A Single Amino Acid Substitution in MIL1 Leads to Activation of Programmed Cell Death and Defense Responses in Rice
Abstract
:1. Introduction
2. Results
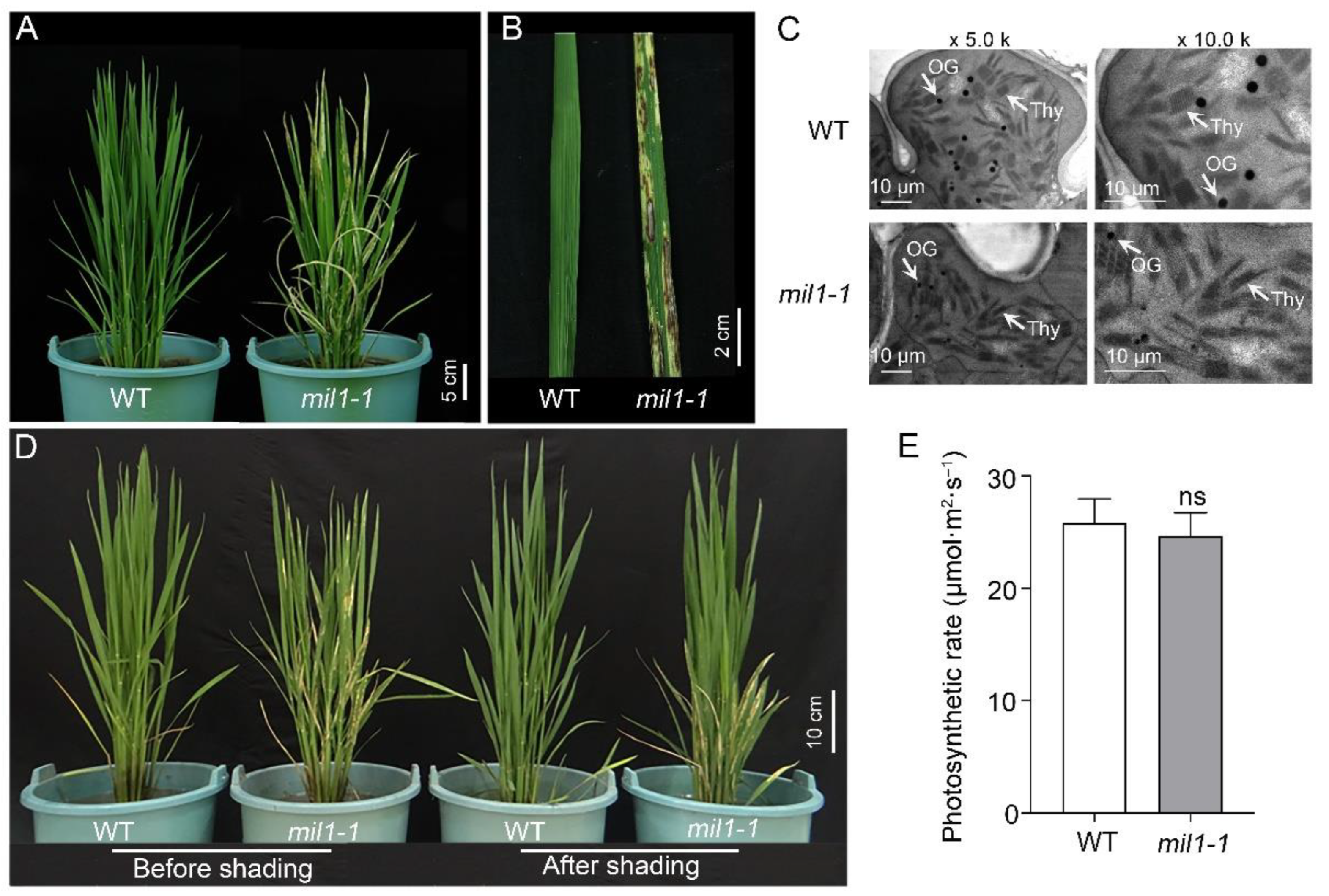
2.1. The Mil1-1 Mutation Confers Spotted Leaf Phenotype in Rice
2.2. Abnormal ROS Accumulation and PCD Occur in Mil1-1
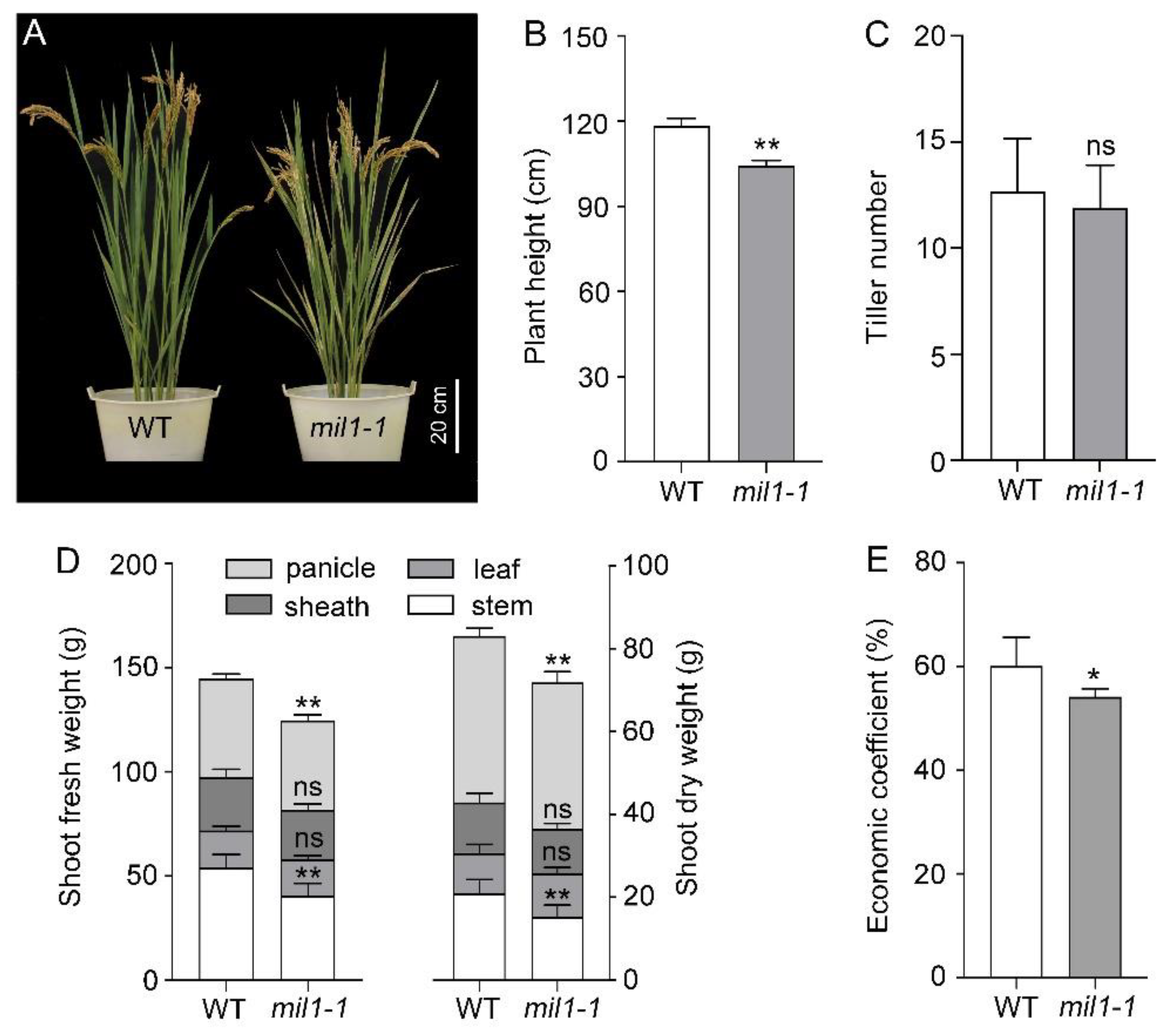
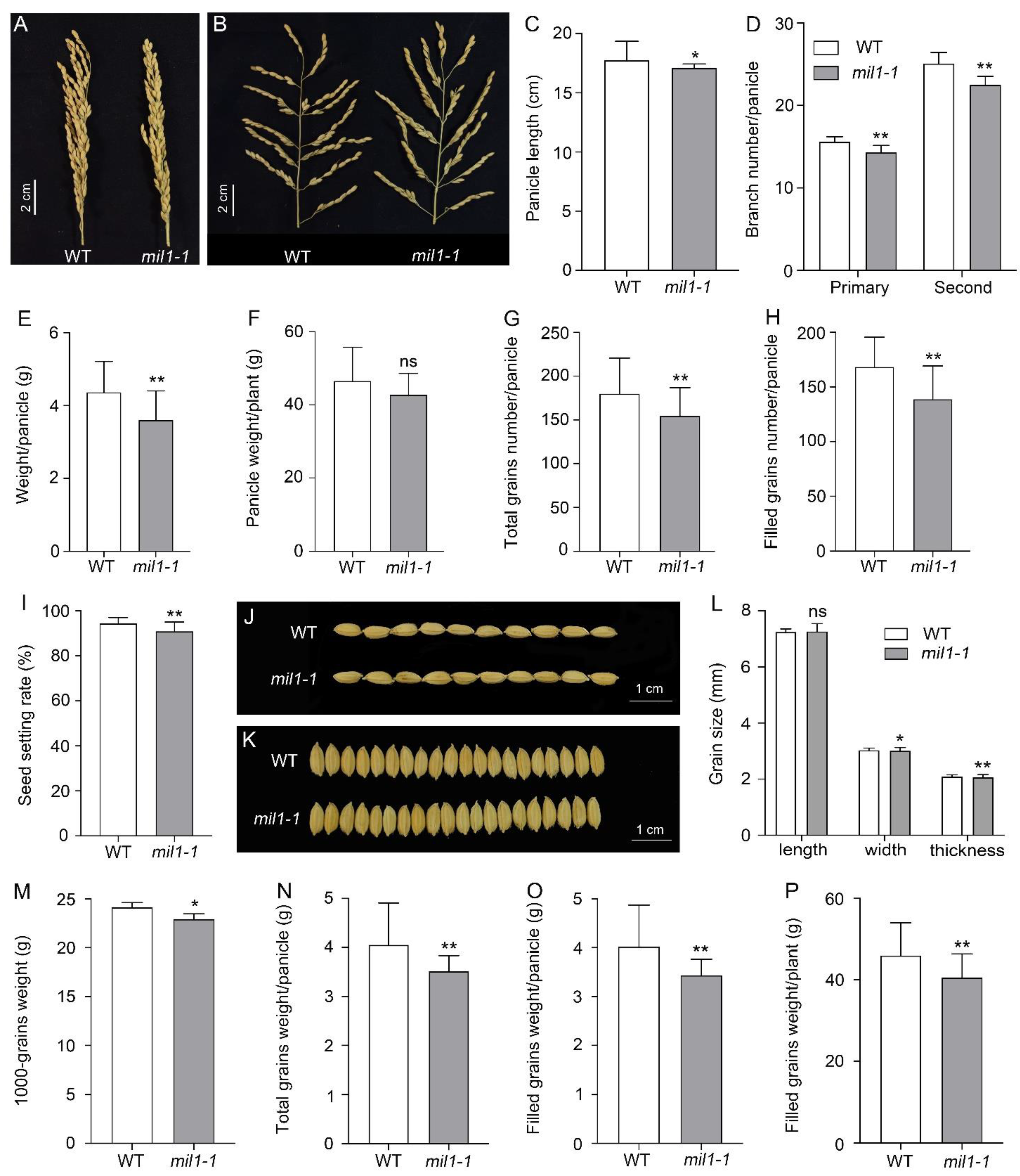
2.3. Mil1-1 Mutation Impacts Pleiotropic Agronomic Traits
2.4. Mutation of MIL1 Enhances Bacterial Blight Resistance
2.5. Mil1-1 Is a Novel Allelic Mutation at OsHPL3 Locus
3. Discussion
3.1. Mutation of MIL1 Increases the Accumulation of ROS Causing Spontaneous Lesions in Leaves
3.2. Mutation of MIL1 Leads to Auto-Activation of Defense Response in Rice
3.3. Mil1-1 Retards Development and Reduces Grain Yield of Rice
3.4. Mutation of MIL1 Is a Novel Missense Point Mutation of OsHPL3
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Agronomic Trait Evaluation
4.2. Histochemical Assay
4.3. Apoptosis Assay
4.4. POD, CAT Activity Detection, and H2O2 Determination
- MAC represents the milli-molar attenuation coefficient and is 12.0 mM−1·cm−1.
- One enzyme activity unit is defined as 1 μg substrate catalyzed per minute by enzymes within 1 mg tissue samples at 37 °C.
- 271 is reciprocal value of slope.
- 1 µmol H2O2 decomposing per mg tissue protein per second is considered as 1 activity unit (U).
4.5. Bacterial Inoculation
4.6. Gene Expression Analysis
4.7. Genetic Analysis of mil1-1 Mutation and Map-Based Positional Cloning of MIL1
4.8. Complementary Test of MIL1
4.9. Hormone and Stress Treatments
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiao, Y.; Jiang, W.; Lee, J.; Park, B.; Choi, M.S.; Piao, R.; Woo, M.O.; Roh, J.H.; Han, L.; Paek, N.C.; et al. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 2010, 185, 258–274. [Google Scholar] [CrossRef] [PubMed]
- Shirsekar, G.S.; Vega-Sanchez, M.E.; Bordeos, A.; Baraoidan, M.; Swisshelm, A.; Fan, J.; Park, C.H.; Leung, H.; Wang, G.L. Identification and characterization of suppressor mutants of spl11- mediated cell death in rice. Mol. Plant-Microbe Interact. 2014, 27, 528–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.V.; Vo, K.T.X.; Rahman, M.M.; Choi, S.H.; Jeon, J.S. Heat stress transcription factor OsSPL7 plays a critical role in reactive oxygen species balance and stress responses in rice. Plant Sci. 2019, 289, 110273. [Google Scholar] [CrossRef] [PubMed]
- Danon, A.; Miersch, O.; Felix, G.; Camp, R.G.; Apel, K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005, 41, 68–80. [Google Scholar] [CrossRef]
- Kang, S.G.; Lee, K.E.; Singh, M.; Kumar, P.; Matin, M.N. Rice Lesion Mimic Mutants (LMM): The Current Understanding of Genetic Mutations in the Failure of ROS Scavenging during Lesion Formation. Plants 2021, 10, 1598. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef]
- Ma, H.; Li, J.; Ma, L.; Wang, P.; Xue, Y.; Yin, P.; Xiao, J.; Wang, S. Pathogen-inducible OsMPKK10.2-OsMPK6 cascade phosphorylates the Raf-like kinase OsEDR1 and inhibits its scaffold function to promote rice disease resistance. Mol. Plant 2021, 14, 620–632. [Google Scholar] [CrossRef]
- Kim, J.A.; Cho, K.; Singh, R.; Jung, Y.H.; Jeong, S.H.; Kim, S.H.; Lee, J.E.; Cho, Y.S.; Agrawal, G.K.; Rakwal, R.; et al. Rice OsACDR1 (Oryza sativa accelerated cell death and resistance 1) is a potential positive regulator of fungal disease resistance. Mol. Cells 2009, 28, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Lim, J.H.; Kim, S.S.; Cho, S.H.; Yoo, S.C.; Koh, H.J.; Sakuraba, Y.; Paek, N.C. Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J. Exp. Bot. 2015, 66, 7045–7059. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Q.; Chen, Y.; Yu, N.; Cao, Y.; Zhan, X.; Cheng, S.; Cao, L. LMM24 Encodes Receptor-Like Cytoplasmic Kinase 109, Which Regulates Cell Death and Defense Responses in Rice. Int. J. Mol. Sci. 2019, 20, 3243. [Google Scholar] [CrossRef] [Green Version]
- Song, G.; Kwon, C.T.; Kim, S.H.; Shim, Y.; Lim, C.; Koh, H.J.; An, G.; Kang, K.; Paek, N.C. The Rice SPOTTED LEAF4 (SPL4) Encodes a Plant Spastin That Inhibits ROS Accumulation in Leaf Development and Functions in Leaf Senescence. Front. Plant Sci. 2018, 9, 1925. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yin, J.; Liang, S.; Liang, R.; Zhou, X.; Chen, Z.; Zhao, W.; Wang, J.; Li, W.; He, M.; et al. The Multivesicular Bodies (MVBs)-Localized AAA ATPase LRD6-6 Inhibits Immunity and Cell Death Likely through Regulating MVBs-Mediated Vesicular Trafficking in Rice. PLoS Genet. 2016, 12, e1006311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fekih, R.; Tamiru, M.; Kanzaki, H.; Abe, A.; Yoshida, K.; Kanzaki, E.; Saitoh, H.; Takagi, H.; Natsume, S.; Undan, J.R.; et al. The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol. Genet. Genom. 2015, 290, 611–622. [Google Scholar] [CrossRef]
- Wang, S.; Lei, C.; Wang, J.; Ma, J.; Tang, S.; Wang, C.; Zhao, K.; Tian, P.; Zhang, H.; Qi, C.; et al. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 2017, 68, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Fan, S.; Deng, L.; Zhong, G.; Zhang, S.; Li, M.; Chen, W.; Wang, G.; Tu, B.; Wang, Y.; et al. LML1, Encoding a Conserved Eukaryotic Release Factor 1 Protein, Regulates Cell Death and Pathogen Resistance by Forming a Conserved Complex with SPL33 in Rice. Plant Cell Physiol. 2018, 59, 887–902. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.L.; et al. OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, L.R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.L. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Wang, Y.; Ma, X.; Meng, L.; Jing, R.; Wang, F.; Wang, S.; Cheng, Z.; Zhang, X.; Jiang, L.; et al. Disruption of gene SPL35, encoding a novel CUE domain-containing protein, leads to cell death and enhanced disease response in rice. Plant Biotechnol. J. 2019, 17, 1679–1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Ye, B.; Yin, J.; Yuan, C.; Zhou, X.; Li, W.; He, M.; Wang, J.; Chen, W.; Qin, P.; et al. Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol. Biochem. 2015, 97, 44–51. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Tang, J.; Lin, A.; Zhang, F.; Fang, J.; Zhang, G.; Chu, C. RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J. Genet. Genom. 2011, 38, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.; Hua, Z.; Zhao, J.; Zhang, B.; Ren, D.; Liu, C.; Yang, S.; Zhang, A.; Jiang, H.; Yu, H.; et al. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnol. J. 2019, 17, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Hong, X.; Hu, D.; Liu, C.; Yang, J.; Li, Y.; Huang, Y.; Feng, Y.; Gong, H.; et al. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 2015, 66, 973–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Chen, T.; Sathe, A.P.; He, Y.; Zhang, X.B.; Wu, J.L. Identification of a Novel Semi-Dominant Spotted-Leaf Mutant with Enhanced Resistance to Xanthomonas oryzae pv. oryzae in Rice. Int. J. Mol. Sci. 2018, 19, 3766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manosalva, P.M.; Bruce, M.; Leach, J.E. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 2011, 68, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, P.; Li, C.; Wang, Y.; Guo, L.; Jiang, G.; Zhai, W. LMM5.1 and LMM5.4, two eukaryotic translation elongation factor 1A-like gene family members, negatively affect cell death and disease resistance in rice. J. Genet. Genom. 2017, 44, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Van Breusegem, F.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabbage, M.; Kessens, R.; Bartholomay, L.C.; Williams, B. The Life and Death of a Plant Cell. Annu. Rev. Plant Biol. 2017, 68, 375–404. [Google Scholar] [CrossRef] [Green Version]
- Mignolet-Spruyt, L.; Xu, E.; Idanheimo, N.; Hoeberichts, F.A.; Muhlenbock, P.; Brosche, M.; Van Breusegem, F.; Kangasjarvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [Green Version]
- Lolle, S.; Stevens, D.; Coaker, G. Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 2020, 62, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, 6395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Rodriguez, E.; El Ghoul, H.; Mundy, J.; Petersen, M. Making sense of plant autoimmunity and ‘negative regulators’. FEBS J. 2016, 283, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, Q.; Raynaud, C.; Benhamed, M.; Delarue, M. To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 2015, 6, 24. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, J.; Ghosh, P.; Das, S. Autoimmunity in plants. Planta 2018, 248, 751–767. [Google Scholar] [CrossRef]
- Liu, X.; Li, F.; Tang, J.; Wang, W.; Zhang, F.; Wang, G.; Chu, J.; Yan, C.; Wang, T.; Chu, C.; et al. Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS ONE 2012, 7, e50089. [Google Scholar] [CrossRef]
- Tong, X.; Qi, J.; Zhu, X.; Mao, B.; Zeng, L.; Wang, B.; Li, Q.; Zhou, G.; Xu, X.; Lou, Y.; et al. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 2012, 71, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Hu, Y.; Tong, A.; Yan, B.; Lv, Y.; Wang, S.; Ma, W.; Cui, Z.; Wang, X. LAZY1 Controls Tiller Angle and Shoot Gravitropism by Regulating the Expression of Auxin Transporters and Signaling Factors in Rice. Plant Cell Physiol. 2021, 61, 2111–2125. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H.; et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wu, F.; Xie, Q.; Wang, H.; Wang, Y.; Yue, Y.; Gahura, O.; Ma, S.; Liu, L.; Cao, Y.; et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 2012, 24, 3278–3295. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Tong, A.; Huo, Y.; Yan, Z.; Yang, W.; Yang, X.; Wang, X.X. SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 2017, 15, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Xu, H.; Sun, X.X.; Cui, Z.B.; Zhang, Y.; Bai, Y.G.; Wang, X.X.; Chen, W.F. Differential transformation efficiency of Japonica rice varieties developed in northern China. Crop Breed. Appl. Biotechnol. 2015, 15, 162–168. [Google Scholar] [CrossRef] [Green Version]



| Cross Combination | Plants with WT Phenotype | Plants with mil1-1 Phenotype | Total Plants | Plants with WT Phenotype: Plants with mil1-1 Phenotype | χ2 |
|---|---|---|---|---|---|
| mil1-1 × WT | 593 | 210 | 803 | 2.82:1 ≈ 3:1 | 0.5683 |
| χ2 = 3.84, p = 0.05, df = 1 | χ2 = 6.64, p = 0.01, df = 1 | χ2 < χ2 (p = 0.05, df = 1) | |||
| Name | Sequences | Note |
|---|---|---|
| OsACT1ReF | CTATGTTCCCTGGCATTGCT | OsACT1, RT-qPCR |
| OsACT1ReR | GGCGATAACAGCTCCTCTTG | |
| PR1AReF | AGGTTATCCTGCTGCTTGCT | PR1A, RT-qPCR |
| PR1AReR | AGTCCGAGTGCTCCAGCTT | |
| PR1BReF | AGAACTACGCCAGCCAGAGA | PR1B, RT-qPCR |
| PR1BReR | GTAGTGCCCGCACACCTT | |
| PBZ1ReF | ATGAAGCTTAACCCTGCCGC | PBZ1, RT-qPCR |
| PBZ1ReR | TCGAGCTCGTACTCCACCTT | |
| POC1ReF | GCTCTCTCAGGCGCGCACA | POC1, RT-qPCR |
| POC1ReR | TTCGACAGCAGGTTGGTGTA | |
| POX1ReF | GCTCTCTCAGGAGCACACAC | POX1, RT-qPCR |
| POX1ReR | CAGCAGGTTGCTGTAGTAGGC | |
| MIL1ReF | AAGGAGGAGGCCATCAACAA | MIL1, RT-qPCR |
| MIL1ReR | CATCCGGAGCACCTCGTAC | |
| MIL1cF | tctagaATGGTGCCGTCGTTCCCG * | 35S:MIL1-GFP vector |
| MIL1cR | ggatccGCTGGGAGTGAGCTCCCGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, B.; Zheng, H.; Sang, Y.; Wang, Y.; Sun, J.; Li, F.; Wang, J.; Wang, X. A Single Amino Acid Substitution in MIL1 Leads to Activation of Programmed Cell Death and Defense Responses in Rice. Int. J. Mol. Sci. 2022, 23, 8853. https://doi.org/10.3390/ijms23168853
Yan B, Zheng H, Sang Y, Wang Y, Sun J, Li F, Wang J, Wang X. A Single Amino Acid Substitution in MIL1 Leads to Activation of Programmed Cell Death and Defense Responses in Rice. International Journal of Molecular Sciences. 2022; 23(16):8853. https://doi.org/10.3390/ijms23168853
Chicago/Turabian StyleYan, Bowen, Haoyu Zheng, Yuwei Sang, Yan Wang, Jian Sun, Fengcheng Li, Jiayu Wang, and Xiaoxue Wang. 2022. "A Single Amino Acid Substitution in MIL1 Leads to Activation of Programmed Cell Death and Defense Responses in Rice" International Journal of Molecular Sciences 23, no. 16: 8853. https://doi.org/10.3390/ijms23168853
APA StyleYan, B., Zheng, H., Sang, Y., Wang, Y., Sun, J., Li, F., Wang, J., & Wang, X. (2022). A Single Amino Acid Substitution in MIL1 Leads to Activation of Programmed Cell Death and Defense Responses in Rice. International Journal of Molecular Sciences, 23(16), 8853. https://doi.org/10.3390/ijms23168853






