Disease Resistance and Molecular Variations in Irradiation Induced Mutants of Two Pea Cultivars
Abstract
1. Introduction
2. Results
2.1. Resistance Variation
2.1.1. Fusarium Wilt
2.1.2. Powdery Mildew
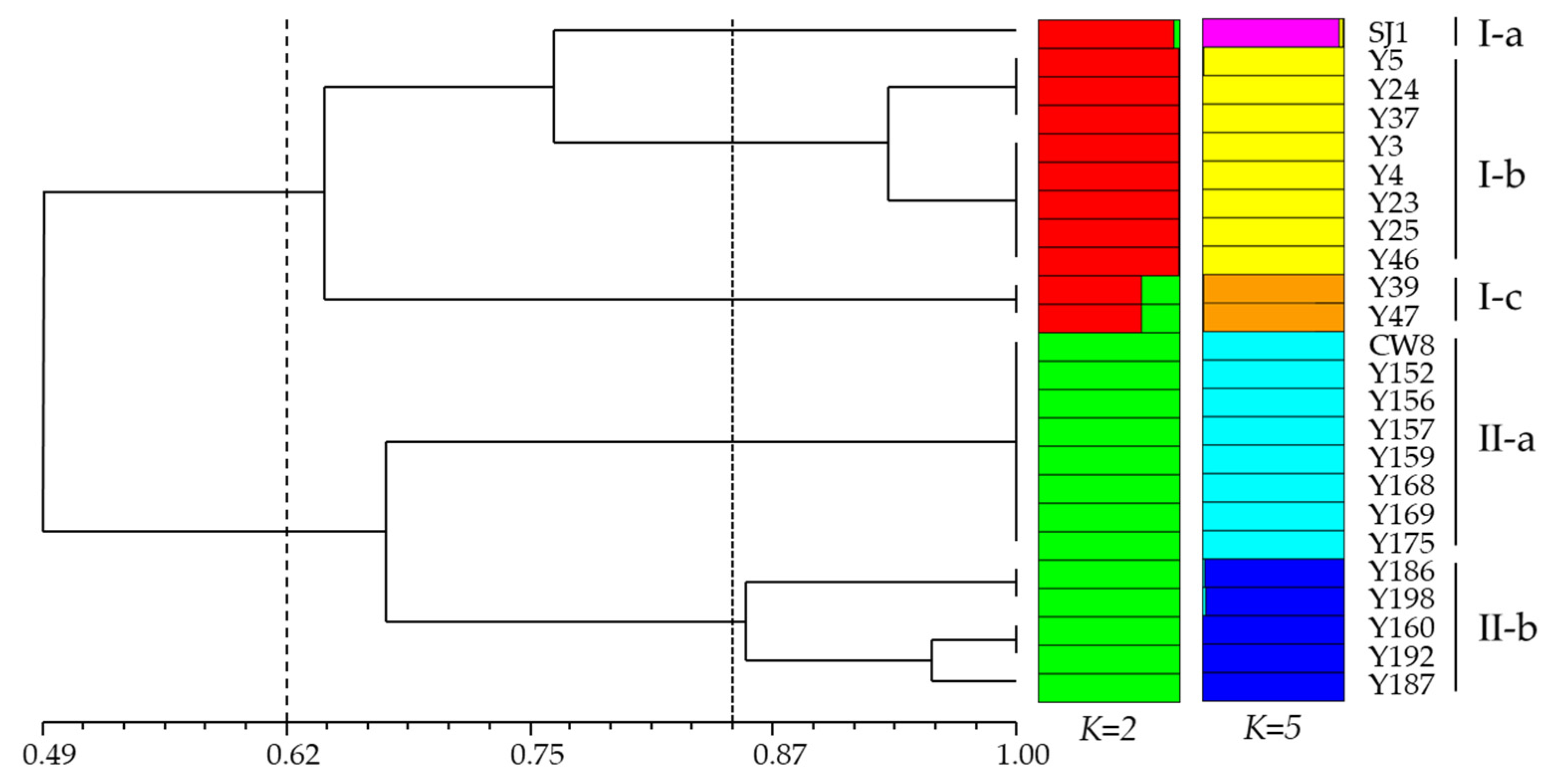
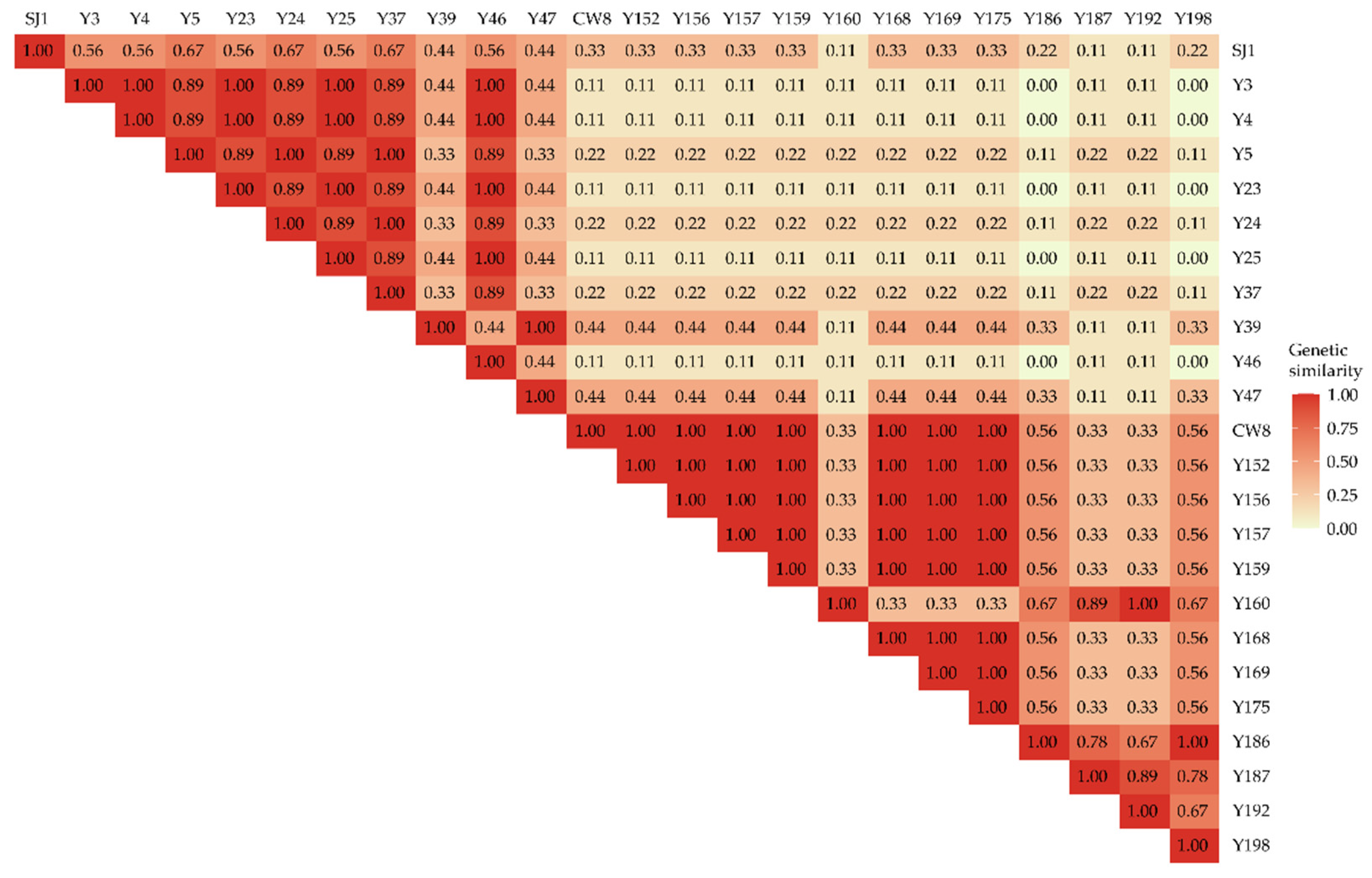
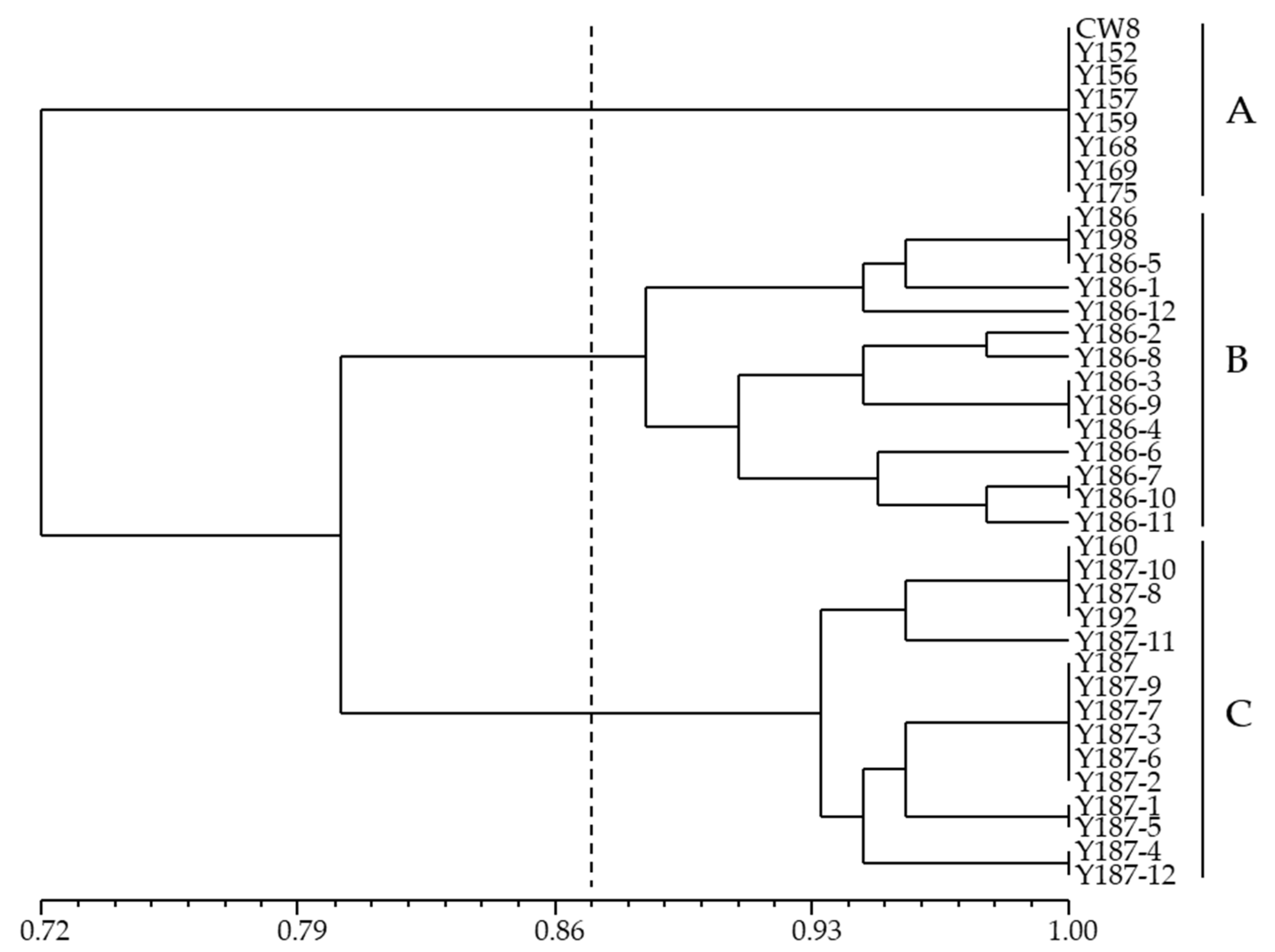
2.2. Molecular Variation Analysis of Mutants
2.3. PsMLO1 Sequence Analysis
3. Discussion
4. Materials and Methods
4.1. Pea Mutants and Pathogen Isolates
4.2. Resistance Evaluation
4.2.1. Fusarium Wilt
4.2.2. Powdery Mildew
4.3. Molecular Variation Analysis of Mutants
4.4. RNA Extraction and PsMLO1 Sequence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cousin, R. Peas (Pisum sativum L.). Field Crops Res. 1997, 53, 111–130. [Google Scholar] [CrossRef]
- Pandey, A.K.; Rubiales, D.; Wang, Y.; Fang, P.; Sun, T.; Liu, N.; Xu, P. Omics resources and omics-enabled approaches for achieving high productivity and improved quality in pea (Pisum sativum L.). Theor. Appl. Genet. 2021, 134, 755–776. [Google Scholar] [CrossRef] [PubMed]
- Guindon, M.F.; Cazzola, F.; Palacios, T.; Gatti, I.; Bermejo, C.; Cointry, E. Biofortification of pea (Pisum sativum L.): A review. J. Sci. Food Agric. 2021, 101, 3551–3563. [Google Scholar] [CrossRef]
- Trinidad, T.P.; Mallillin, A.C.; Loyola, A.S.; Sagum, R.S.; Encabo, R.R. The potential health benefits of legumes as a good source of dietary fibre. Br. J. Nutr. 2010, 103, 569–574. [Google Scholar] [CrossRef]
- Dahl, W.J.; Foster, L.M.; Tyler, R.T. Review of the health benefits of peas (Pisum sativum L.). Br. J. Nutr. 2012, 108 (Suppl. 1), S3–S10. [Google Scholar] [CrossRef]
- FAOSTAT. FAOSTAT Crop Statistics. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 17 February 2022).
- Kraft, J.M.; Pfleger, F.L. Compendium of Pea Diseases and Pests, 2nd ed.; American Phytopathological Society (APS Press): St. Paul, MN, USA, 2001; pp. 13–14. [Google Scholar]
- Sharma, A.; Rathour, R.; Plaha, P.; Katoch, V.; Khalsa, G.S.; Patial, V.; Singh, Y.; Pathania, N.K. Induction of Fusarium wilt (Fusarium oxysporum f. sp. pisi) resistance in garden pea using induced mutagenesis and in vitro selection techniques. Euphytica 2010, 173, 345–356. [Google Scholar] [CrossRef]
- Fondevilla, S.; Cubero, J.I.; Rubiales, D. Confirmation that the Er3 gene, conferring resistance to Erysiphe pisi in pea, is a different gene from er1 and er2 genes. Plant Breed. 2011, 130, 281–282. [Google Scholar] [CrossRef]
- Sun, S.; He, Y.; Dai, C.; Duan, C.; Zhu, Z. Two major er1 alleles confer powdery mildew resistance in three pea cultivars bred in Yunnan Province, China. Crop J. 2016, 4, 353–359. [Google Scholar] [CrossRef]
- Jain, S.; Weeden, N.F.; Kumar, A.; Chittem, K.; McPhee, K. Functional codominant marker for selecting the Fw gene conferring resistance to Fusarium wilt race 1 in pea. Crop Sci. 2015, 55, 2639–2646. [Google Scholar] [CrossRef]
- Sun, S.; Deng, D.; Wang, Z.; Duan, C.; Wu, X.; Wang, X.; Zong, X.; Zhu, Z. A novel er1 allele and the development and validation of its functional marker for breeding pea (Pisum sativum L.) resistance to powdery mildew. Theor. Appl. Genet. 2016, 129, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Horáček, J.; Dostálová, R.; Hýbl, M. Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J. Appl. Genet. 2008, 49, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Coyne, C.J.; Kumar, S.; Wettberg, E.J.B.; Marques, E.; Berger, J.D.; Redden, R.J.; Ellis, T.H.N.; Brus, J.; Zablatzká, L.; Smýkal, P. Potential and limits of exploitation of crop wild relatives for pea, lentil, and chickpea improvement. Legume Sci. 2020, 2, e36. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 2002, 124, 163–180. [Google Scholar] [CrossRef]
- Rimbaud, L.; Papaix, J.; Rey, J.F.; Barrett, L.G.; Thrall, P.H. Assessing the durability and efficiency of landscape-based strategies to deploy plant resistance to pathogens. PLoS Comput. Biol. 2018, 14, e1006067. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, T.; Liu, R.; Redden, B.; Maalouf, F.; Zong, X. Food legume production in China. Crop J. 2017, 5, 115–126. [Google Scholar] [CrossRef]
- Abdel-Hamid, A.M.; Salem, K.F. Breeding Strategies of Garden Pea (Pisum sativum L.). In Advances in Plant Breeding Strategies: Vegetable Crops; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2021; pp. 331–377. [Google Scholar] [CrossRef]
- Jacobsen, E.; Feenstra, W.J. A new pea mutant with efficient nodulation in the presence of nitrate. Plant Sci. Lett. 1984, 33, 337–344. [Google Scholar] [CrossRef]
- Sinjushin, A.; Semenova, E.; Vishnyakova, M. Usage of morphological mutations for improvement of a garden pea (Pisum sativum): The experience of breeding in Russia. Agronomy 2022, 12, 544. [Google Scholar] [CrossRef]
- Sax, K. The stimulation of plant growth by ionizing radiation. Radiat. Bot. 1963, 3, 179–186. [Google Scholar] [CrossRef]
- Jaranowski, J.; Mickle, A. Mutation breeding in peas. Mutation Breeding Rev. 1985, 2, 1–23. [Google Scholar]
- Sinjushin, A. Mutation genetics of pea (Pisum sativum L.): What is done and what is left to do. Ratar. Povrt. 2013, 50, 36–43. [Google Scholar]
- Fadl, F.A.M. Induced mutations in beans and peas for resistance to rust. In Research Co-Ordination Meeting on Induced Mutations for Disease Resistance in Crop Plants II; International Atomic Energy Agency (IAEA): Risoe, Denmark, 1983; pp. 163–170. [Google Scholar]
- Pereira, G.; Marques, C.; Ribeiro, R.; Formiga, S.; Dâmaso, M.; Tavares Sousa, M.; Farinhó, M.; Leitão, J.M. Identification of DNA markers linked to an induced mutated gene conferring resistance to powdery mildew in pea (Pisum sativum L.). Euphytica 2009, 171, 327–335. [Google Scholar] [CrossRef]
- Weller, J.L.; Reid, J.B.; Taylor, S.A.; Murfet, I.C. The genetic control of flowering in pea. Trends Plant Sci. 1997, 2, 412–418. [Google Scholar] [CrossRef]
- Moreau, C.; Warren, F.J.; Rayner, T.; Perez-Moral, N.; Lawson, D.M.; Wang, T.L.; Domoney, C. An allelic series of starch-branching enzyme mutants in pea (Pisum sativum L.) reveals complex relationships with seed starch phenotypes. Carbohydr. Polym. 2022, 288, 119386. [Google Scholar] [CrossRef]
- Tsyganov, V.E.; Tsyganova, A.V. Symbiotic regulatory genes controlling nodule development in Pisum sativum L. Plants 2020, 9, 1741. [Google Scholar] [CrossRef] [PubMed]
- Moreau, C.; Hofer, J.M.I.; Eleouet, M.; Sinjushin, A.; Ambrose, M.; Skot, K.; Blackmore, T.; Swain, M.; Hegarty, M.; Balanza, V.; et al. Identification of Stipules reduced, a leaf morphology gene in pea (Pisum sativum). New Phytol. 2018, 220, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Harland, S.C. Inheritance of immunity to mildew in peruvian forms of Pisum sativum. Heredity 1948, 2, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Humphry, M.; Consonni, C.; Panstruga, R. mlo-based powdery mildew immunity: Silver bullet or simply non-host resistance? Mol. Plant Pathol. 2006, 7, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Marcotrigiano, A.R.; Cillo, F.; Visser, R.G.; Bai, Y.; Lotti, C.; Ricciardi, L. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 2011, 123, 1425–1431. [Google Scholar] [CrossRef]
- Santo, T.; Rashkova, M.; Alabaça, C.; Leitão, J. The ENU-induced powdery mildew resistant mutant pea (Pisum sativum L.) lines S(er1mut1) and F(er1mut2) harbour early stop codons in the PsMLO1 gene. Mol. Breed. 2013, 32, 723–727. [Google Scholar] [CrossRef]
- Du, L.F.; Gu, Z.L.; Zhong, H.; Dong, Y.; Wu, M.P.; Pan, C.G. Mutation effect of plasma immersion N+ ion implantation on plumule cells of M1 in pea seeds. Hereditas 2000, 22, 398–400. (In Chinese) [Google Scholar]
- Wang, B.F.; Fu, J.F.; Dong, L.F. Inducing autotetraploid pea with colchicine and DMSO. J. Nucl. Agric. Sci. 2009, 23, 203. (In Chinese) [Google Scholar]
- Ouyang, Y.Y.; Yang, M.; Xiang, C.; Yu, D.M. Multiple analysis of agronomic characters and yield of pea M2 induced by 60Coγ. Chin. Agric. Sci. Bull. 2018, 34, 33–40. (In Chinese) [Google Scholar]
- Xu, D.P.; Feng, H.Y.; Pan, J.B.; Yao, Z.E.; Wang, J.R. Radiation dose effects on the morphological development of M1 generation pea (Pisum sativum). Nucl. Sci. Tech. 2021, 32, 124. [Google Scholar] [CrossRef]
- Xu, D.P.; Yao, Z.E.; Pan, J.B.; Feng, H.Y.; Guo, Z.Q.; Lu, X.L. Study on the multiple characteristics of M3 generation of pea mutants obtained by neutron irradiation. Nucl. Sci. Tech. 2020, 31, 67. [Google Scholar] [CrossRef]
- Deng, D.; Sun, S.; Wu, W.; Zong, X.; Yang, X.; Zhang, X.; He, Y.; Duan, C.; Zhu, Z. Screening for pea germplasms resistant to Fusarium wilt race 5. Agronomy 2022, 12, 1354. [Google Scholar] [CrossRef]
- Suprasanna, P.; Mirajkar, S.J.; Bhagwat, S.G. Induced mutations and crop improvement. In Plant Biology and Biotechnology; Bahadur, B., Venkat Rajam, M., Sahijram, L., Krishnamurthy, K., Eds.; Springer: New Delhi, India, 2015; Volume 1, pp. 593–617. [Google Scholar] [CrossRef]
- Ahmar, S.; Gill, R.A.; Jung, K.H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and molecular techniques from simple breeding to speed breeding in crop plants: Recent advances and future outlook. Int. J. Mol. Sci. 2020, 21, 2950. [Google Scholar] [CrossRef] [PubMed]
- Arora, L.; Narula, A. Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant. Sci. 2017, 8, 1932. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, B.; Chen, W.; Qiu, T.; Wang, L.; Yang, S. Space breeding in modern agriculture. Am. J. Agric. Res. 2020, 5, 81. [Google Scholar] [CrossRef][Green Version]
- Mohanta, T.K.; Mishra, A.K.; Mohanta, Y.K.; Al-Harrasi, A. Space Breeding: The Next-Generation Crops. Front. Plant. Sci. 2021, 12, 771985. [Google Scholar] [CrossRef]
- Sigurbjörnsson, B. Induced mutations 3rd ed. In Crop Breeding; Wood, D.R., Ed.; Soil Science Society of America: Madison, WI, USA; pp. 153–176. [CrossRef]
- Al-Choboq, J.; Ferlazzo, M.L.; Sonzogni, L.; Granzotto, A.; El-Nachef, L.; Maalouf, M.; Berthel, E.; Foray, N. Usher syndrome belongs to the genetic diseases associated with radiosensitivity: Influence of the ATM protein kinase. Int. J. Mol. Sci. 2022, 23, 1570. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Coradini, A.L.; Shen, A.; Ehrenreich, I.M. Layers of cryptic genetic variation underlie a yeast complex trait. Genetics 2019, 211, 1469–1482. [Google Scholar] [CrossRef] [PubMed]
- Raina, A.; Laskar, R.A.; Tantray, Y.R.; Khursheed, S.; Wani, M.R.; Khan, S. Characterization of induced high yielding cowpea mutant lines using physiological, biochemical and molecular markers. Sci. Rep. 2020, 10, 3687. [Google Scholar] [CrossRef]
- Ramchander, S.; Leon, M.; Souframanien, J.; Arumugam Pillai, M. Genetic diversity, allelic variation and marker trait associations in gamma irradiated mutants of rice (Oryza sativa L.). Int. J. Radiat. Biol. 2022, 98, 90–99. [Google Scholar] [CrossRef]
- Yang, T.; Fang, L.; Zhang, X.; Hu, J.; Bao, S.; Hao, J.; Li, L.; He, Y.; Jiang, J.; Wang, F.; et al. High-throughput development of SSR markers from pea (Pisum sativum L.) based on next generation sequencing of a purified Chinese commercial variety. PLoS ONE 2015, 10, e0139775. [Google Scholar] [CrossRef]
- Javaid, A.; Ghafoor, A.; Rabbani, M.A. Analysis of genetic diversity among local and exotic Pisum sativum genotypes through RAPD and SSR markers. Pak. J. Bot. 2022, 54, 903–909. [Google Scholar] [CrossRef]
- Muñoz-Falcón, J.E.; Vilanova, S.; Plazas, M.; Prohens, J. Diversity, relationships, and genetic fingerprinting of the Listada de Gandía eggplant landrace using genomic SSRs and EST-SSRs. Sci. Hortic. 2011, 129, 238–246. [Google Scholar] [CrossRef]
- Anupam, A.; Imam, J.; Quatadah, S.M.; Siddaiah, A.; Prasad Das, S.; Variar, M.; Prasad Mandal, N. Genetic diversity analysis of rice germplasm in Tripura State of Northeast India using drought and blast linked markers. Rice Sci. 2017, 24, 10–20. [Google Scholar] [CrossRef]
- Ge, H.; Liu, Y.; Jiang, M.; Zhang, J.; Han, H.; Chen, H. Analysis of genetic diversity and structure of eggplant populations (Solanum melongena L.) in China using simple sequence repeat markers. Sci. Hortic. 2013, 162, 71–75. [Google Scholar] [CrossRef]
- Cheng, X.; Chai, L.; Chen, Z.; Xu, L.; Zhai, H.; Zhao, A.; Peng, H.; Yao, Y.; You, M.; Sun, Q.; et al. Identification and characterization of a high kernel weight mutant induced by gamma radiation in wheat (Triticum aestivum L.). BMC Genet. 2015, 16, 127. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Feng, H.; Du, Y.; Luo, S.; Li, W.; Yu, L.; Feng, Z.; Cui, T.; Zhou, L. Genetic polymorphisms in mutagenesis progeny of Arabidopsis thaliana irradiated by carbon-ion beams and gamma-rays irradiations. Int. J. Radiat. Biol. 2020, 96, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Theerawitaya, C.; Triwitayakorn, K.; Kirdmanee, C.; Smith, D.R.; Supaibulwatana, K. Genetic variations associated with salt tolerance detected in mutants of kdml105 (‘oryza sativa L. Spp. Indica’) rice. Aust. J. Crop. Sci. 2011, 5, 1475–1480. [Google Scholar]
- Humphry, M.; Reinstadler, A.; Ivanov, S.; Bisseling, T.; Panstruga, R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 2011, 12, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Sulima, A.S.; Zhukov, V.A. War and Peas: Molecular bases of resistance to powdery mildew in pea (Pisum sativum L.) and other legumes. Plants 2022, 11, 339. [Google Scholar] [CrossRef]
- Cooper, J.L.; Till, B.J.; Laport, R.G.; Darlow, M.C.; Kleffner, J.M.; Jamai, A.; El-Mellouki, T.; Liu, S.; Ritchie, R.; Nielsen, N.; et al. TILLING to detect induced mutations in soybean. BMC Plant Biol. 2008, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Hirakawa, H.; Nunome, T.; Tabata, S.; Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 2016, 14, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Burstin, J.; Kreplak, J.; Macas, J.; Lichtenzveig, J. Pisum sativum (Pea). Trends Genet. 2020, 36, 312–313. [Google Scholar] [CrossRef]
- Grandbastien, M.A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998, 3, 181–187. [Google Scholar] [CrossRef]
- Gowda, M.V.C.; Bhat, R.S.; Sujay, V.; Kusuma, P.; Varshakumari; Bhat, S.; Varshney, R.K. Characterization of AhMITE1 transposition and its association with the mutational and evolutionary origin of botanical types in peanut (Arachis spp.). Plant Syst. Evol. 2010, 291, 153–158. [Google Scholar] [CrossRef]
- Negi, P.; Rai, A.N.; Suprasanna, P.M. Moving through the stressed genome: Emerging regulatory roles for transposons in plant stress response. Front. Plant Sci. 2016, 7, 1448. [Google Scholar] [CrossRef]
- Hung, N.N.; Kim, D.G.; Lyu, J.I.; Park, K.C.; Kim, J.M.; Kim, J.B.; Ha, B.K.; Kwon, S.J. Detecting genetic mobility using a transposon-based marker system in gamma-ray irradiated soybean mutants. Plants 2021, 10, 373. [Google Scholar] [CrossRef]
- Sen, A. Retrotransposon insertion variations in doubled haploid bread wheat mutants. Plant Growth Regul. 2016, 81, 325–333. [Google Scholar] [CrossRef]
- Moreau, C.; Ambrose, M.J.; Turner, L.; Hill, L.; Ellis, T.H.; Hofer, J.M. The b gene of pea encodes a defective flavonoid 3′,5′-hydroxylase, and confers pink flower color. Plant Physiol. 2012, 159, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Sun, S.; Zhu, L.; Duan, C.; Zhu, Z. Confirmation of Fusarium oxysporum as a causal agent of mung bean wilt in China. Crop Prot. 2019, 117, 77–85. [Google Scholar] [CrossRef]
- Haglund, W.A. A rapid method for inoculating pea seedlings with Fusarium oxysporum f. sp. pisi. Plant Dis. 1989, 73, 457–458. [Google Scholar] [CrossRef]
- Liu, R.; Wang, F.; Fang, L.; Yang, T.; Zhang, H.; Huang, Y.; Wang, D.; Ji, Y.; Xu, D.; Li, G.; et al. An integrated high-density SSR genetic linkage map from two F2 population in Chinese pea. Acta Agron. Sin. 2020, 46, 1496–1506. (In Chinese) [Google Scholar]
- Liu, R.; Fang, L.; Yang, T.; Zhang, X.; Hu, J.; Zhang, H.; Han, W.; Hua, Z.; Hao, J.; Zong, X. Marker-trait association analysis of frost tolerance of 672 worldwide pea (Pisum sativum L.) collections. Sci. Rep. 2017, 7, 5919. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.J.; Ye, Z.H.; Mao, J.X. Popgene, the User-friendly Shareware for Population Genetic Analysis; Molecular Biology and Biotechnology Centre, University of Alberta: Edmonton, AB, Canada, 1997. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker V3.25: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 9, 2128–2129. [Google Scholar] [CrossRef]
- Rohlf, F.J. Ntsys-pc: Numerical Taxonomy and Multivariate Analysis System; Department of Ecology and Evolution, State University of New York: Stony Brook, NY, USA, 1992. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]


| Line 1 | Parent | Generation | Fusarium Wilt | Powdery Mildew | ||
|---|---|---|---|---|---|---|
| Disease Index | Reaction 2 | Infection Type | Reaction 3 | |||
| SJ1 | - | - | 20.00 | R | 4 | S |
| CW8 | - | - | 20.00 | R | 4 | S |
| Y3 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y4 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y23 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y25 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y37 | SJ1 | M3 | 40.00 | M | 4 | S |
| Y38 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y39 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y46 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y47 | SJ1 | M3 | 100.00 | HS | 4 | S |
| Y160 | CW8 | M3 | 100.00 | HS | 0, 1 | I, R |
| Y186 | CW8 | M3 | 66.67 | M | 0, 4 | I, S |
| Y187 | CW8 | M3 | 20.00 | R | 0, 4 | I, S |
| Y192 | CW8 | M3 | 100.00 | HS | 0 | I |
| Y198 | CW8 | M3 | 86.67 | S | 0, 1 | I, R |
| Y4-1 | Y4 | M4 | 100.00 | HS | 4 | S |
| Y4-2 | Y4 | M4 | 100.00 | HS | 4 | S |
| Y37-1 | Y37 | M4 | 20.00 | R | 4 | S |
| Y37-2 | Y37 | M4 | 20.00 | R | 4 | S |
| Y37-5 | Y37 | M4 | 20.00 | R | 4 | S |
| Y160-1 | Y160 | M4 | 100.00 | HS | 0 | I |
| Y186-1 | Y186 | M4 | 20.00 | R | 4 | S |
| Y186-2 | Y186 | M4 | 96.67 | HS | 0, 1 | I, R |
| Y186-5 | Y186 | M4 | 20.00 | R | 0 | I |
| Y186-8 | Y186 | M4 | 20.00 | R | 0 | I |
| Y187-5 | Y187 | M4 | 20.00 | R | 0 | I |
| Y187-9 | Y187 | M4 | 20.00 | R | 0 | I |
| Y187-10 | Y187 | M4 | 20.00 | R | 0 | I |
| Y187-11 | Y187 | M4 | 20.00 | R | 0 | I |
| Y187-12 | Y187 | M4 | 20.00 | R | 0, 4 | I, S |
| Y192-1 | Y192 | M4 | 100.00 | HS | 0 | I |
| Y198-1 | Y198 | M4 | 36.00 | M | 0 | I |
| Y198-2 | Y198 | M4 | 36.00 | M | 0 | I |
| Y198-3 | Y198 | M4 | 52.00 | M | 0 | I |
| Y198-4 | Y198 | M4 | 20.00 | R | 1 | R |
| Y198-5 | Y198 | M4 | 76.00 | S | 0 | I |
| Y186-1-1 | Y186-1 | M5 | 20.00 | R | 4 | S |
| Y186-1-2 | Y186-1 | M5 | 20.00 | R | 4 | S |
| Y186-1-4 | Y186-1 | M5 | 20.00 | R | 4 | S |
| Y186-1-6 | Y186-1 | M5 | 20.00 | R | 4 | S |
| Y186-1-8 | Y186-1 | M5 | 20.00 | R | 4 | S |
| Y186-2-1 | Y186-2 | M5 | 100.00 | HS | 1 | R |
| Y186-2-2 | Y186-2 | M5 | 88.00 | S | 1 | R |
| Y186-2-3 | Y186-2 | M5 | 100.00 | HS | 0 | I |
| Y186-2-4 | Y186-2 | M5 | 100.00 | HS | 0 | I |
| Y186-2-6 | Y186-2 | M5 | 100.00 | HS | 1 | R |
| Y186-5-2 | Y186-5 | M5 | 20.00 | R | 0 | I |
| Y186-5-3 | Y186-5 | M5 | 20.00 | R | 0 | I |
| Y186-5-4 | Y186-5 | M5 | 20.00 | R | 0 | I |
| Y186-5-5 | Y186-5 | M5 | 20.00 | R | 0 | I |
| Y186-5-6 | Y186-5 | M5 | 20.00 | R | 0 | I |
| Y186-8-2 | Y186-8 | M5 | 20.00 | R | 0 | I |
| Y186-8-4 | Y186-8 | M5 | 20.00 | R | 0 | I |
| Y186-8-5 | Y186-8 | M5 | 20.00 | R | 0 | I |
| Y186-8-6 | Y186-8 | M5 | 20.00 | R | 0 | I |
| Y186-8-7 | Y186-8 | M5 | 20.00 | R | 0 | I |
| Y187-5-1 | Y187-5 | M5 | 20.00 | R | 4 | S |
| Y187-5-4 | Y187-5 | M5 | 20.00 | R | 4 | S |
| Y187-5-5 | Y187-5 | M5 | 20.00 | R | 4 | S |
| Y187-5-6 | Y187-5 | M5 | 20.00 | R | 4 | S |
| Y187-5-8 | Y187-5 | M5 | 20.00 | R | 4 | S |
| Y187-9-1 | Y187-9 | M5 | 20.00 | R | 0 | I |
| Y187-9-2 | Y187-9 | M5 | 20.00 | R | 0 | I |
| Y187-9-3 | Y187-9 | M5 | 20.00 | R | 0 | I |
| Y187-9-4 | Y187-9 | M5 | 20.00 | R | 0 | I |
| Y187-9-5 | Y187-9 | M5 | 20.00 | R | 0 | I |
| Y187-10-2 | Y187-10 | M5 | 20.00 | R | 0 | I |
| Y187-10-3 | Y187-10 | M5 | 20.00 | R | 0 | I |
| Y187-10-4 | Y187-10 | M5 | 20.00 | R | 0 | I |
| Y187-10-5 | Y187-10 | M5 | 20.00 | R | 0 | I |
| Y187-10-6 | Y187-10 | M5 | 20.00 | R | 0 | I |
| Y187-11-2 | Y187-11 | M5 | 20.00 | R | 0 | I |
| Y187-11-3 | Y187-11 | M5 | 20.00 | R | 0 | I |
| Y187-11-4 | Y187-11 | M5 | 20.00 | R | 0 | I |
| Y187-11-7 | Y187-11 | M5 | 20.00 | R | 0 | I |
| Y187-11-1 | Y187-11 | M5 | 20.00 | R | 0 | I |
| Y187-12-1 | Y187-12 | M5 | 20.00 | R | 0, 4 | I, S |
| Y187-12-2 | Y187-12 | M5 | 20.00 | R | 0, 4 | I, S |
| Y187-12-3 | Y187-12 | M5 | 20.00 | R | 0, 4 | I, S |
| Y187-12-6 | Y187-12 | M5 | 20.00 | R | 0 | I |
| Y187-12-7 | Y187-12 | M5 | 20.00 | R | 0 | I |
| Lines 1 | Parent | Generation | Powdery Mildew | |
|---|---|---|---|---|
| Infection Type | Reaction 2 | |||
| SJ1 | - | - | 4 | S |
| CW8 | - | - | 4 | S |
| Y4 | SJ1 | M3 | 4 | S |
| Y37 | SJ1 | M3 | 4 | S |
| Y160 | CW8 | M3 | 0 | I |
| Y186 | CW8 | M3 | 0, 4 | I, S |
| Y187 | CW8 | M3 | 0, 4 | I, S |
| Y192 | CW8 | M3 | 0 | I |
| Y198 | CW8 | M3 | 0 | I |
| Y160-1 | Y160 | M4 | 0 | I |
| Y186-1 | Y186 | M4 | 4 | S |
| Y186-2 | Y186 | M4 | 0 | I |
| Y186-5 | Y186 | M4 | 0 | I |
| Y186-8 | Y186 | M4 | 0 | I |
| Y187-9 | Y187 | M4 | 0 | I |
| Y187-10 | Y187 | M4 | 0 | I |
| Y187-11 | Y187 | M4 | 0 | I |
| Y187-12 | Y187 | M4 | 0, 4 | I, S |
| Y192-1 | Y192 | M4 | 0 | I |
| Y198-1 | Y198 | M4 | 0 | I |
| Y198-2 | Y198 | M4 | 0 | I |
| Marker | Na 1 | Ne | MAF | H | PIC |
|---|---|---|---|---|---|
| 25986 | 6 | 3.03 | 0.42 | 0.67 | 0.61 |
| 26117 | 3 | 1.99 | 0.54 | 0.50 | 0.37 |
| 25433 | 5 | 1.88 | 0.63 | 0.47 | 0.36 |
| EST921 | 3 | 1.70 | 0.71 | 0.41 | 0.33 |
| EST709 | 7 | 3.56 | 0.33 | 0.72 | 0.67 |
| PSGAPA1 | 6 | 2.46 | 0.50 | 0.59 | 0.51 |
| 24407 | 4 | 1.54 | 0.79 | 0.35 | 0.32 |
| 24575 | 7 | 3.31 | 0.38 | 0.70 | 0.64 |
| AD147 | 4 | 1.99 | 0.54 | 0.50 | 0.37 |
| Mean | 5 | 2.38 | 0.54 | 0.55 | 0.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, D.; Sun, S.; Wu, W.; Xiang, C.; Duan, C.; Yu, D.; Wu, X.; Zhu, Z. Disease Resistance and Molecular Variations in Irradiation Induced Mutants of Two Pea Cultivars. Int. J. Mol. Sci. 2022, 23, 8793. https://doi.org/10.3390/ijms23158793
Deng D, Sun S, Wu W, Xiang C, Duan C, Yu D, Wu X, Zhu Z. Disease Resistance and Molecular Variations in Irradiation Induced Mutants of Two Pea Cultivars. International Journal of Molecular Sciences. 2022; 23(15):8793. https://doi.org/10.3390/ijms23158793
Chicago/Turabian StyleDeng, Dong, Suli Sun, Wenqi Wu, Chao Xiang, Canxing Duan, Dongmei Yu, Xuehong Wu, and Zhendong Zhu. 2022. "Disease Resistance and Molecular Variations in Irradiation Induced Mutants of Two Pea Cultivars" International Journal of Molecular Sciences 23, no. 15: 8793. https://doi.org/10.3390/ijms23158793
APA StyleDeng, D., Sun, S., Wu, W., Xiang, C., Duan, C., Yu, D., Wu, X., & Zhu, Z. (2022). Disease Resistance and Molecular Variations in Irradiation Induced Mutants of Two Pea Cultivars. International Journal of Molecular Sciences, 23(15), 8793. https://doi.org/10.3390/ijms23158793







