High-Depth Transcriptome Reveals Differences in Natural Haploid Ginkgo biloba L. Due to the Effect of Reduced Gene Dosage
Abstract
:1. Introduction
2. Results
2.1. Haploid Ginkgo Exhibits Weak Growth, Biomass Accumulation and Secondary Metabolic Synthesis
2.2. Transcriptomic Analysis Overview
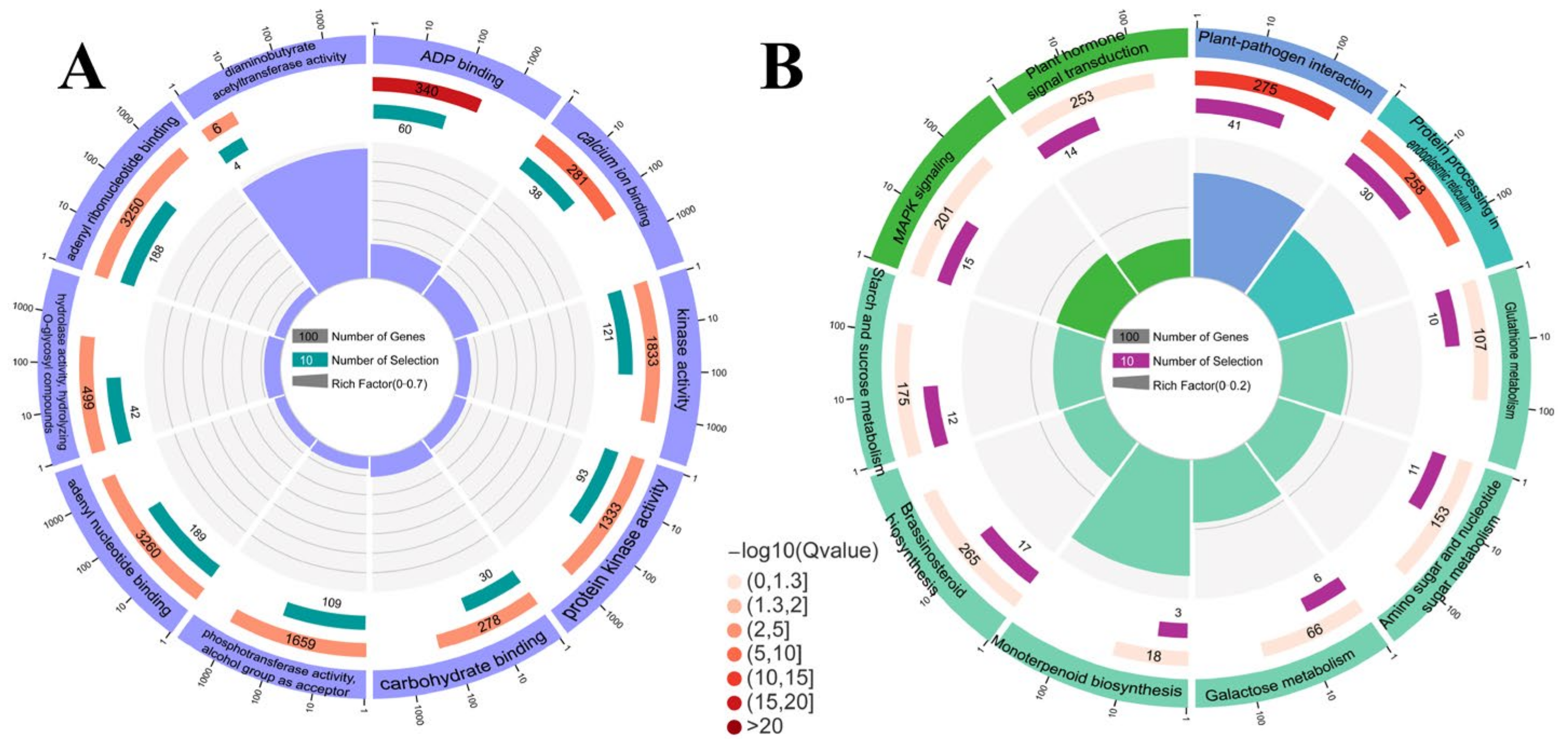
2.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis of DEGs
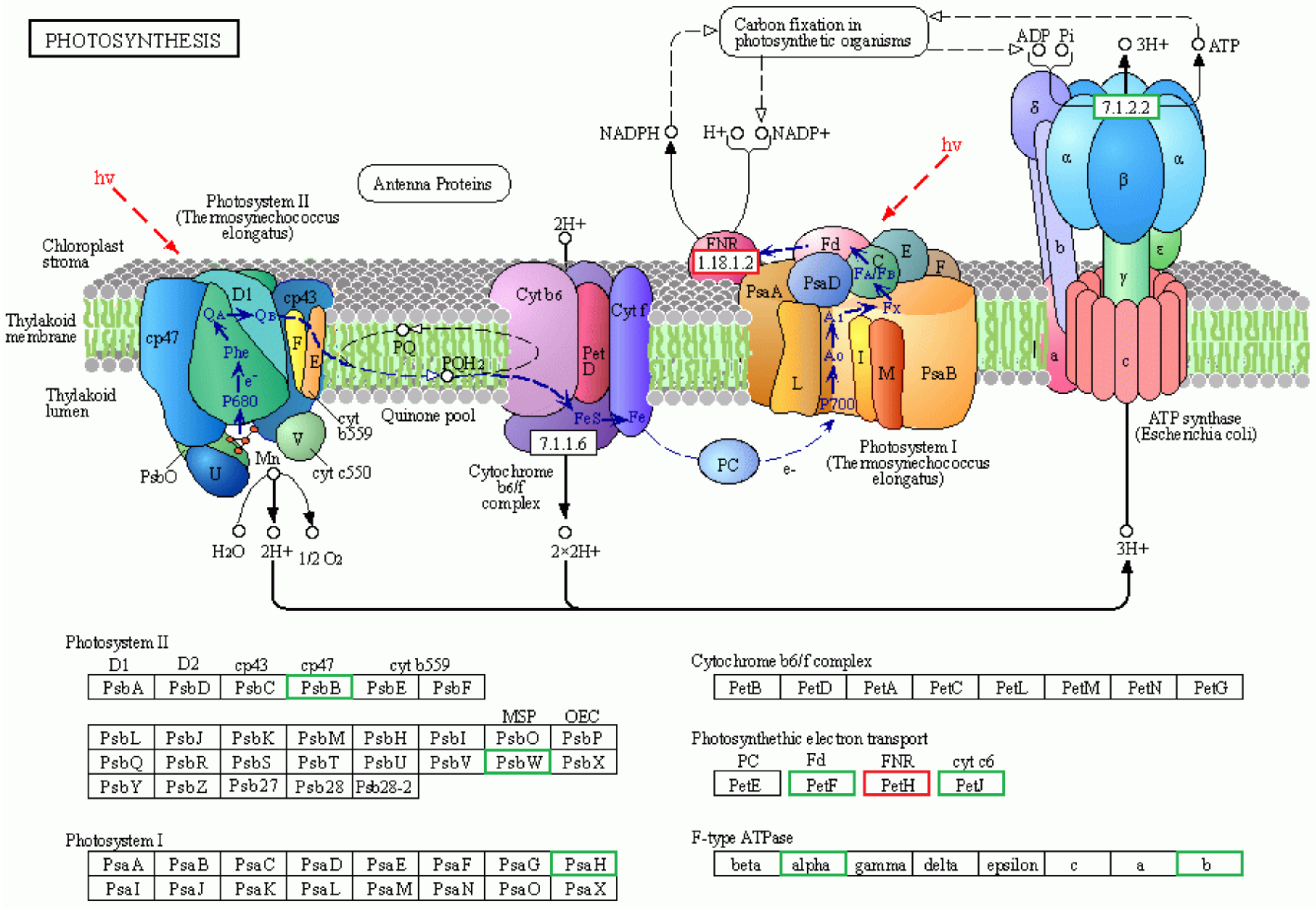
2.4. Differential Analysis of Photosynthesis Metabolic Pathway
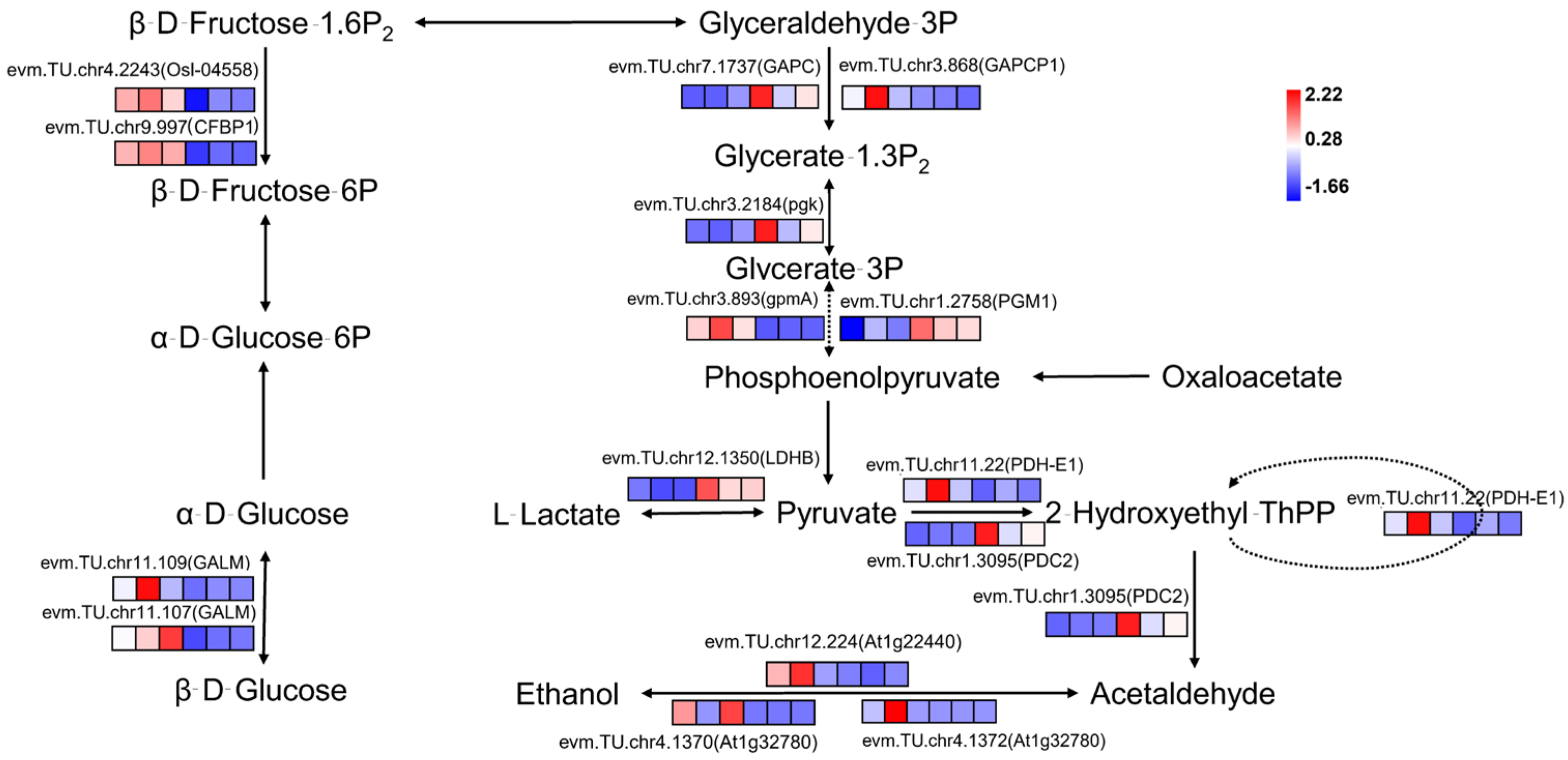
2.5. Differential Analysis of Glycolysis/Gluconeogenesis Pathway
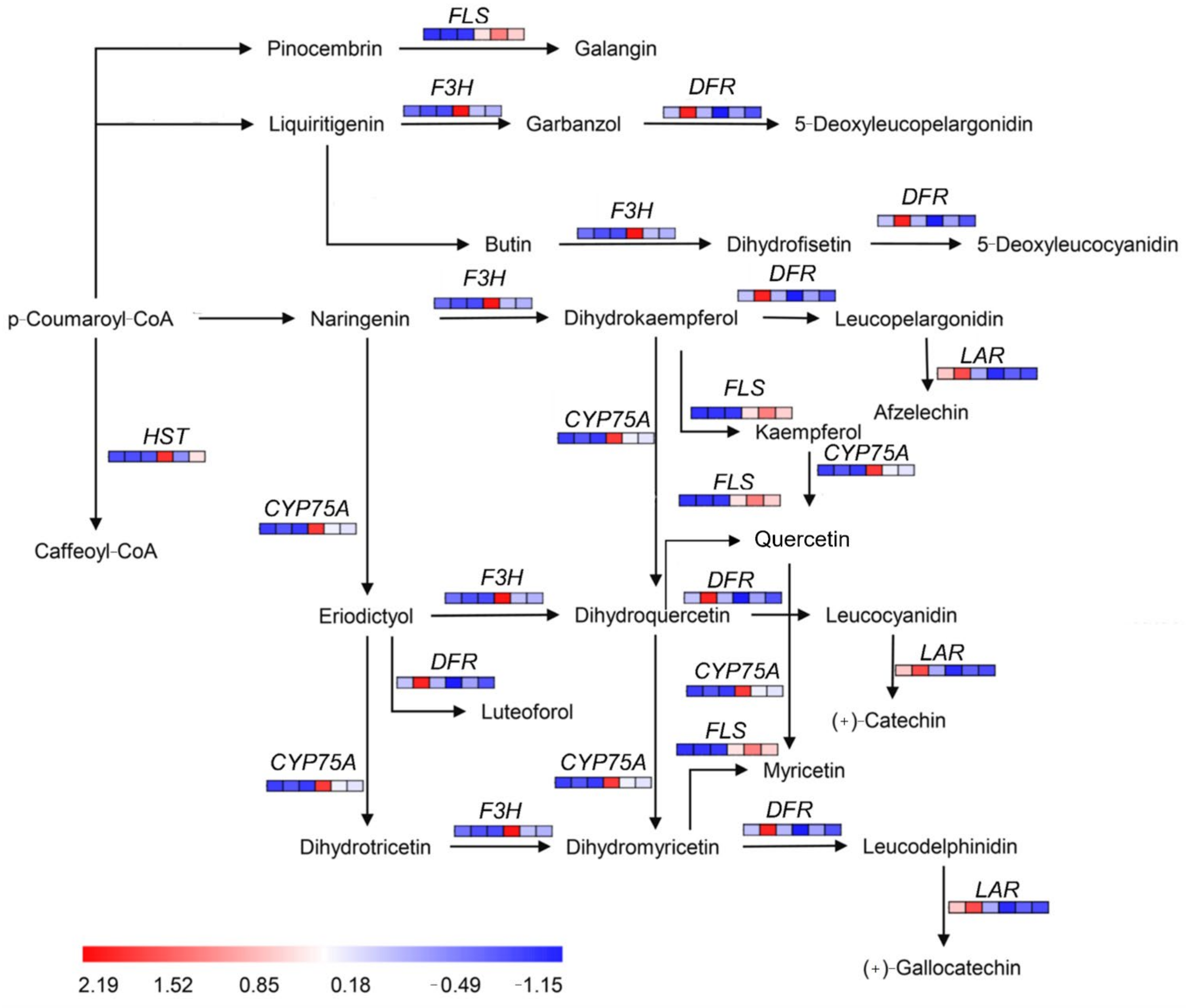
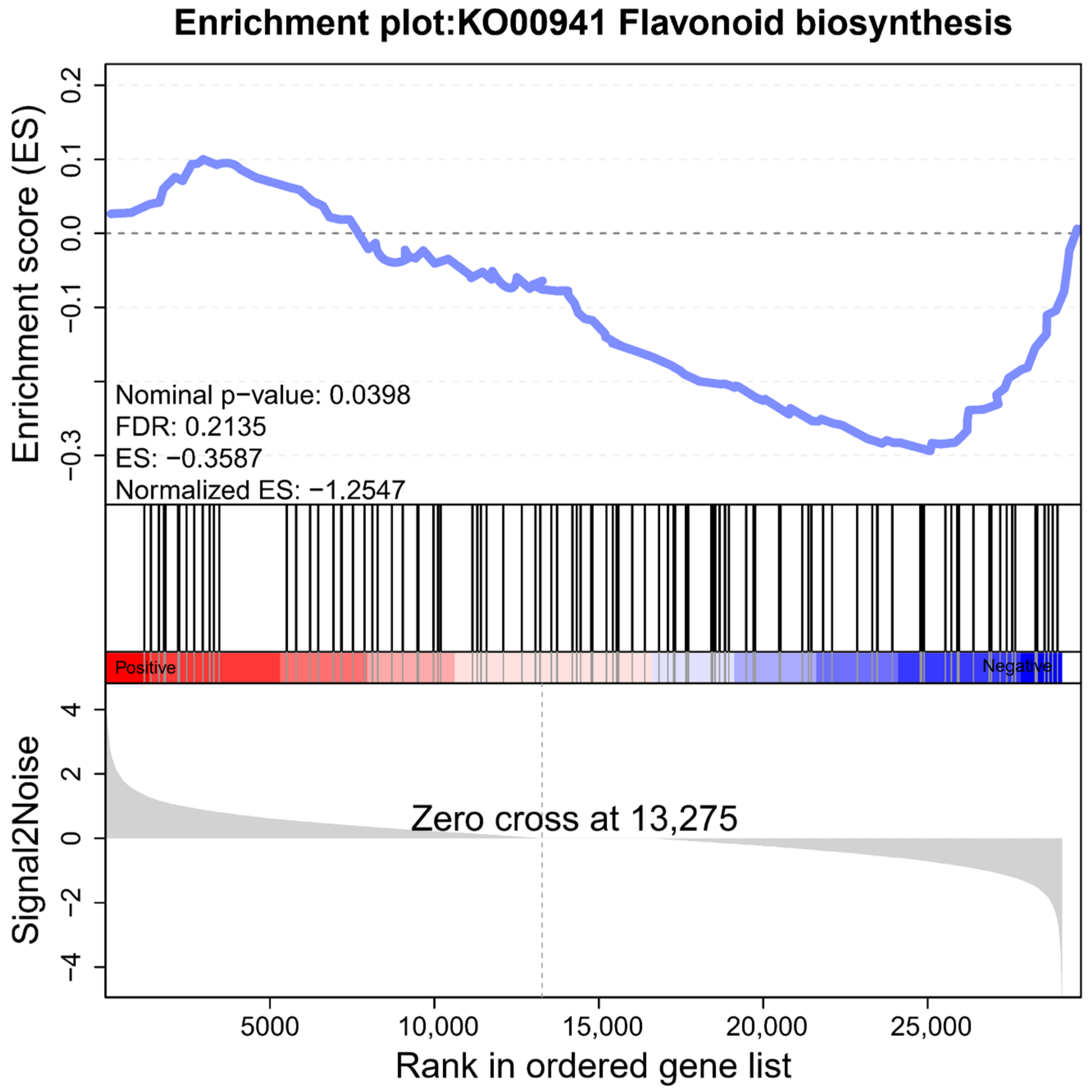
2.6. Differential Analysis of Flavonoid Biosynthesis Pathway
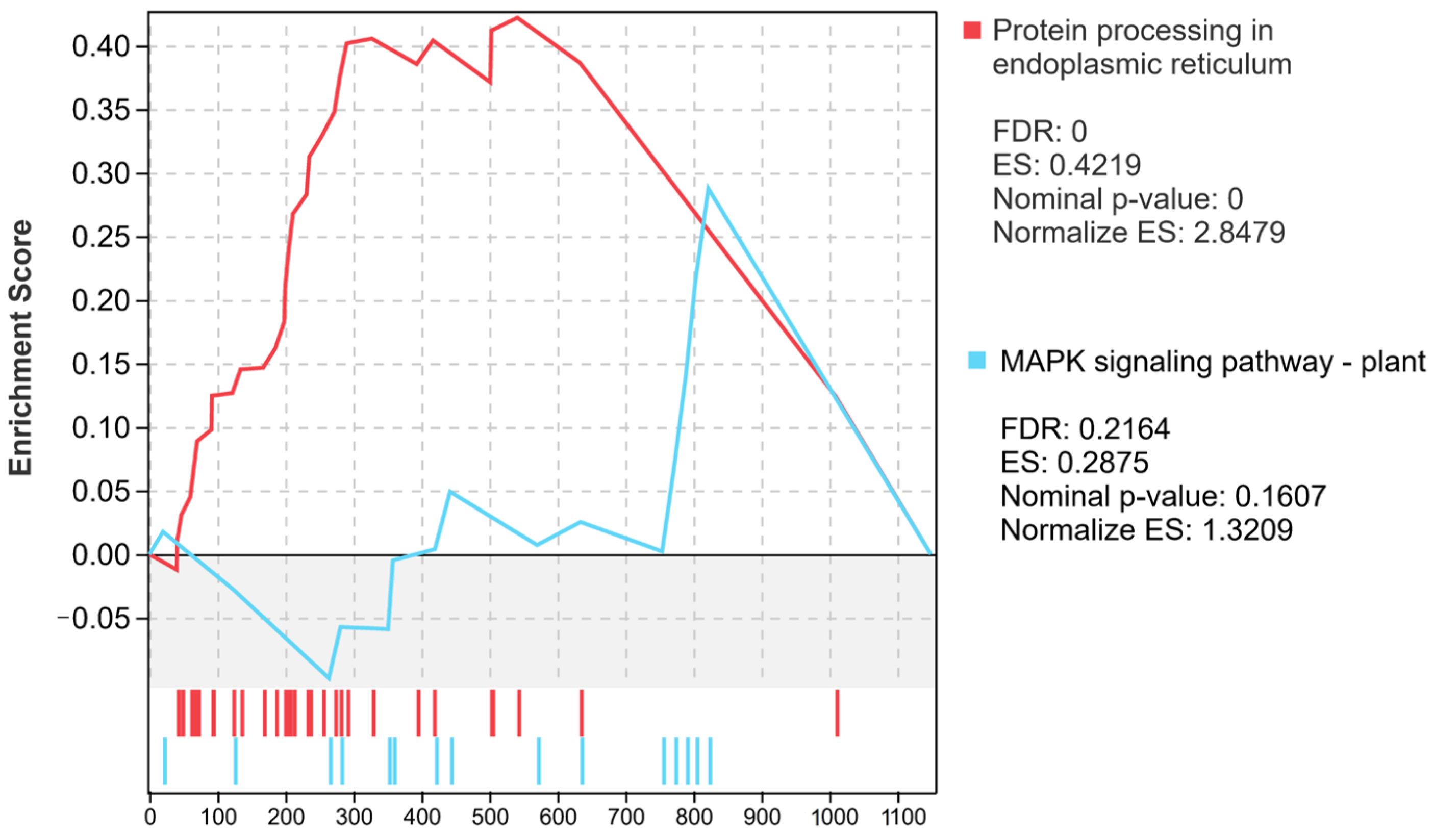
2.7. Enrichment Analysis of Up-Regulated DEGs in Haploid Ginkgo
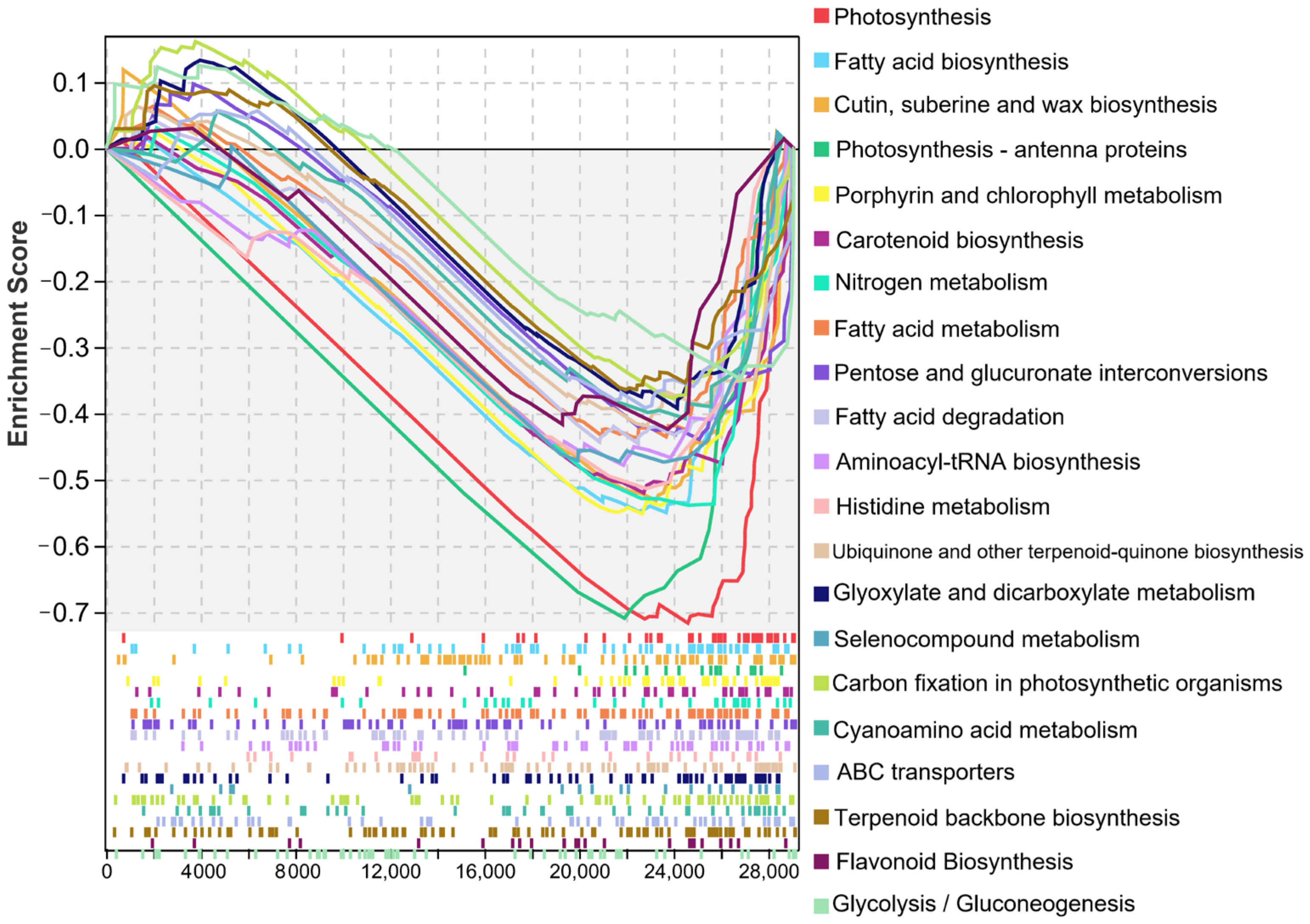
2.8. An Overall Trend of Down-Regulated Expression of DEGs in Haploid Ginkgo
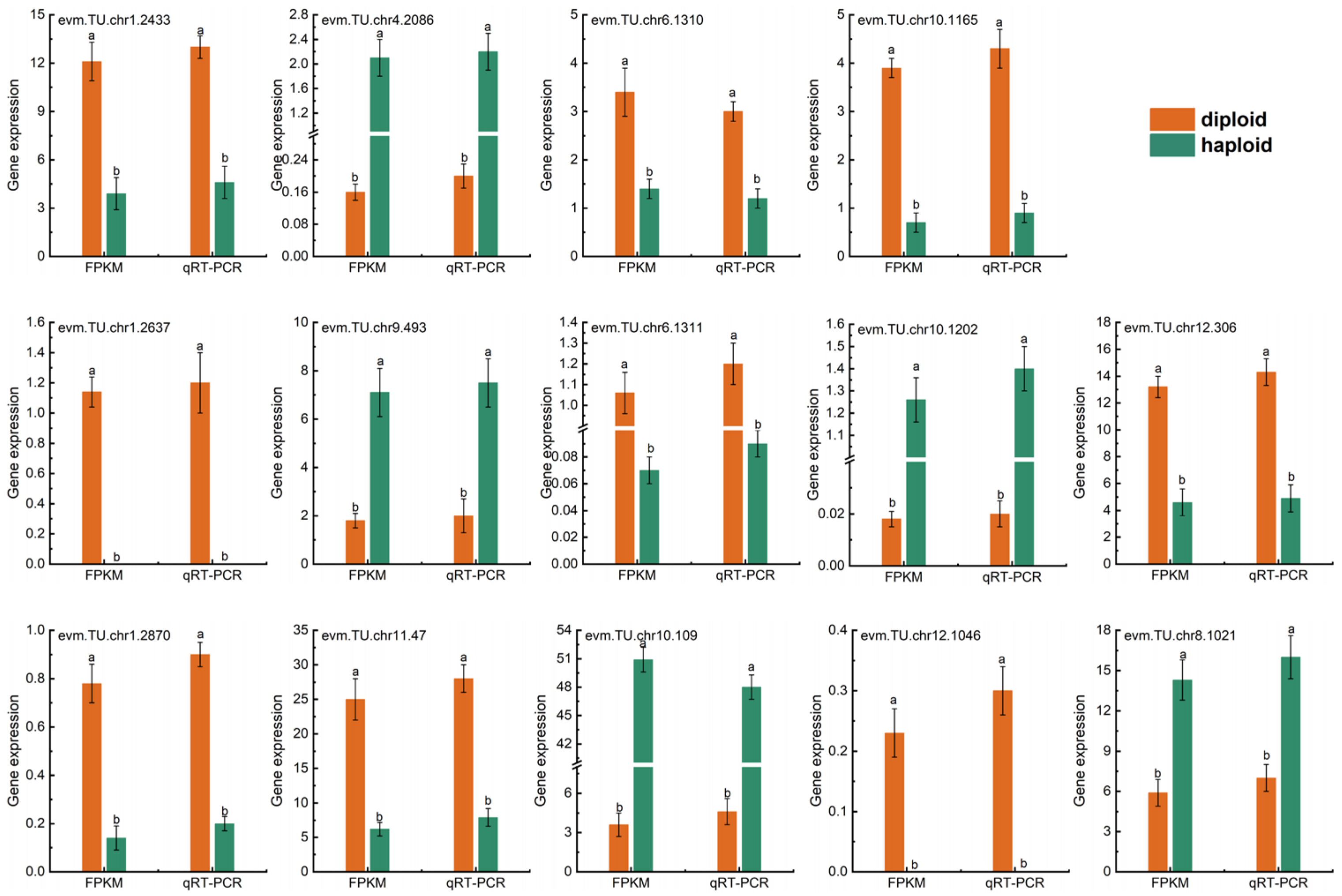
2.9. Transcript Level and Quantitative Real-Time PCR (qRT-PCR) Validation of mRNA in Haploid and Diploid Ginkgo
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sample Preparation
4.2. RNA Sequencing and Data Analysis
4.3. Determination of Flavonoid Content and Photosynthetic Rate of Ginkgo
4.4. Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.Y.; Kim, C.Y.; Min, J.S.; Lee, D.-J.; Naing, A.H.; Chung, J.D.; Kim, C.K. In vitro induction of tetraploids in an interspecific hybrid of Calanthe (Calanthe discolor x Calanthe sieboldii) through colchicine and oryzalin treatments. Plant Biotechnol. Rep. 2014, 8, 251–257. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.; Song, L.; Qi, Q.; Du, K.; Han, Q.; Kang, X. Study of variation in the growth, photosynthesis, and content of secondary metabolites in Eucommia triploids. Trees-Struct. Funct. 2019, 33, 817–826. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, D.; Hu, H.; Zuo, X.; Xia, T.; Xie, J. A comparative study on morphological and fruit quality traits of diploid and polyploid carambola (Averrhoa carambola L.) genotypes. Sci. Hortic. 2021, 277, 109843. [Google Scholar] [CrossRef]
- Zhang, W.; Hao, H.; Ma, L.; Zhao, C.; Yu, X. Tetraploid muskmelon alters morphological characteristics and improves fruit quality. Sci. Hortic. 2010, 125, 396–400. [Google Scholar] [CrossRef]
- Guo, H.; Mendrikahy, J.N.; Xie, L.; Deng, J.; Lu, Z.; Wu, J.; Li, X.; Shahid, M.Q.; Liu, X. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 2017, 7, 40139. [Google Scholar] [CrossRef]
- Luo, G.; Xue, L.; Xu, W.; Zhao, J.; Wang, J.; Ding, Y.; Luan, K.; Lei, J. Breeding decaploid strawberry with improved cold resistance and fruit quality. Sci. Hortic. 2019, 251, 1–8. [Google Scholar] [CrossRef]
- Weber, D.F. Today’s Use of Haploids in Corn Plant Breeding. In Advances in Agronomy; Spark, D.L., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 123, pp. 123–144. [Google Scholar]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Rodomiro, O. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. Int. Rev. J. 2015, 33, 812–829. [Google Scholar] [CrossRef]
- Dunwell, J.M. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol. J. 2010, 8, 377–424. [Google Scholar] [CrossRef] [PubMed]
- Isakov, Y.N.; Butorina, A.K.; Muraya, L.S. Discovery of spontaneous haploids in pinus silvestris and the prospects of their using in forest genetics and selection. Genetika 1981, 17, 701–707. [Google Scholar]
- Blakeslee, A.F.; Belling, J.; Farnham, M.E.; Bergner, A.D. A Haploid Mutant in the Jimson Weed, “Datura Stramonium”. Science 1922, 55, 646–647. [Google Scholar] [CrossRef] [PubMed]
- Von Dassow, P.; Ogata, H.; Probert, I.; Wincker, P.; Da Silva, C.; Audic, S.; Claverie, J.M.; de Vargas, C. Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol. 2009, 10, R114. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, H.; Yang, J.; Du, K.; Li, J.; Zhang, Y.; Qiu, T.; Liu, Z.; Ren, Y.; Song, L.; et al. High-quality de novo assembly of the Eucommia ulmoides haploid genome provides new insights into evolution and rubber biosynthesis. Hortic. Res. 2020, 7, 183. [Google Scholar] [CrossRef]
- Zhou, Q.; Tang, D.; Huang, W.; Yang, Z.; Zhang, Y.; Hamilton, J.P.; Visser, R.G.F.; Bachem, C.W.B.; Robin Buell, C.; Zhang, Z.; et al. Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat. Genet. 2020, 52, 1018–1023. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Wang, N.; Xia, X.; Jiang, T.; Li, L.; Zhang, P.; Niu, L.; Cheng, H.; Wang, K.; Lin, H. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, B.; Wang, D.; Zhu, X.; Li, M.; Zhang, J.; Chen, M.; Wang, M.; Riksen, T.; Liu, J.; et al. In vivo maternal haploid induction in tomato. Plant Biotechnol. J. 2022, 20, 250–252. [Google Scholar] [CrossRef]
- Sun, Y.H.; Wang, Y.R.; Li, Y.Y.; Jiang, J.Z.; Yang, N.; Niu, C.; Li, Y. Effects of colchicine treatment on the microtubule cytoskeleton and total protein during microsporogenesis in Ginkgo biloba L. Pak. J. Bot. 2015, 47, 159–170. [Google Scholar]
- Zhao, Y.; Paule, J.; Fu, C.; Koch, M.A. Out of China: Distribution history of Ginkgo biloba L. Taxon 2010, 59, 495–504. [Google Scholar] [CrossRef]
- McElwain, J.C. Ginkgo The Tree That Time Forgot. Science 2013, 340, 812–813. [Google Scholar] [CrossRef]
- Efferth, T.; Koch, E. Complex Interactions between Phytochemicals. The Multi-Target Therapeutic Concept of Phytotherapy. Curr. Drug Targets 2011, 12, 122–132. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, J.; Xie, Y. Improvement strategies for the oral bioavailability of poorly water-soluble flavonoids: An overview. Int. J. Pharm. 2019, 570, 118642. [Google Scholar] [CrossRef]
- Wu, D.; Feng, J.; Lai, M.; Ouyang, D.; Yu, W.; Wang, G.; Cao, F.; Jacobs, D.F.; Zeng, S. Combined application of bud and leaf growth fertilizer improves leaf flavonoids yield of Ginkgo biloba. Ind. Crops Prod. 2020, 150, 112379. [Google Scholar] [CrossRef]
- Jun, N.; Juan, H.; Zhifang, J.; Xiaori, Z.; Lixiang, D.; Xiuli, Y.; Zhehang, S.; Wenya, X.; Zhikun, W.; Maojun, X. NaCl Induces Flavonoid Biosynthesis through a Putative Novel Pathway in Post-harvest Ginkgo Leaves. Front. Plant Sci. 2017, 8, 920. [Google Scholar]
- Wang, G.; Cao, F.; Chang, L.; Guo, X.; Wang, J. Temperature has more effects than soil moisture on biosynthesis of flavonoids in Ginkgo (Ginkgo biloba L.) leaves. New For. 2014, 45, 797–812. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, G.; Cao, F.; Zhu, C.; Wang, G.; El-Kassaby, Y.A. Light intensity affects the growth and flavonol biosynthesis of Ginkgo (Ginkgo biloba L.). New For. 2014, 45, 765–776. [Google Scholar] [CrossRef]
- Ying, G.; Cga, B.; Mwa, B.; Fffa, B.; Ek, C.; Tw, C.; Gwa, B. Metabolome and transcriptome analyses reveal flavonoids biosynthesis differences in Ginkgo biloba associated with environmental conditions—ScienceDirect. Ind. Crops Prod. 2020, 158, 112963. [Google Scholar]
- Ni, J.; Dong, L.; Jiang, Z.; Yang, X.; Chen, Z.; Wu, Y.; Xu, M. Comprehensive transcriptome analysis and flavonoid profiling of Ginkgo leaves reveals flavonoid content alterations in day–night cycles. PLoS ONE 2018, 13, e0193897. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Qi, Z.; Xin, Y.; Wang, G.; Xu, L.A. De novo transcriptome analysis revealed genes involved in flavonoid biosynthesis, transport and regulation in Ginkgo biloba. Ind. Crops Prod. 2018, 124, 226–235. [Google Scholar] [CrossRef]
- Smarda, P.; Vesely, P.; Smerda, J.; Bures, P.; Knapek, O.; Chytra, M. Polyploidy in a “living fossil’ Ginkgo biloba. New Phytol. 2016, 212, 11–14. [Google Scholar] [CrossRef]
- Smarda, P.; Horova, L.; Knapek, O.; Dieck, H.; Dieck, M.; Razna, K.; Hrubik, P.; Orloci, L.; Papp, L.; Vesela, K.; et al. Multiple haploids, triploids, and tetraploids found in modern-day “living fossil” Ginkgo biloba. Hortic. Res. 2018, 5, 55. [Google Scholar] [CrossRef]
- Lin, Z.; Pu, H.; Duan, Z.; Li, Y.; Liu, B.; Zhang, Q.; Li, W.; Jean-David, R.; Liu, L.; Peng, L. Nucleus-Encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor 1. Plant Cell 2018, 30, 1770–1788. [Google Scholar]
- Tamayo, P.; Steinhardt, G.; Liberzon, A.; Mesirov, J.P. The limitations of simple gene set enrichment analysis assuming gene independence. Stat. Methods Med. Res. 2016, 25, 472–487. [Google Scholar] [CrossRef]
- Tilford, C.A.; Siemers, N.O. Gene Set Enrichment Analysis. In Protein Networks and Pathway Analysis; Nikolsky, Y., Bryant, J., Eds.; Methods in Molecular Biology: New York, NY, USA, 2009; Volume 563, pp. 99–121. [Google Scholar]
- Chen, T.T.; Sheng, Y.; Hao, Z.D.; Long, X.F.; Fu, F.F.; Liu, Y.; Tang, Z.H.; Ali, A.; Peng, Y.; Lu, L.; et al. Transcriptome and proteome analysis suggest enhanced photosynthesis in tetraploid Liriodendron sino-americanum. Tree Physiol. 2021, 41, 1953–1971. [Google Scholar] [CrossRef]
- Tokumoto, Y.; Kajiura, H.; Takeno, S.; Harada, Y.; Suzuki, N.; Hosaka, T.; Gyokusen, K.; Nakazawa, Y. Induction of tetraploid hardy rubber tree, Eucommia ulmoides, and phenotypic differences from diploid. Plant Biotechnol. 2016, 33, 51–57. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, Y.; Sun, S.; Li, G.; Wang, J.; Wang, B.; Lin, X.Y.; Huang, M.; Gong, Z.Y.; Sanguinet, K.A.; et al. Aneuploidization under segmental allotetraploidy in rice and its phenotypic manifestation. Theor. Appl. Genet. 2018, 131, 1273–1285. [Google Scholar] [CrossRef]
- Mo, L.; Chen, J.H.; Chen, F.; Xu, Q.W.; Tong, Z.K.; Huang, H.H.; Dong, R.H.; Lou, X.Z.; Lin, E.P. Induction and characterization of polyploids from seeds of Rhododendron fortunei Lindl. J. Integr. Agric. 2020, 19, 2016–2026. [Google Scholar] [CrossRef]
- Shmeit, Y.H.; Fernandez, E.; Novy, P.; Kloucek, P.; Orosz, M.; Kokoska, L. Autopolyploidy effect on morphological variation and essential oil content in Thymus vulgaris L. Sci. Hortic. 2020, 263. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.Q.; Wang, F.; Zhang, Z.H. Involvement of Auxin and Brassinosteroid in Dwarfism of Autotetraploid Apple (Malus x domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Tadeo, F.; Froelicher, Y.; Talon, M.; Navarro, L.; Ollitrault, P.; Morillon, R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 2011, 62, 2507–2519. [Google Scholar] [CrossRef]
- Adams, K.L. Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 2007, 98, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Udall, J.A.; Wendel, J.F. Polyploidy and crop improvement. Crop Sci. 2006, 46, S3–S14. [Google Scholar] [CrossRef]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005, 10, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Wang, G.; Cui, P.; Wu, S.; Ai, C.; Hu, N.; Li, A.; He, B.; Shao, X.; et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Hasing, T.; Tang, H.; Brym, M.; Khazi, F.; Huang, T.; Chambers, A.H. A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat. Food 2020, 1, 811–819. [Google Scholar] [CrossRef]
- Tian, Y.; Thrimawithana, A.; Ding, T.; Guo, J.; Gleave, A.; Chagne, D.; Ampomah-Dwamena, C.; Ireland, H.S.; Schaffer, R.J.; Luo, Z.; et al. Transposon insertions regulate genome-wide allele-specific expression and underpin flower colour variations in apple (Malus spp.). Plant Biotechnol. J. 2022, 20, 1285–1297. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia Commission. 2015. Available online: http://www.chp.org.cn (accessed on 15 September 2021).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2013, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, T.; Yang, X.; Fu, F.; Wang, G.; Cao, F. se1ection of suitab1e reference genes based on transcriptomic data in ginkgo bi1oba under different experimenta1 conditions. Forests 2020, 11, 1217. [Google Scholar] [CrossRef]




| Sample | Clean Reads | Clean Data (bp) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| H1 | 84,053,070 | 12,606,818,190 | 97.84 | 94.66 | 44.72 |
| H2 | 74,996,246 | 10,743,266,079 | 97.76 | 94.37 | 44.65 |
| H3 | 73,313,140 | 10,496,771,635 | 97.87 | 94.66 | 44.55 |
| D1 | 89,428,662 | 13,348,479,805 | 97.80 | 94.55 | 44.39 |
| D2 | 82,482,584 | 12,311,759,601 | 97.79 | 94.46 | 44.48 |
| D3 | 82,336,494 | 12,322,466,518 | 97.91 | 94.79 | 44.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Šmarda, P.; Liu, G.; Wang, B.; Gao, X.; Guo, Q. High-Depth Transcriptome Reveals Differences in Natural Haploid Ginkgo biloba L. Due to the Effect of Reduced Gene Dosage. Int. J. Mol. Sci. 2022, 23, 8958. https://doi.org/10.3390/ijms23168958
Hu Y, Šmarda P, Liu G, Wang B, Gao X, Guo Q. High-Depth Transcriptome Reveals Differences in Natural Haploid Ginkgo biloba L. Due to the Effect of Reduced Gene Dosage. International Journal of Molecular Sciences. 2022; 23(16):8958. https://doi.org/10.3390/ijms23168958
Chicago/Turabian StyleHu, Yaping, Petr Šmarda, Ganping Liu, Beibei Wang, Xiaoge Gao, and Qirong Guo. 2022. "High-Depth Transcriptome Reveals Differences in Natural Haploid Ginkgo biloba L. Due to the Effect of Reduced Gene Dosage" International Journal of Molecular Sciences 23, no. 16: 8958. https://doi.org/10.3390/ijms23168958






