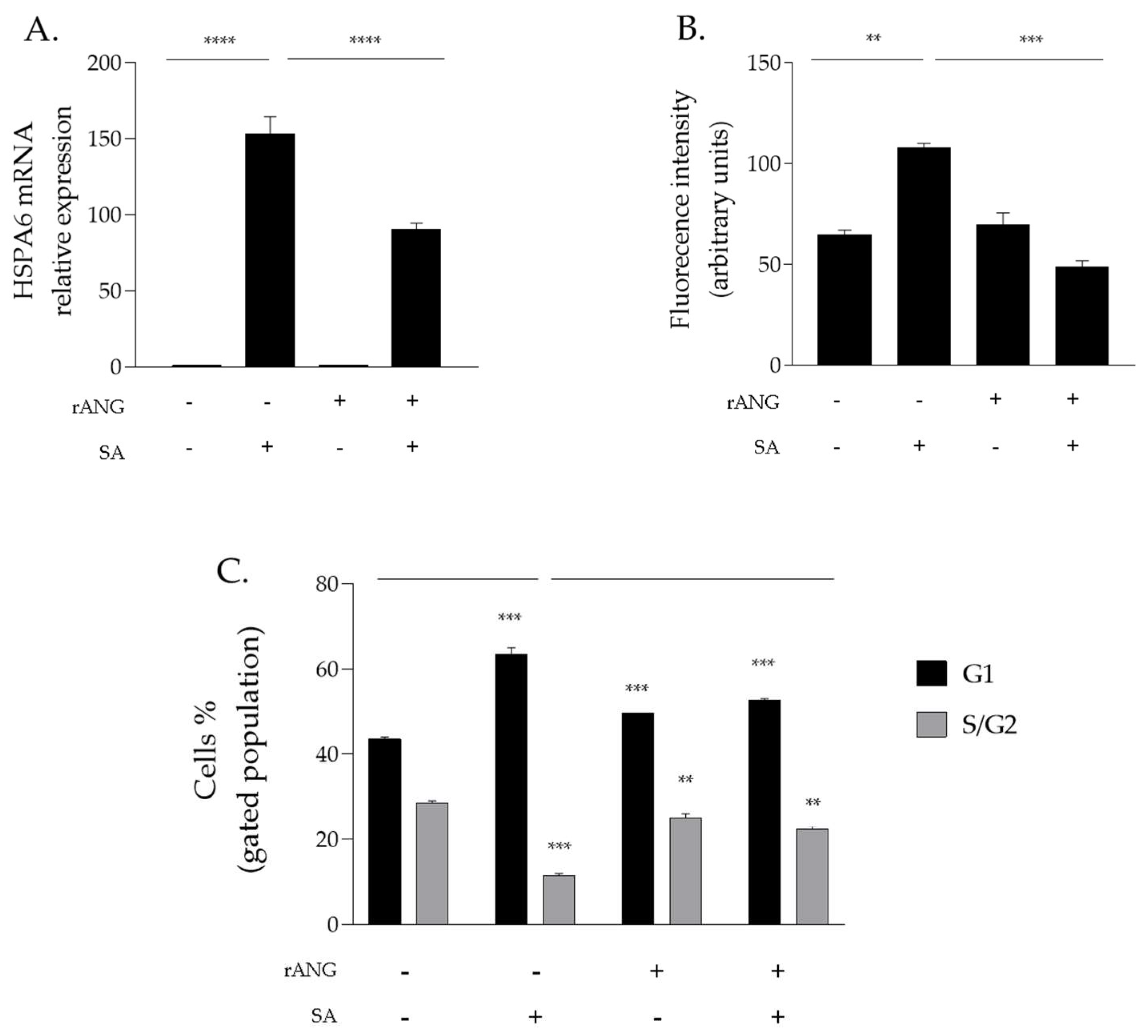
2.2. Effects of rANG on Viability and Homeostasis of HaCaT Cells
2.3. Internalization of rANG6xHis and Its Colocalization with SGs
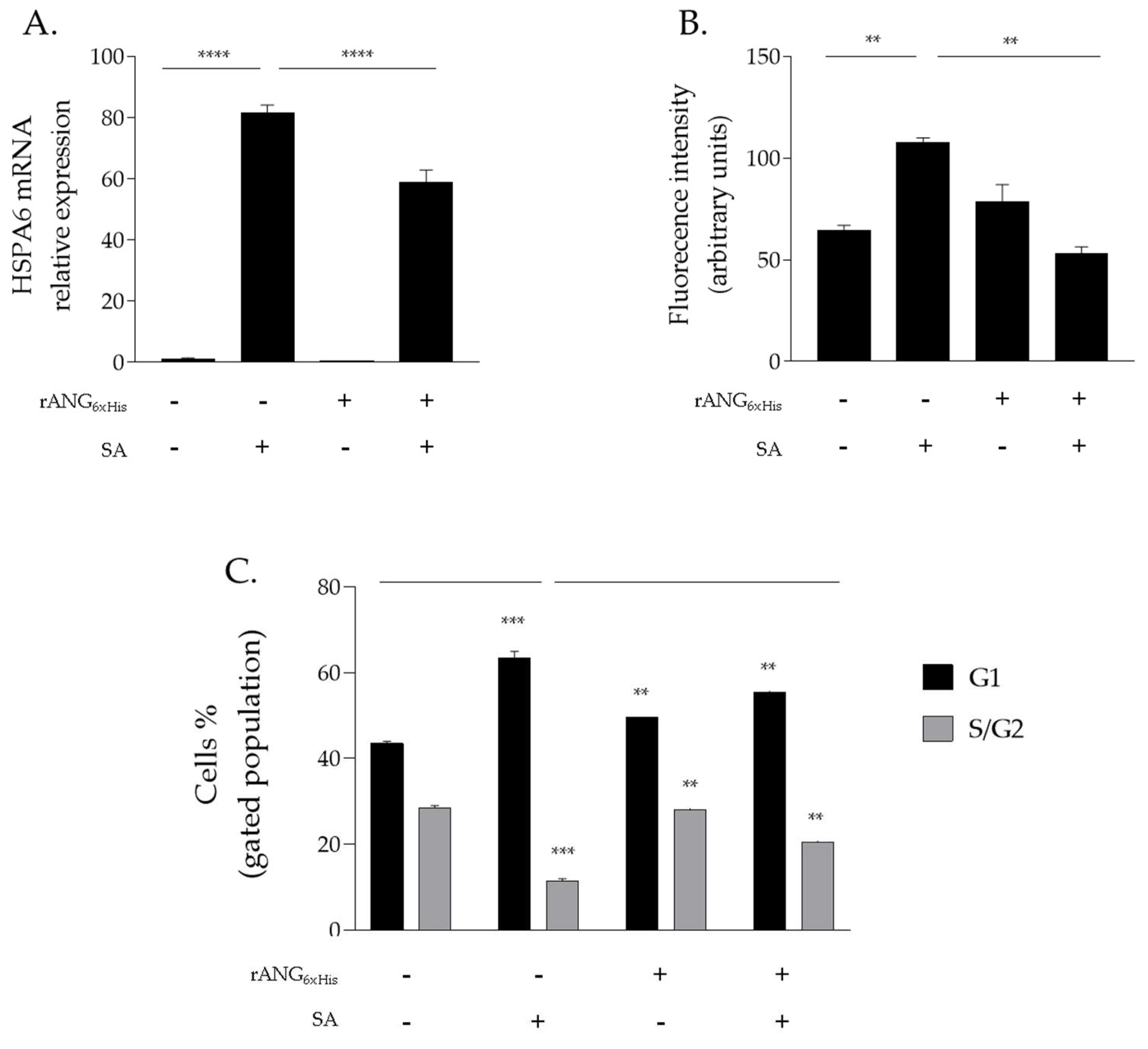
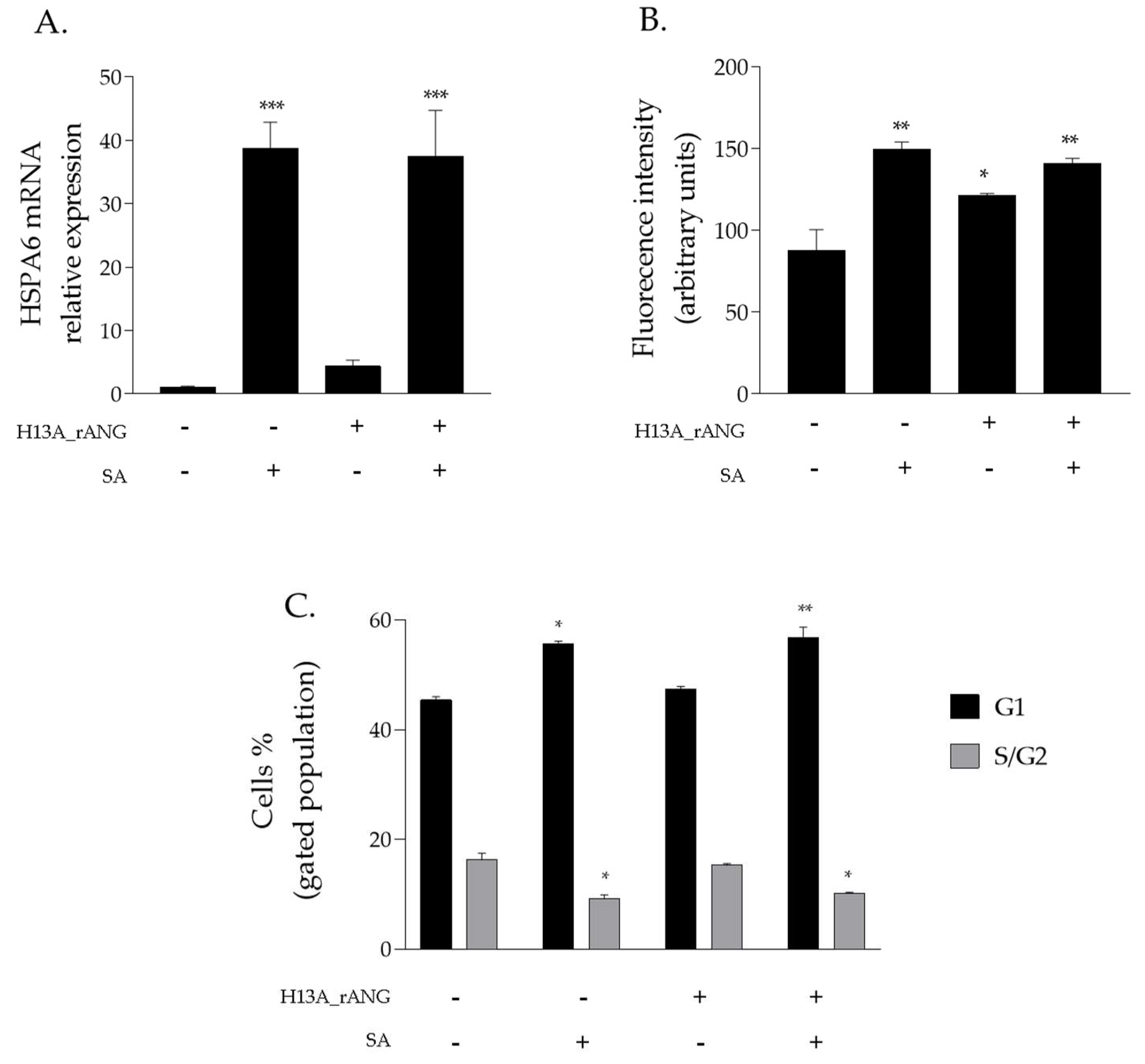
2.4. Effects of H13A_rANG Variant on SA-Stressed HaCaT Cells
3. Discussion
4. Materials and Methods
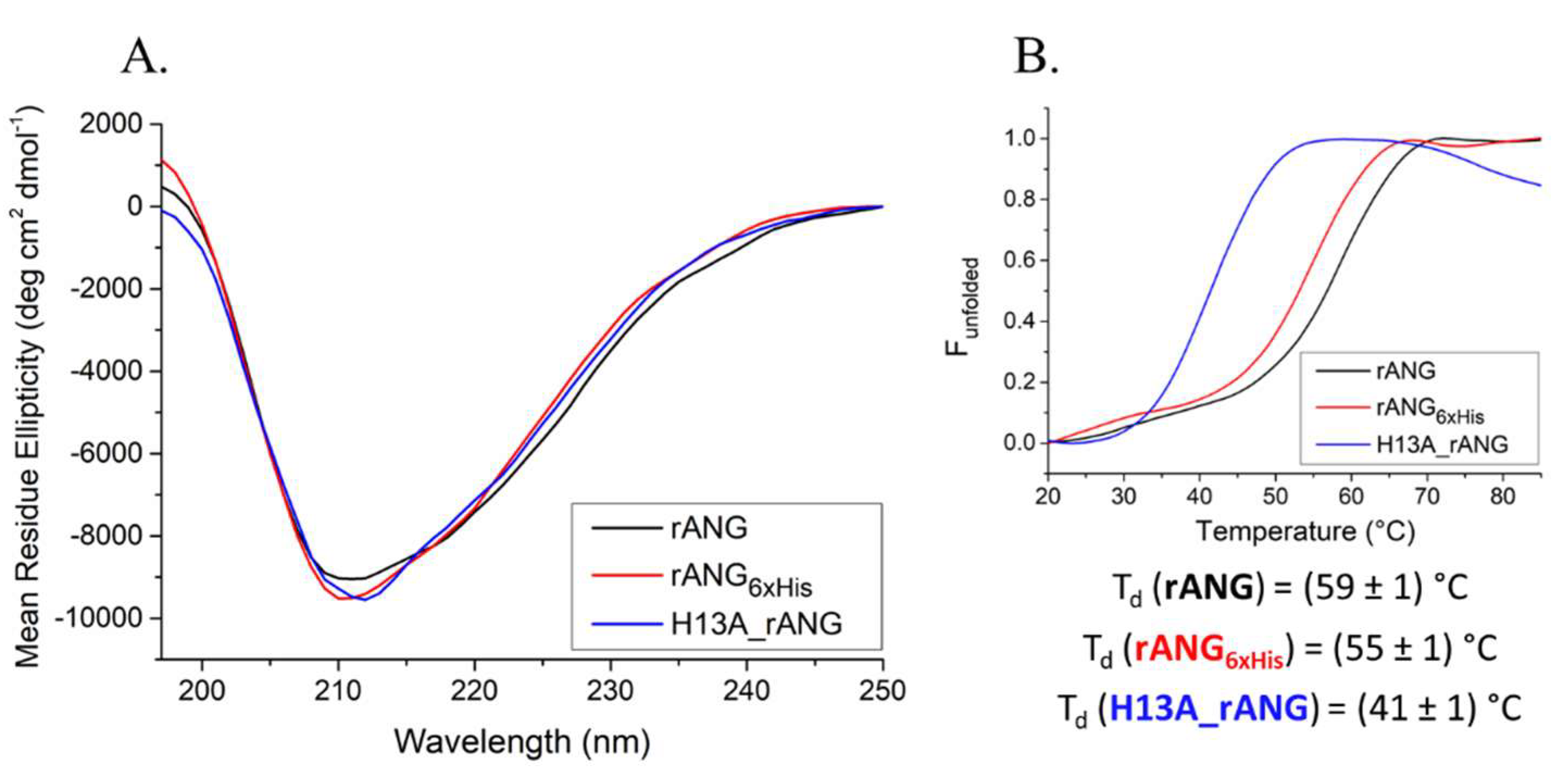
4.1. Heterologous Proteins Production and Purification
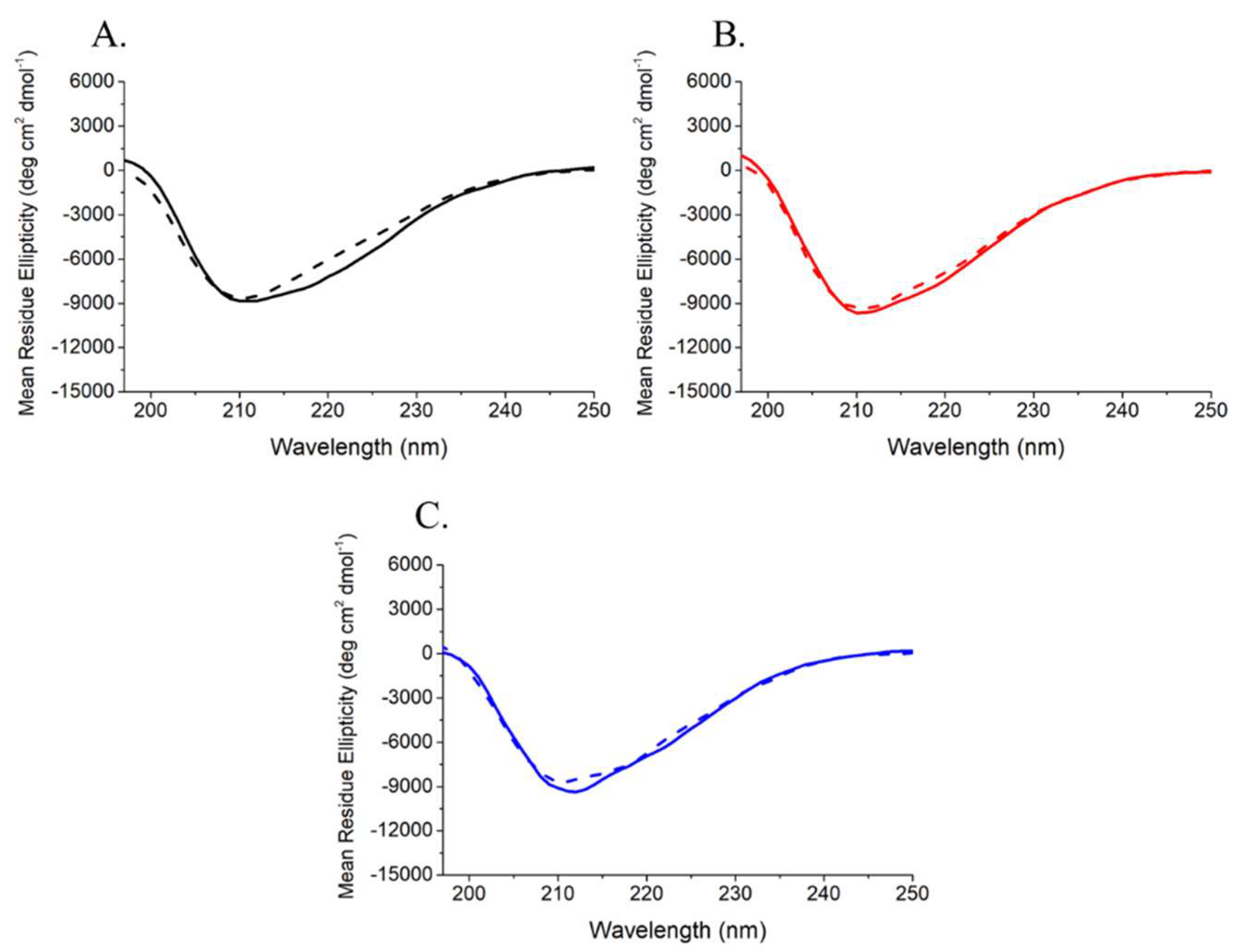
4.2. Circular Dichroism Analysis
4.3. Ribonucleolytic Activity Assays
4.4. Cell Culture and Treatments
4.5. Cellular Extract
4.6. Western Blot Analysis
4.7. Densitometric Analysis
4.8. Cell Viability Assay
4.9. DCFH-DA Assay
4.10. Real-Time Quantitative PCR (RT-qPCR)
4.11. Immunofluorescence
4.12. Cell Cycle Analysis
4.13. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational Regulation of Gene Expression during Conditions of Cell Stress. Mol. Cell 2010, 40, 228–237. [Google Scholar] [CrossRef]
- Yamasaki, S.; Anderson, P. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008, 20, 222–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Bohlen, J.; Roiuk, M.; Teleman, A.A. Phosphorylation of ribosomal protein S6 differentially affects mRNA translation based on ORF length. Nucleic Acids Res. 2021, 49, 13062–13074. [Google Scholar] [CrossRef]
- Clemens, M.J. Initiation Factor e1F2a Phosphorylation in Stress Responses and Apoptosis. In Progress in Molecular and Subcellular Biology; Springer: Berlin/Heidelberg, Germany, 2001; Volume 27, pp. 57–89. [Google Scholar]
- Martínez, A.; Sesé, M.; Losa, J.H.; Robichaud, N.; Sonenberg, N.; Aasen, T.; Ramón, Y.; Cajal, S. Phosphorylation of eIF4E confers resistance to cellular stress and DNA-damaging agents through an interaction with 4E-T: A Rationale for novel therapeutic approaches. PLoS ONE 2015, 10, e0123352. [Google Scholar] [CrossRef] [Green Version]
- Scadden, A.D.J. Inosine-Containing dsRNA Binds a Stress-Granule-like Complex and Downregulates Gene Expression In trans. Mol. Cell 2007, 28, 491–500. [Google Scholar] [CrossRef]
- Shedlovskiy, D.; Zinskie, J.A.; Gardner, E.; Pestov, D.G.; Shcherbik, N. Endonucleolytic cleavage in the expansion segment 7 of 25S rRNA is an early marker of low-level oxidative stress in yeast. J. Biol. Chem. 2017, 292, 18469–18485. [Google Scholar] [CrossRef] [Green Version]
- Thompson, D.M.; Parker, R. Stressing Out over tRNA Cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Pizzo, E.; Sarcinelli, C.; Sheng, J.; Fusco, S.; Formiggini, F.; Netti, P.; Yu, W.; D’Alessio, G.; Hu, G.F. Ribonuclease/angiogenin inhibitor 1 regulates stressinduced subcellular localization of angiogenin to control growth and survival. J. Cell Sci. 2013, 126, 4308–4319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, J.; Xu, Z. Three decades of research on angiogenin: A review and perspective. Acta Biochim. Biophys. Sin. 2016, 48, 399–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leland, P.A.; Staniszewski, K.E.; Park, C.; Kelemen, B.R.; Raines, R.T. The ribonucleolytic activity of angiogenin. Biochemistry 2002, 41, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, K.; Liu, S.; Tsuji, T.; Olson, K.A.; Hu, G.F. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene 2005, 24, 445–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fett, J.W.; Strydom, D.J.; Lobb, R.R.; Alderman, E.M.; Bethune, J.L.; Riordan, J.F.; Vallee, B.L. Isolation and Characterization of Angiogenin, an Angiogenic Protein from Human Carcinoma Cells. Biochemistry 1985, 24, 5480–5486. [Google Scholar] [CrossRef]
- Yu, W.; Goncalves, K.A.; Li, S.; Kishikawa, H.; Sun, G.; Yang, H.; Vanli, N.; Wu, Y.; Jiang, Y.; Hu, M.G.; et al. Plexin-B2 Mediates Physiologic and Pathologic Functions of Angiogenin. Cell 2017, 171, 849–864.e25. [Google Scholar] [CrossRef] [Green Version]
- Greenway, M.J.; Andersen, P.M.; Russ, G.; Ennis, S.; Cashman, S.; Donaghy, C.; Patterson, V.; Swingler, R.; Kieran, D.; Prehn, J.; et al. ANG mutations segregate with familial and “sporadic” amyotrophic lateral sclerosis. Nat. Genet. 2006, 38, 411–413. [Google Scholar] [CrossRef]
- Prehn, J.H.M.; Jirström, E. Angiogenin and tRNA fragments in Parkinson’s disease and neurodegeneration. Acta Pharmacol. Sin. 2020, 41, 442–446. [Google Scholar] [CrossRef]
- Kao, R.Y.T.; Jenkins, J.L.; Olson, K.A.; Key, M.E.; Fett, J.W.; Shapiro, R. A small-molecule inhibitor of the ribonucleolytic activity of human angiogenin that possesses antitumor activity. Proc. Natl. Acad. Sci. USA 2002, 99, 10066–10071. [Google Scholar] [CrossRef] [Green Version]
- Kishikawa, H.; Wu, D.; Hu, G.F. Targeting angiogenin in therapy of amyotropic lateral sclerosis. Expert Opin. Ther. Targets 2008, 12, 1229–1242. [Google Scholar] [CrossRef] [Green Version]
- Moroianu, J.; Riordan, J.F. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc. Natl. Acad. Sci. USA 1994, 91, 1677–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Kim, K.W.; Min, K.M.; Kim, K.W.; Chang, S.I.; Kim, J.C. Angiogenin reduces immune inflammation via inhibition of tank-binding kinase 1 expression in human corneal fibroblast cells. Mediators Inflamm. 2014, 2014, 861435. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003, 4, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Cucci, L.M.; Satriano, C.; Marzo, T.; La Mendola, D. Angiogenin and copper crossing in wound healing. Int. J. Mol. Sci. 2021, 22, 10704. [Google Scholar] [CrossRef]
- Skorupa, A.; King, M.A.; Aparicio, I.M.; Dussmann, H.; Coughlan, K.; Breen, B.; Kieran, D.; Concannon, C.G.; Marin, P.; Prehn, J.H.M. Motoneurons secrete angiogenin to induce RNA cleavage in astroglia. J. Neurosci. 2012, 32, 5024–5038. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Tao, E.W.; Cheng, W.Y.; Li, W.L.; Yu, J.; Gao, Q.Y. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J. Cell. Physiol. 2020, 235, 683–690. [Google Scholar] [CrossRef]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar] [CrossRef] [Green Version]
- Lyons, S.M.; Fay, M.M.; Akiyama, Y.; Anderson, P.J.; Ivanov, P. RNA biology of angiogenin: Current state and perspectives. RNA Biol. 2017, 14, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Tao, E.W.; Wang, H.L.; Cheng, W.Y.; Liu, Q.Q.; Chen, Y.X.; Gao, Q.Y. A specific tRNA half, 5′tiRNA-His-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. J. Exp. Clin. Cancer Res. 2021, 40, 67. [Google Scholar] [CrossRef]
- Li, S.; Shi, X.; Chen, M.; Xu, N.; Sun, D.; Bai, R.; Chen, H.; Ding, K.; Sheng, J.; Xu, Z. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int. J. Cancer 2019, 145, 1395–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, G.K.; Kligman, A.M. Barrier functions of human skin: A holistic view. Skin Pharmacol. Physiol. 2009, 22, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Yurina, N.V.; Ageeva, T.A.; Goryachkin, A.M.; Varaksin, N.A.; Ryabicheva, T.G.; Ostanin, A.A.; Chernykh, E.R.; Romashchenko, A.V.; Proskurina, A.S.; Bogachev, S.; et al. Effects of recombinant angiogenin on collagen fiber formation and angiogenesis in the dermis of wistar rats. Clin. Cosmet. Investig. Dermatol. 2021, 14, 187–196. [Google Scholar] [CrossRef]
- Pizzo, E.; Buonanno, P.; Di Maro, A.; Ponticelli, S.; De Falco, S.; Quarto, N.; Cubellis, M.V.; D’Alessio, G. Ribonucleases and angiogenins from fish. J. Biol. Chem. 2006, 281, 27454–27460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crabtree, B.; Thiyagarajan, N.; Prior, S.H.; Wilson, P.; Iyer, S.; Ferns, T.; Shapiro, R.; Brew, K.; Subramanian, V.; Acharya, K.R. Characterization of Human Angiogenin Variants Implicated in Amyotrophic Lateral. Biochemistry 2007, 46, 11810–11818. [Google Scholar] [CrossRef] [Green Version]
- Pizzo, E.; Merlino, A.; Turano, M.; Russo Krauss, I.; Coscia, F.; Zanfardino, A.; Varcamonti, M.; Furia, A.; Giancola, C.; Mazzarella, L.; et al. A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish. Biochem. J. 2011, 433, 345–355. [Google Scholar] [CrossRef]
- Ramirez, V.P.; Stamatis, M.; Shmukler, A.; Aneskievich, B.J. Basal and stress-inducible expression of HSPA6 in human keratinocytes is regulated by negative and positive promoter regions. Cell Stress Chaperones 2015, 20, 95–107. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, R.; Vallee, B.L. Site-Directed Mutagenesis of Histidine-13 and Histidine-114 of Human Angiogenin. Alanine Derivatives Inhibit Angiogenin-Induced Angiogenesis. Biochemistry 1989, 28, 7401–7408. [Google Scholar] [CrossRef]
- Fasoli, S.; Bettin, I.; Montioli, R.; Fagagnini, A.; Peterle, D.; Laurents, D.V.; Gotte, G. Dimerization of human angiogenin and of variants involved in neurodegenerative diseases. Int. J. Mol. Sci. 2021, 22, 10068. [Google Scholar] [CrossRef]
- Leonidas, D.D.; Shapiro, R.; Allen, S.C.; Subbarao, G.V.; Veluraja, K.; Acharya, K.R. Refined crystal structures of native human angiogenin and two active site variants: Implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth. J. Mol. Biol. 1999, 285, 1209–1233. [Google Scholar] [CrossRef]
- Sica, F.; Russo Krauss, I.; Troisi, R.; Bosso, A.; Culurciello, R.; Carluccio, C.; Trapani, M.; Merlino, A.; Mazzarella, L.; Pizzo, E. The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition. Int. J. Biol. Macromol. 2021, 182, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Varcamonti, M.; Di Maro, A.; Zanfardino, A.; Giancola, C.; D’Alessio, G. Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon. FEBS J. 2008, 275, 1283–1295. [Google Scholar] [CrossRef]
- Kelemen, B.R.; Klink, T.A.; Behlke, M.A.; Eubanks, S.R.; Leland, P.A.; Raines, R.T. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999, 27, 3696–3701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emara, M.M.; Fujimura, K.; Sciaranghella, D.; Ivanova, V.; Ivanov, P.; Anderson, P. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 2012, 423, 763–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018, 2018, 469–471. [Google Scholar] [CrossRef]
- Aranda, A.; Sequedo, L.; Tolosa, L.; Quintas, G.; Burello, E.; Castell, J.V.; Gombau, L. Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay: A quantitative method for oxidative stress assessment of nanoparticle-treated cells. Toxicol. Vitr. 2013, 27, 954–963. [Google Scholar] [CrossRef]



| kcat/KM (M−1 s−1) | |
|---|---|
| RNase A | (4.6 ± 0.5) × 107 |
| rANG | (5.2 ± 0.3) × 103 |
| rANG6xHis | (1.6 ± 0.2) × 102 |
| Genes | Forward | Reverse |
|---|---|---|
| GAPDH | 5′-CACCACACTGAATCTCCCCT-3′ | 5′-TGGTTGAGCACAGGGTACTT-3′ |
| ANG | 5′-CACTTCCTGACCCAGCACTA-3′ | 5′-ATGTCTTTGCAGGGTGAGGT-3′ |
| HSPA6 | 5′-TGCAAGAGGAAAGCCTTAGGGACA-3′ | 5′-TTTGCTCCAGCTCCCTCT TCTGAT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Culurciello, R.; Bosso, A.; Troisi, R.; Barrella, V.; Di Nardo, I.; Borriello, M.; Gaglione, R.; Pistorio, V.; Aceto, S.; Cafaro, V.; et al. Protective Effects of Recombinant Human Angiogenin in Keratinocytes: New Insights on Oxidative Stress Response Mediated by RNases. Int. J. Mol. Sci. 2022, 23, 8781. https://doi.org/10.3390/ijms23158781
Culurciello R, Bosso A, Troisi R, Barrella V, Di Nardo I, Borriello M, Gaglione R, Pistorio V, Aceto S, Cafaro V, et al. Protective Effects of Recombinant Human Angiogenin in Keratinocytes: New Insights on Oxidative Stress Response Mediated by RNases. International Journal of Molecular Sciences. 2022; 23(15):8781. https://doi.org/10.3390/ijms23158781
Chicago/Turabian StyleCulurciello, Rosanna, Andrea Bosso, Romualdo Troisi, Valentina Barrella, Ilaria Di Nardo, Margherita Borriello, Rosa Gaglione, Valeria Pistorio, Serena Aceto, Valeria Cafaro, and et al. 2022. "Protective Effects of Recombinant Human Angiogenin in Keratinocytes: New Insights on Oxidative Stress Response Mediated by RNases" International Journal of Molecular Sciences 23, no. 15: 8781. https://doi.org/10.3390/ijms23158781
APA StyleCulurciello, R., Bosso, A., Troisi, R., Barrella, V., Di Nardo, I., Borriello, M., Gaglione, R., Pistorio, V., Aceto, S., Cafaro, V., Notomista, E., Sica, F., Arciello, A., & Pizzo, E. (2022). Protective Effects of Recombinant Human Angiogenin in Keratinocytes: New Insights on Oxidative Stress Response Mediated by RNases. International Journal of Molecular Sciences, 23(15), 8781. https://doi.org/10.3390/ijms23158781









