CDC42-IQGAP Interactions Scrutinized: New Insights into the Binding Properties of the GAP-Related Domain
Abstract
:1. Introduction
2. Results and Discussion
2.1. GRD Is Not the Prominent Binding Domain for High IQGAP-CDC42 Affinity
2.1.1. GRD Binds to CDC42 with Very Low Affinity in a Nucleotide-Independent Manner
2.1.2. Endogenous IQGAP1 also Binds CDC42 GDP
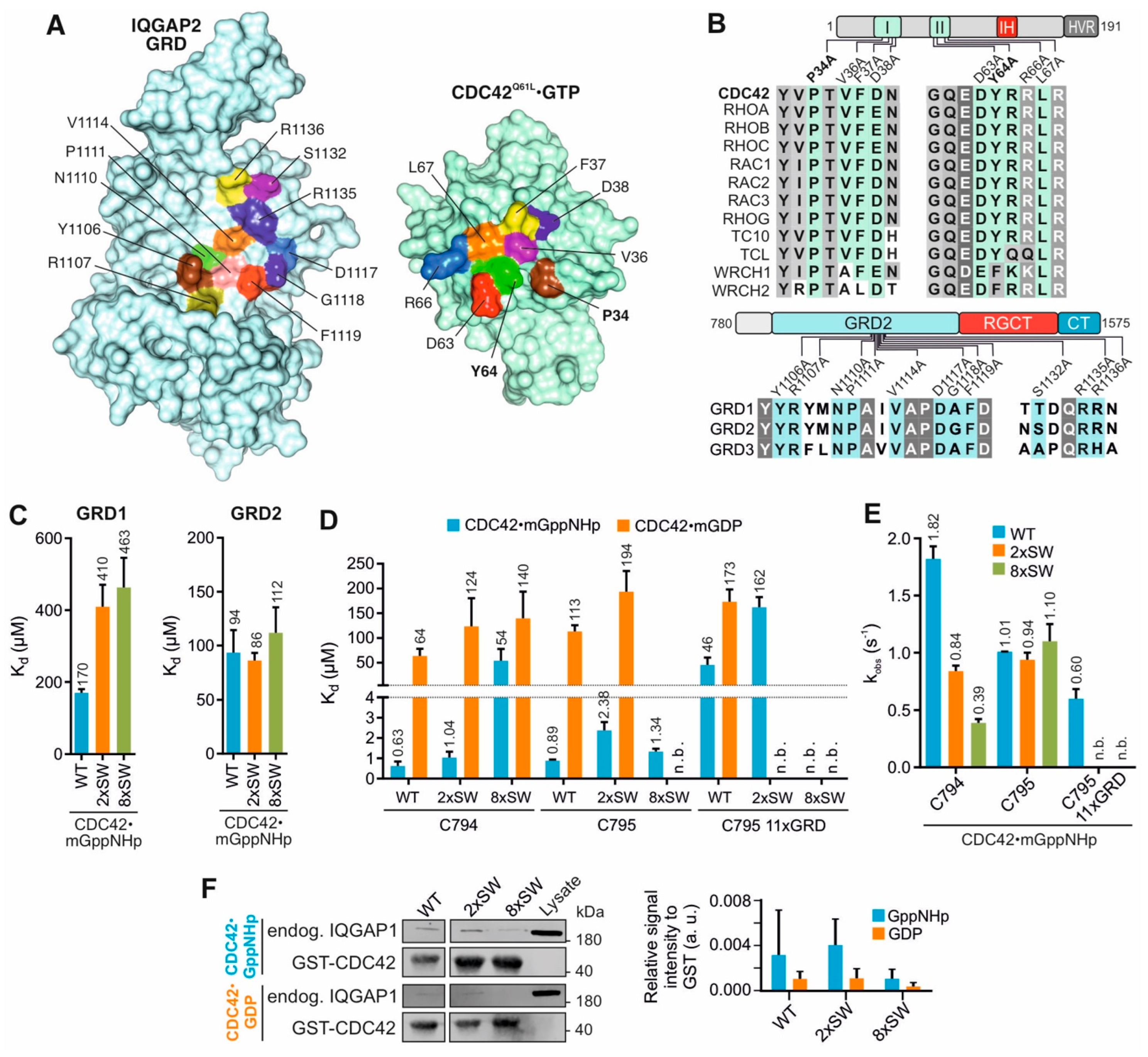
2.2. Switch Regions of CDC42 Are Not the Main Binding Sites for the GRDs
2.2.1. Mutations in CDC42 Switch Regions Only Mildly Affect GRD Binding
2.2.2. IQGAP C794/C795 Binding Is Impaired by Switch Region and GRD Mutations
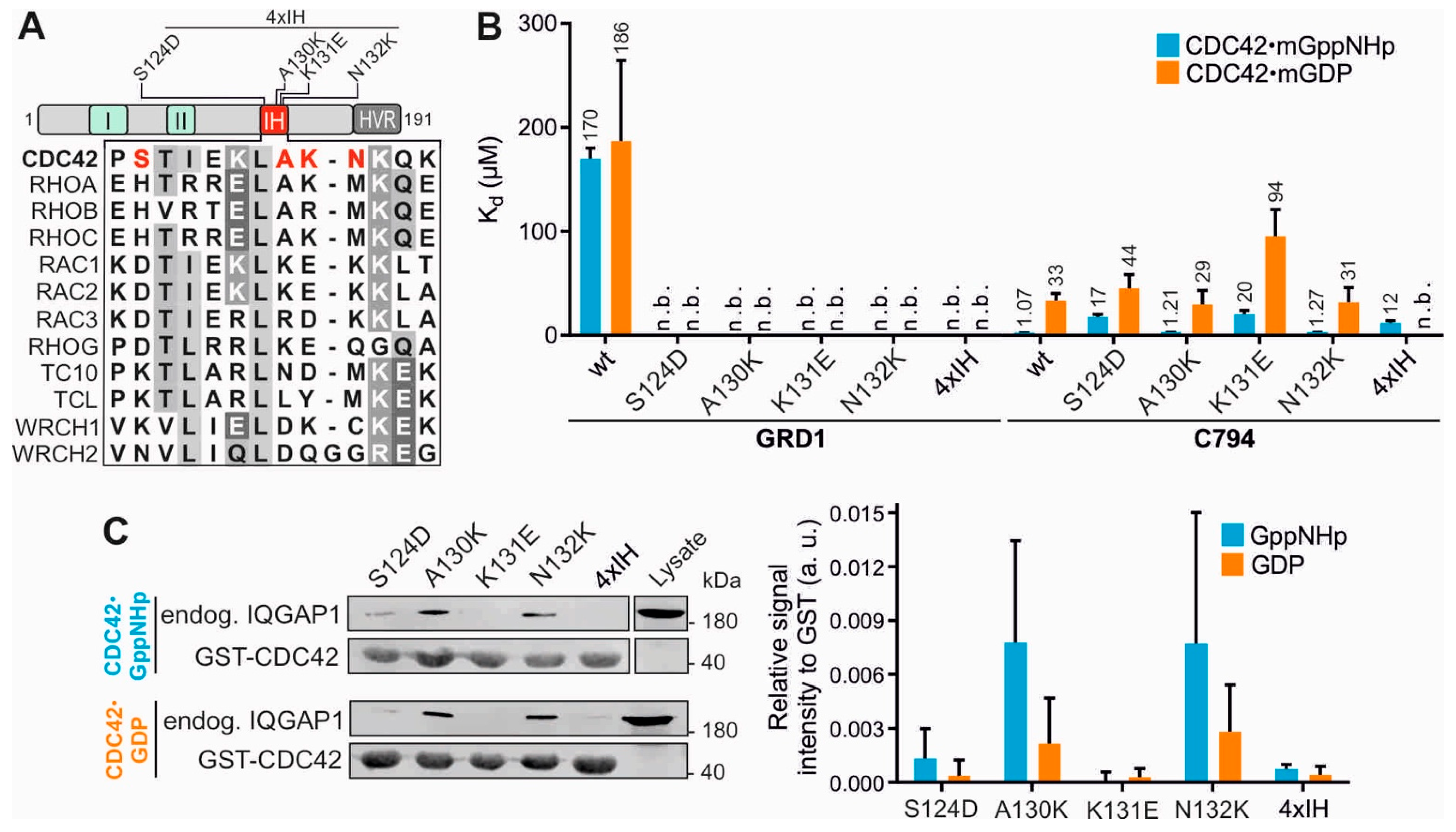
2.3. Insert Helix Contributes to the Binding Affinity of CDC42 for IQGAP1 GRD
2.4. Q61L Variant Is Not a Wildtype Equivalent for CDC42-IQGAP Interactions
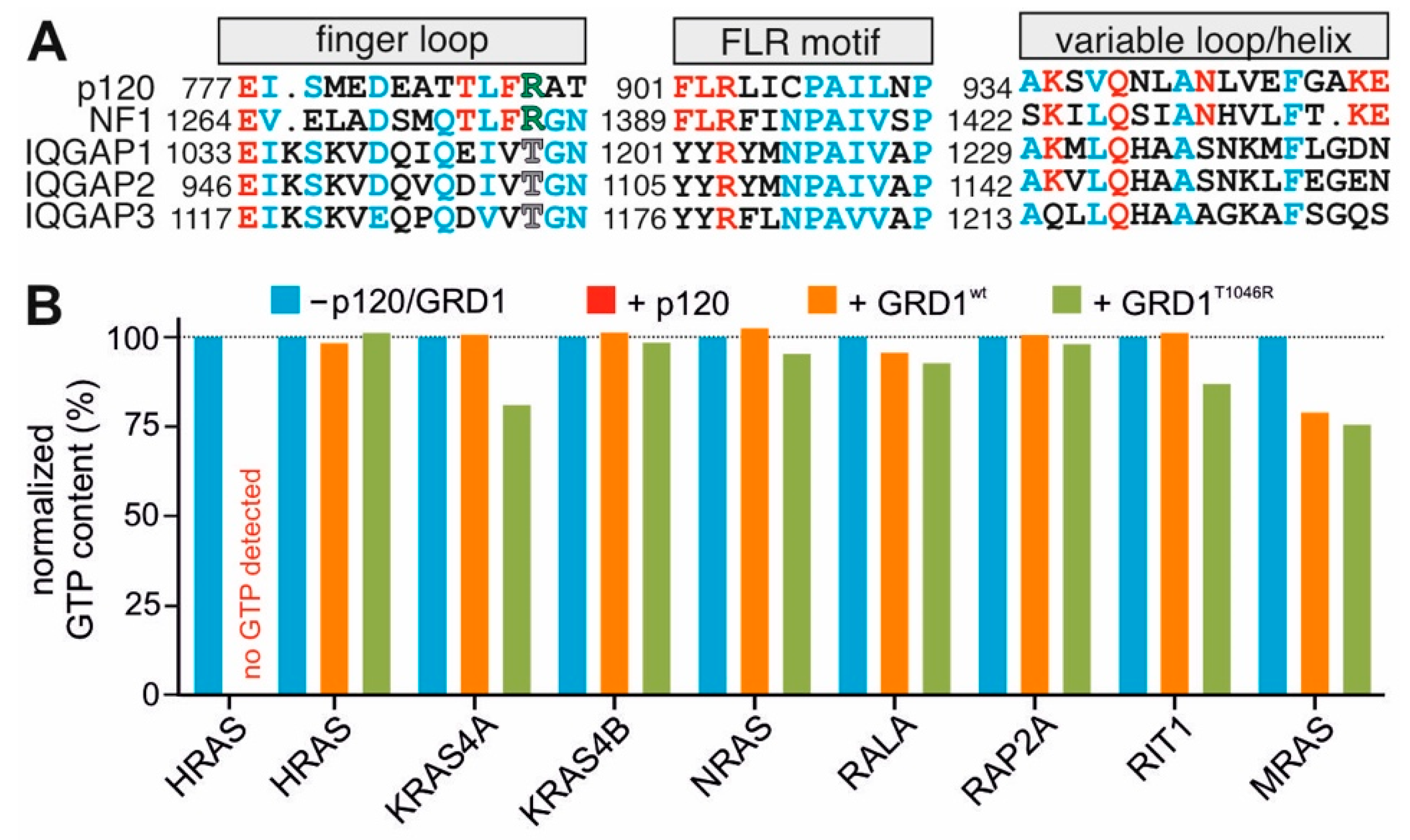
2.5. GRD Lacks the Structural Fingerprints to Induce the GAP Activity
3. Material and Methods
3.1. Constructs
3.2. Circular Dichroism (CD) Spectrometry
3.3. Cell Culture and Lysis
3.4. GST-Pull-Down
3.5. Fluorescence Stopped-Flow Spectrometry
3.6. Fluorescence Polarization
3.7. GTP Hydrolysis Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmadian, M.R.; Jaiswal, M.; Fansa, E.K.; Dvorsky, R. New insight into the molecular switch mechanism of human Rho family proteins: Shifting a paradigm. Biol. Chem. 2013, 394, 89–95. [Google Scholar] [CrossRef]
- Mosaddeghzadeh, N.; Ahmadian, M.R. The RHO Family GTPases: Mechanisms of Regulation and Signaling. Cells 2021, 10, 1831. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Manan, N.; Aghazadeh, B.; Liu, G.A.; Majumdar, A.; Ouerfelli, O.; Simlnovitch, K.A.; Rosen, M.K. Structure of Cdc42 in complex with the GTPase-binding domain of the “Wiskott-Aldrich syndrome” protein. Nature 1999, 399, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Mott, H.R.; Owen, D.; Nietlispach, D.; Lowe, P.N.; Manser, E.; Lim, L.; Laue, E.D. Structure of the small G protein Cdc42 bound to the GTpasebinding domain of ACK. Nature 1999, 399, 384–388. [Google Scholar] [CrossRef]
- Nouri, K.; Timson, D.J.; Ahmadian, M.R. New model for the interaction of IQGAP1 with CDC42 and RAC1. Small GTPases 2020, 11, 16–22. [Google Scholar] [CrossRef]
- Morreale, A.; Venkatesan, M.; Mott, H.R.; Owen, D.; Nietlispach, D.; Lowe, P.N.; Laue, E.D. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 2000, 7, 384–388. [Google Scholar] [CrossRef]
- Dvorsky, R.; Ahmadian, M.R. Always look on the bright site of Rho: Structural implications for a conserved intermolecular interface. EMBO Rep. 2004, 5, 1130–1136. [Google Scholar] [CrossRef] [Green Version]
- Dvorsky, R.; Blumenstein, L.; Vetter, I.R.; Ahmadian, M.R. Structural Insights into the Interaction of ROCKI with the Switch Regions of RhoA. J. Biol. Chem. 2004, 279, 7098–7104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemsath, L.; Dvorsky, R.; Fiegen, D.; Carlier, M.F.; Ahmadian, M.R. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell 2005, 20, 313–324. [Google Scholar] [CrossRef]
- Rose, R.; Weyand, M.; Lammers, M.; Ishizaki, T.; Ahmadian, M.R.; Wittinghofer, A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 2005, 435, 513–518. [Google Scholar] [CrossRef]
- Hall, A. Rho family GTPases. In Biochemical Society Transactions; Portland Press: London, UK, 2012; Volume 40, pp. 1378–1382. [Google Scholar]
- LeCour, L.; Boyapati, V.K.; Liu, J.; Li, Z.; Sacks, D.B.; Worthylake, D.K. The Structural Basis for Cdc42-Induced Dimerization of IQGAPs. Structure 2016, 24, 1499–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, D.; Mott, H.R. CRIB effector disorder: Exquisite function from chaos. Biochem. Soc. Trans. 2018, 46, 1289–1302. [Google Scholar] [CrossRef] [PubMed]
- Mott, H.R.; Owen, D. Structures of Ras superfamily effector complexes: What have we learnt in two decades? Crit. Rev. Biochem. Mol. Biol. 2015, 50, 85–133. [Google Scholar] [CrossRef] [PubMed]
- Thapar, R.; Karnoub, A.E.; Campbell, S.L. Structural and Biophysical Insights into the Role of the Insert Region in Rac1 Function. Biochemistry 2002, 41, 3875–3883. [Google Scholar] [CrossRef] [PubMed]
- Haspel, N.; Jang, H.; Nussinov, R. Active and Inactive Cdc42 Differ in Their Insert Region Conformational Dynamics. Biophys. J. 2021, 120, 306. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.J.; Leonard, D.A.; Cerione, R.A.; Manor, D. Interaction between Cdc42Hs and RhoGDI Is Mediated through the Rho Insert Region. J. Biol. Chem. 1997, 272, 26153–26158. [Google Scholar] [CrossRef] [Green Version]
- Nassar, N.; Hoffman, G.R.; Manor, D.; Clardy, J.C.; Cerione, R.A. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998, 5, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Lammers, M.; Meyer, S.; Kühlmann, D.; Wittinghofer, A. Specificity of Interactions between mDia Isoforms and Rho Proteins. J. Biol. Chem. 2008, 283, 35236–35246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kühn, S.; Erdmann, C.; Kage, F.; Block, J.; Schwenkmezger, L.; Steffen, A.; Rottner, K.; Geyer, M. The structure of FMNL2–Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat. Commun. 2015, 6, 7088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, S.J.; Brown, H.A. Specificity of Rho insert-mediated activation of phospholipase D1. J. Biol. Chem. 2002, 277, 26260–26267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnoub, A.E.; Der, C.J.; Campbell, S.L. The Insert Region of Rac1 Is Essential for Membrane Ruffling but Not Cellular Transformation. Mol. Cell. Biol. 2001, 21, 2847–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, H.; Kaibuchi, K.; Quilliam, L.A. The Insert Region of RhoA Is Essential for Rho Kinase Activation and Cellular Transformation. Mol. Cell. Biol. 2001, 21, 5287–5298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Wang, S.; Noritake, J.; Sato, K.; Fukata, M.; Takefuji, M.; Nakagawa, M.; Izumi, N.; Akiyama, T.; Kaibuchi, K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell 2004, 7, 871–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosaddeghzadeh, N.; Nouri, K.; Krumbach, O.H.F.; Amin, E.; Dvorsky, R.; Ahmadian, M.R. Selectivity determinants of rho gtpase binding to iqgaps. Int. J. Mol. Sci. 2021, 22, 12596. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 Binds ERK2 and Modulates Its Activity. J. Biol. Chem. 2004, 279, 17329–17337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 Is a Scaffold for Mitogen-Activated Protein Kinase Signaling. Mol. Cell. Biol. 2005, 25, 7940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.G.; Li, Z.; Sacks, D.B. IQGAP1 modulates activation of B-Raf. Proc. Natl. Acad. Sci. USA 2007, 104, 10465–10469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benseñor, L.B.; Kan, H.M.; Wang, N.; Wallrabe, H.; Davidson, L.A.; Cai, Y.; Schafer, D.A.; Bloom, G.S. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J. Cell Sci. 2007, 120, 658–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Clainche, C.; Schlaepfer, D.; Ferrari, A.; Klingauf, M.; Grohmanova, K.; Veligodskiy, A.; Didry, D.; Le, D.; Egile, C.; Carlier, M.F.; et al. IQGAP1 stimulates actin assembly through the N-wasp-Arp2/3 pathway. J. Biol. Chem. 2007, 282, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Kaur, R.; Yuan, X.; Lu, M.L.; Balk, S.P. Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate 2008, 68, 1510–1516. [Google Scholar] [CrossRef]
- Usatyuk, P.V.; Gorshkova, I.A.; He, D.; Zhao, Y.; Kalari, S.K.; Garcia, J.G.N.; Natarajan, V. Phospholipase D-mediated Activation of IQGAP1 through Rac1 regulates hyperoxia-induced p47phox translocation and reactive oxygen species generation in lung endothelial cells. J. Biol. Chem. 2009, 284, 15339–15352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelikan-Conchaudron, A.; Le Clainche, C.; Didry, D.; Carlier, M.F. The IQGAP1 protein is a calmodulin-regulated barbed end capper of actin filaments: Possible implications in its function in cell migration. J. Biol. Chem. 2011, 286, 35119–35128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pudewell, S.; Wittich, C.; Kazemein Jasemi, N.S.; Bazgir, F.; Ahmadian, M.R. Accessory proteins of the RAS-MAPK pathway: Moving from the side line to the front line. Commun. Biol. 2021, 4, 696. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Wang, S.; Kaibuchi, K. IQGAPs as Key Regulators of Actin-cytoskeleton Dynamics Mini-review and Review. Cell Struct. Funct. 2015, 40, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Abel, A.M.; Schuldt, K.M.; Rajasekaran, K.; Hwang, D.; Riese, M.J.; Rao, S.; Thakar, M.S.; Malarkannan, S. IQGAP1: Insights into the function of a molecular puppeteer. Mol. Immunol. 2015, 65, 336–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedman, A.C.; Smith, J.M.; Sacks, D.B. The biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Rep. 2015, 16, 427–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.M.; Hedman, A.C.; Sacks, D.B. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015, 25, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Anderson, R.A. IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv. Biol. Regul. 2016, 60, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanos, B.E.; Yeaman, C.; Rodriguez-Boulan, E. An emerging role for IQGAP1 in tight junction control. Small GTPases 2018, 9, 375–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussinov, R.; Zhang, M.; Tsai, C.J.; Jang, H. Calmodulin and IQGAP1 activation of PI3Kα and Akt in KRAS, HRAS and NRAS-driven cancers. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2304–2314. [Google Scholar] [CrossRef] [PubMed]
- White, C.D.; Li, Z.; Dillon, D.A.; Sacks, D.B. IQGAP1 Protein Binds Human Epidermal Growth Factor Receptor 2 (HER2) and Modulates Trastuzumab Resistance. J. Biol. Chem. 2011, 286, 29734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Billadeau, D.D.; Abdelhakim, H.; Leof, E.; Kaibuchi, K.; Bernabeu, C.; Bloom, G.S.; Yang, L.; Boardman, L.; Shah, V.H.; et al. IQGAP1 suppresses TβRII-mediated myofibroblastic activation and metastatic growth in liver. J. Clin. Investig. 2013, 123, 1138–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jameson, K.L.; Mazur, P.K.; Zehnder, A.M.; Zhang, J.; Zarnegar, B.; Sage, J.; Khavari, P.A. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase–driven tumors. Nat. Med. 2013, 19, 626–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.; Hedman, A.C.; Sayedyahossein, S.; Thapa, N.; Sacks, D.B.; Anderson, R.A. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat. Cell Biol. 2016, 18, 1324–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Wang, T.; Gao, H.; Yue, X.; Bian, W.; Mei, J.; Zhang, Y. The interplay between IQGAP1 and small GTPases in cancer metastasis. Biomed. Pharmacother. 2021, 135, 111243. [Google Scholar] [CrossRef]
- Wei, T.; Lambert, P.F. Role of IQGAP1 in Carcinogenesis. Cancers 2021, 13, 3940. [Google Scholar] [CrossRef]
- Rotoli, D.; Díaz-Flores, L.; Gutiérrez, R.; Morales, M.; Ávila, J.; Martín-Vasallo, P. AmotL2, IQGAP1, and FKBP51 Scaffold Proteins in Glioblastoma Stem Cell Niches. J. Histochem. Cytochem. 2022, 70, 9–16. [Google Scholar] [CrossRef]
- Elliott, S.F. Biochemical analysis of the interactions of IQGAP1 C-terminal domain with CDC42. World J. Biol. Chem. 2012, 3, 53. [Google Scholar] [CrossRef] [Green Version]
- Nouri, K.; Fansa, E.K.; Amin, E.; Dvorsky, R.; Gremer, L.; Willbold, D.; Schmitt, L.; Timson, D.J.; Ahmadian, M.R. IQGAP1 interaction with RHO family proteins revisited kinetic and equilibrium evidence for multiple distinct binding sites. J. Biol. Chem. 2016, 291, 26364–26376. [Google Scholar] [CrossRef] [Green Version]
- Swart-Mataraza, J.M.; Li, Z.; Sacks, D.B. IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J. Biol. Chem. 2002, 277, 24753–24763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Debreceni, B.; Jia, B.; Gao, Y.; Tigyi, G.; Zheng, Y. Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of Cdc42. J. Biol. Chem. 1999, 274, 29648–29654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sila Ozdemir, E.; Jang, H.; Gursoy, A.; Keskin, O.; Li, Z.; Sacks, D.B.; Nussinov, R. Unraveling the molecular mechanism of interactions of the Rho GTPases Cdc42 and Rac1 with the scaffolding protein IQGAP2. J. Biol. Chem. 2018, 293, 3685–3699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheffzek, K.; Ahmadian, M.R.; Kabsch, W.; Wiesmüller, L.; Lautwein, A.; Schmitz, F.; Wittinghofer, A. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic ras mutants. Science 1997, 277, 333–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, D.; Campbell, L.J.; Littlefield, K.; Evetts, K.A.; Li, Z.; Sacks, D.B.; Lowe, P.N.; Mott, H.R. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: Interfaces differ between the complexes. J. Biol. Chem. 2008, 283, 1692–1704. [Google Scholar] [CrossRef] [Green Version]
- Kurella, V.B.; Richard, J.M.; Parke, C.L.; LeCour, L.F.; Bellamy, H.D.; Worthylake, D.K. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J. Biol. Chem. 2009, 284, 14857–14865. [Google Scholar] [CrossRef] [Green Version]
- Gorisse, L.; Li, Z.; Wagner, C.D.; Worthylake, D.K.; Zappacosta, F.; Hedman, A.C.; Annan, R.S.; Sacks, D.B. Ubiquitination of the scaffold protein IQGAP1 diminishes its interaction with and activation of the Rho GTPase CDC42. J. Biol. Chem. 2020, 295, 4822–4835. [Google Scholar] [CrossRef]
- McCallum, S.J.; Wu, W.J.; Cerione, R.A. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J. Biol. Chem. 1996, 271, 21732–21737. [Google Scholar] [CrossRef] [Green Version]
- Hart, M.J.; Callow, M.G.; Souza, B.; Polakis, P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996, 15, 2997–3005. [Google Scholar] [CrossRef]
- Zhang, B.; Chernoff, J.; Zheng, Y. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J. Biol. Chem. 1998, 273, 8776–8782. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, S.; Fukata, M.; Nakagawa, M.; Fujii, K.; Nakamura, T.; Ookubo, T.; Izawa, I.; Nagase, T.; Nomura, N.; Tani, H.; et al. Cdc42 and Rac1 Regulate the Interaction of IQGAP1 with Î2-Catenin. J. Biol. Chem. 1999, 274, 26044–26050. [Google Scholar] [CrossRef] [Green Version]
- Grohmanova, K.; Schlaepfer, D.; Hess, D.; Gutierrez, P.; Beck, M.; Kroschewski, R. Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of Rho-GTPase regulator. J. Biol. Chem. 2004, 279, 48495–48504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mataraza, J.M.; Briggs, M.W.; Li, Z.; Frank, R.; Sacks, D.B. Identification and characterization of the Cdc42-binding site of IQGAP1. Biochem. Biophys. Res. Commun. 2003, 305, 315–321. [Google Scholar] [CrossRef]
- Scheffzek, K.; Ahmadian, M.R.; Wittinghofer, A. GTPase-activating proteins: Helping hands to complement an active site. Trends Biochem. Sci. 1998, 23, 257–262. [Google Scholar] [CrossRef]
- Chen, S.; Shu, L.; Zhao, R.; Zhao, Y. Molecular dynamics simulations reveal the activation mechanism of mutations G12V and Q61L of Cdc42. Proteins Struct. Funct. Bioinform. 2022, 90, 1376–1389. [Google Scholar] [CrossRef]
- Ahmadian, M.R.; Stege, P.; Scheffzek, K.; Wittinghofer, A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997, 4, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.R.; Kiel, C.; Stege, P.; Scheffzek, K. Structural fingerprints of the Ras-GTPase activating proteins neurofibromin and p120GAP. J. Mol. Biol. 2003, 329, 699–710. [Google Scholar] [CrossRef]
- Scheffzek, K.; Shivalingaiah, G. Ras-specific gtpase-activating proteins— structures, mechanisms, and interactions. Cold Spring Harb. Perspect. Med. 2019, 9, a031500. [Google Scholar] [CrossRef]
- Scheffzek, K.; Ahmadian, M.R. GTPase activating proteins: Structural and functional insights 18 years after discovery. Cell. Mol. Life Sci. 2005, 62, 3014–3038. [Google Scholar] [CrossRef]
- Jaiswal, M.; Dubey, B.N.; Koessmeier, K.T.; Gremer, L.; Ahmadian, M.R. Biochemical assays to characterize Rho GTPases. Methods Mol. Biol. 2012, 827, 37–58. [Google Scholar] [CrossRef]
- Hemsath, L.; Ahmadian, M.R. Fluorescence approaches for monitoring interactions of Rho GTPases with nucleotides, regulators, and effectors. Methods 2005, 37, 173–182. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosaddeghzadeh, N.; Pudewell, S.; Bazgir, F.; Kazemein Jasemi, N.S.; Krumbach, O.H.F.; Gremer, L.; Willbold, D.; Dvorsky, R.; Ahmadian, M.R. CDC42-IQGAP Interactions Scrutinized: New Insights into the Binding Properties of the GAP-Related Domain. Int. J. Mol. Sci. 2022, 23, 8842. https://doi.org/10.3390/ijms23168842
Mosaddeghzadeh N, Pudewell S, Bazgir F, Kazemein Jasemi NS, Krumbach OHF, Gremer L, Willbold D, Dvorsky R, Ahmadian MR. CDC42-IQGAP Interactions Scrutinized: New Insights into the Binding Properties of the GAP-Related Domain. International Journal of Molecular Sciences. 2022; 23(16):8842. https://doi.org/10.3390/ijms23168842
Chicago/Turabian StyleMosaddeghzadeh, Niloufar, Silke Pudewell, Farhad Bazgir, Neda S. Kazemein Jasemi, Oliver H. F. Krumbach, Lothar Gremer, Dieter Willbold, Radovan Dvorsky, and Mohammad R. Ahmadian. 2022. "CDC42-IQGAP Interactions Scrutinized: New Insights into the Binding Properties of the GAP-Related Domain" International Journal of Molecular Sciences 23, no. 16: 8842. https://doi.org/10.3390/ijms23168842





