Genome-Wide Identification and Analysis of the Growth-Regulating Factor Family in Zanthoxylum armatum DC and Functional Analysis of ZaGRF6 in Leaf Size and Longevity Regulation
Abstract
:1. Introduction
2. Results
2.1. Identification of the GRF Gene Family in Z. armatum
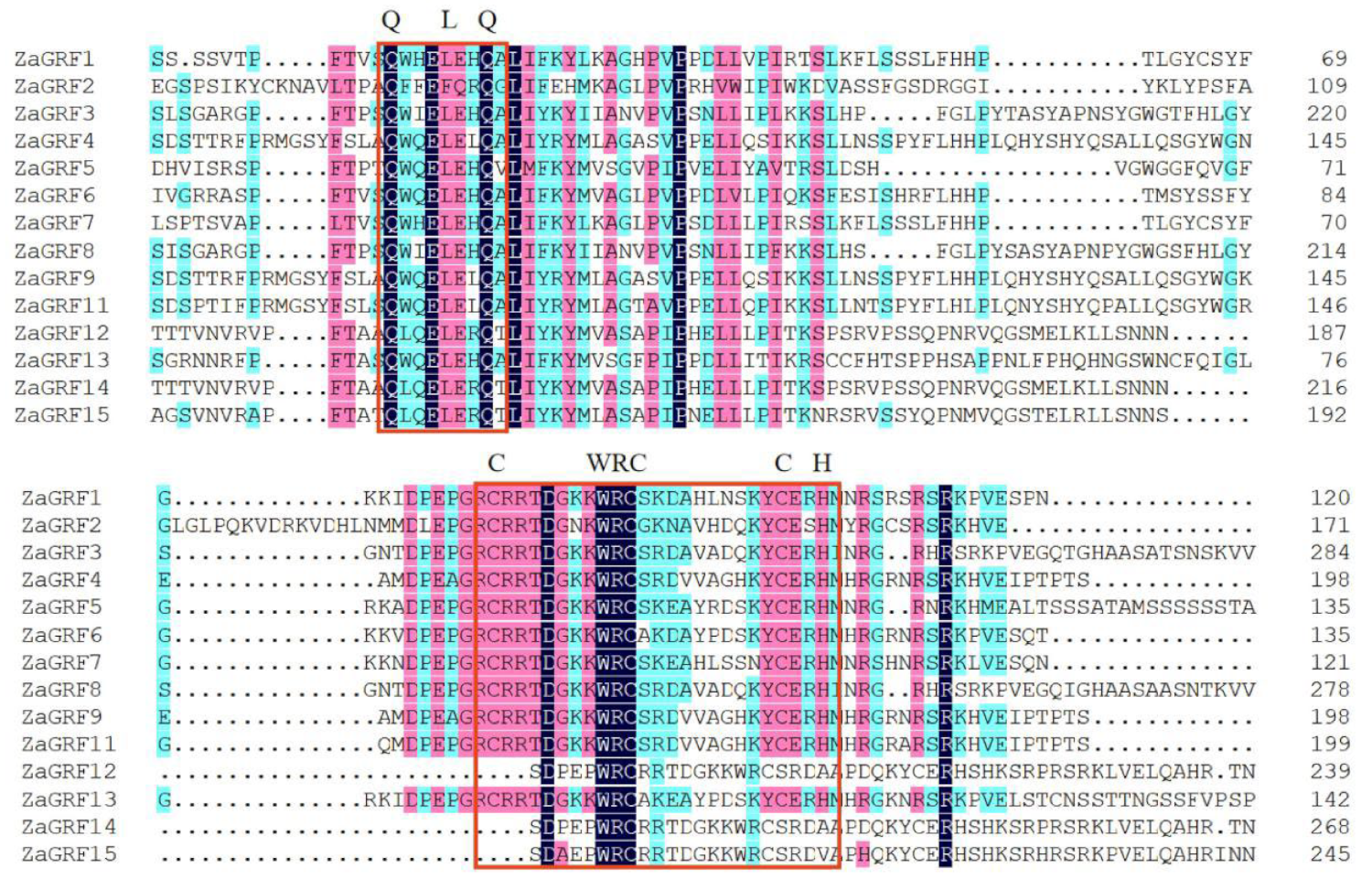
2.2. The Protein Structure, Subcellular Localization Prediction, and Post-Translational Modification of ZaGRFs
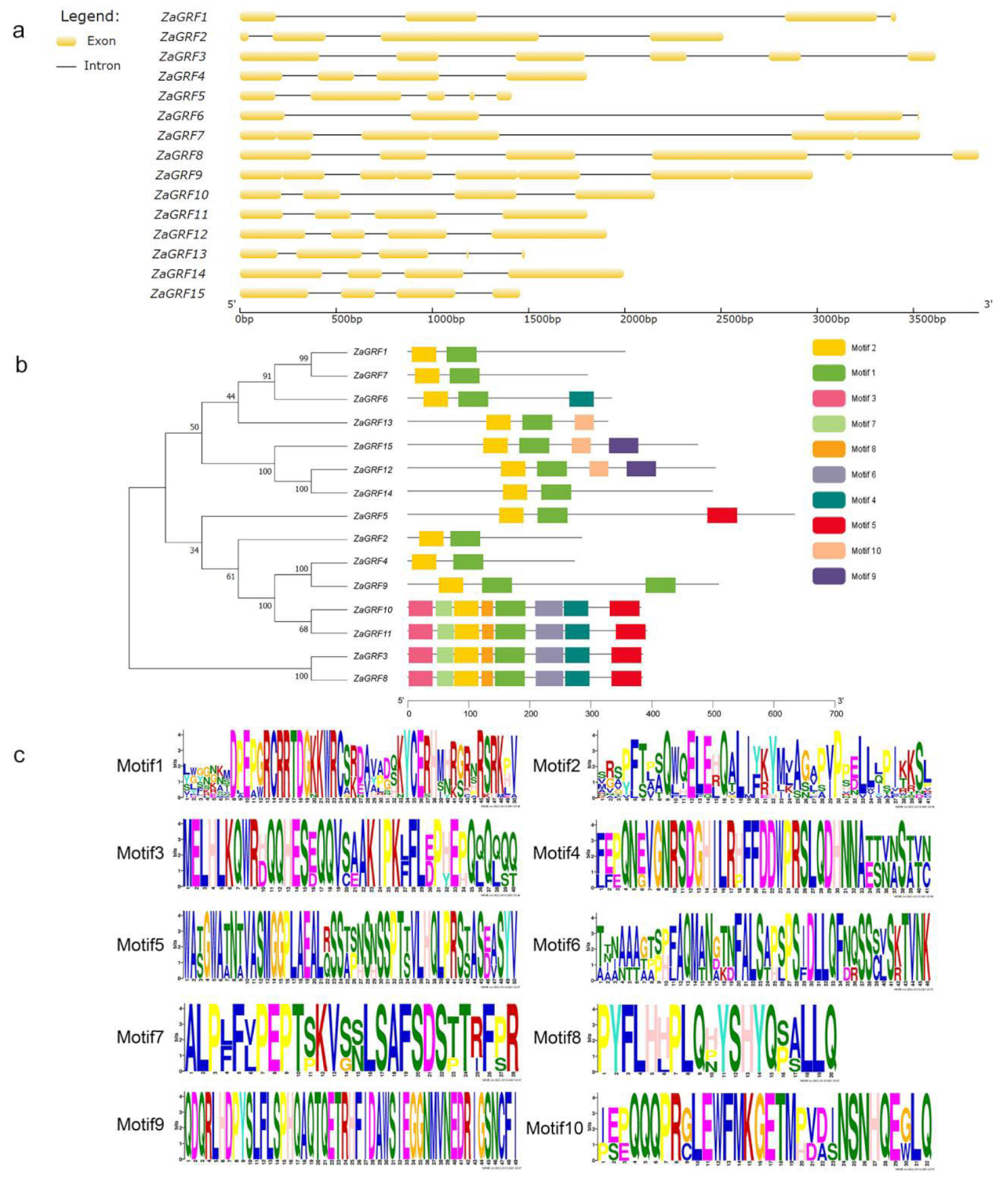
2.3. Phylogenetic Analysis
2.4. Gene Structure, Motif Distribution, and Conserved Domain Alignment
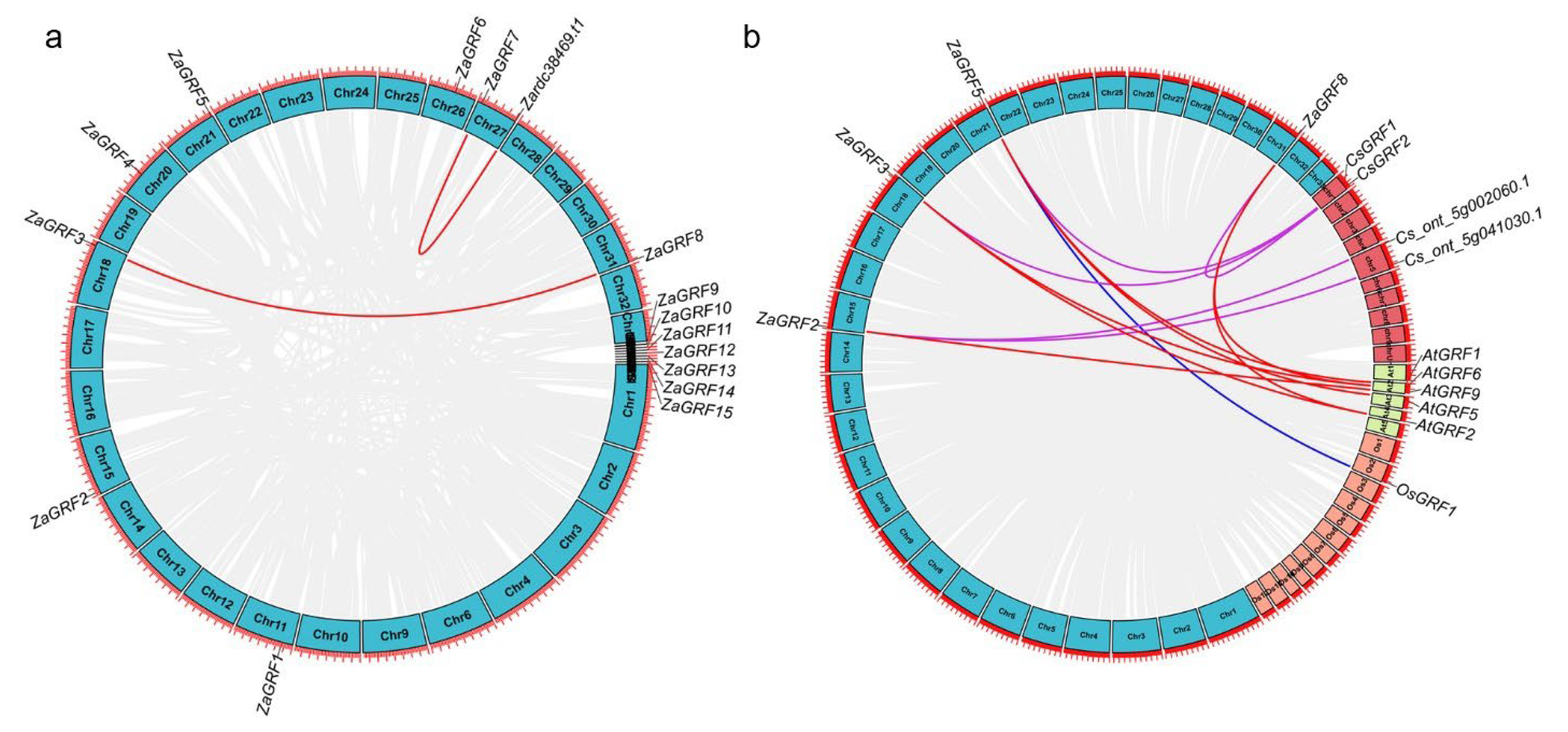
2.5. Chromosome Localization and Synteny Analysis
2.6. Promoter Analysis
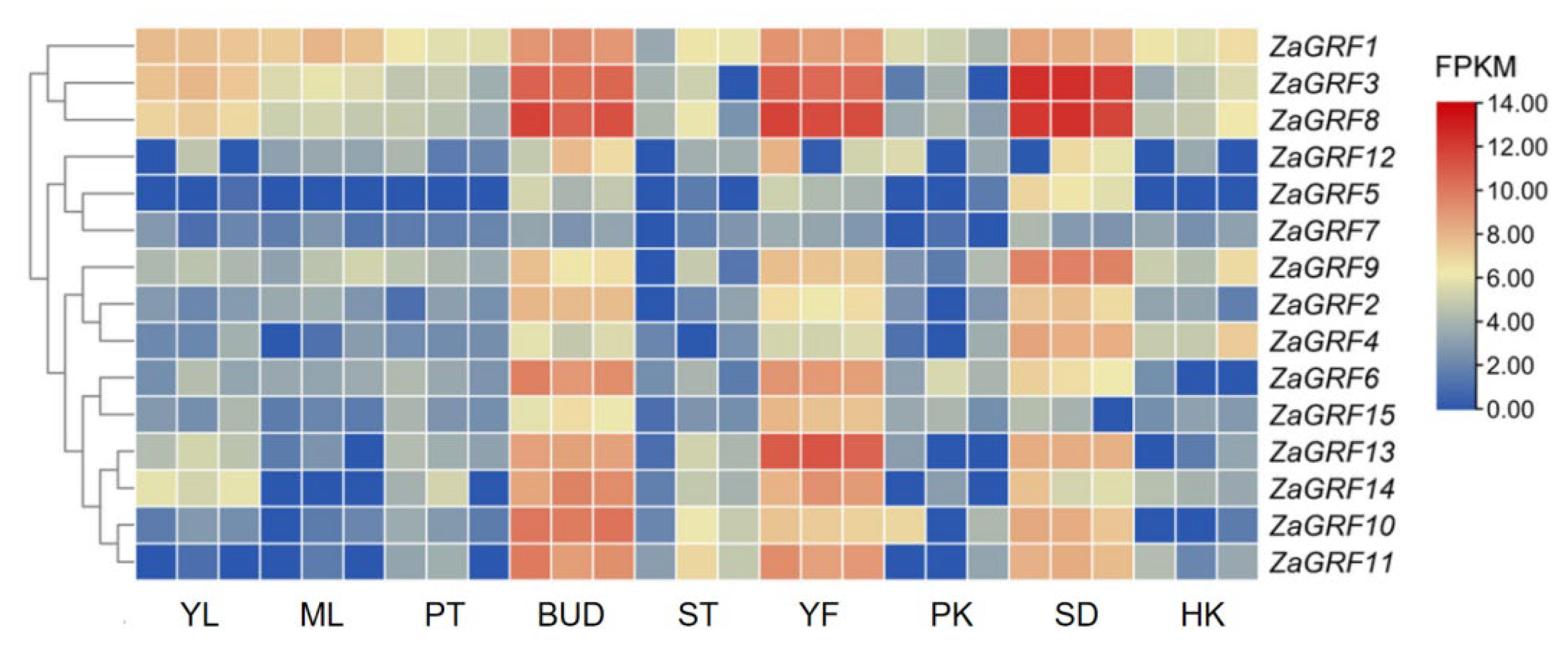
2.7. In Silico Tissue-Specific Expression of ZaGRF Genes
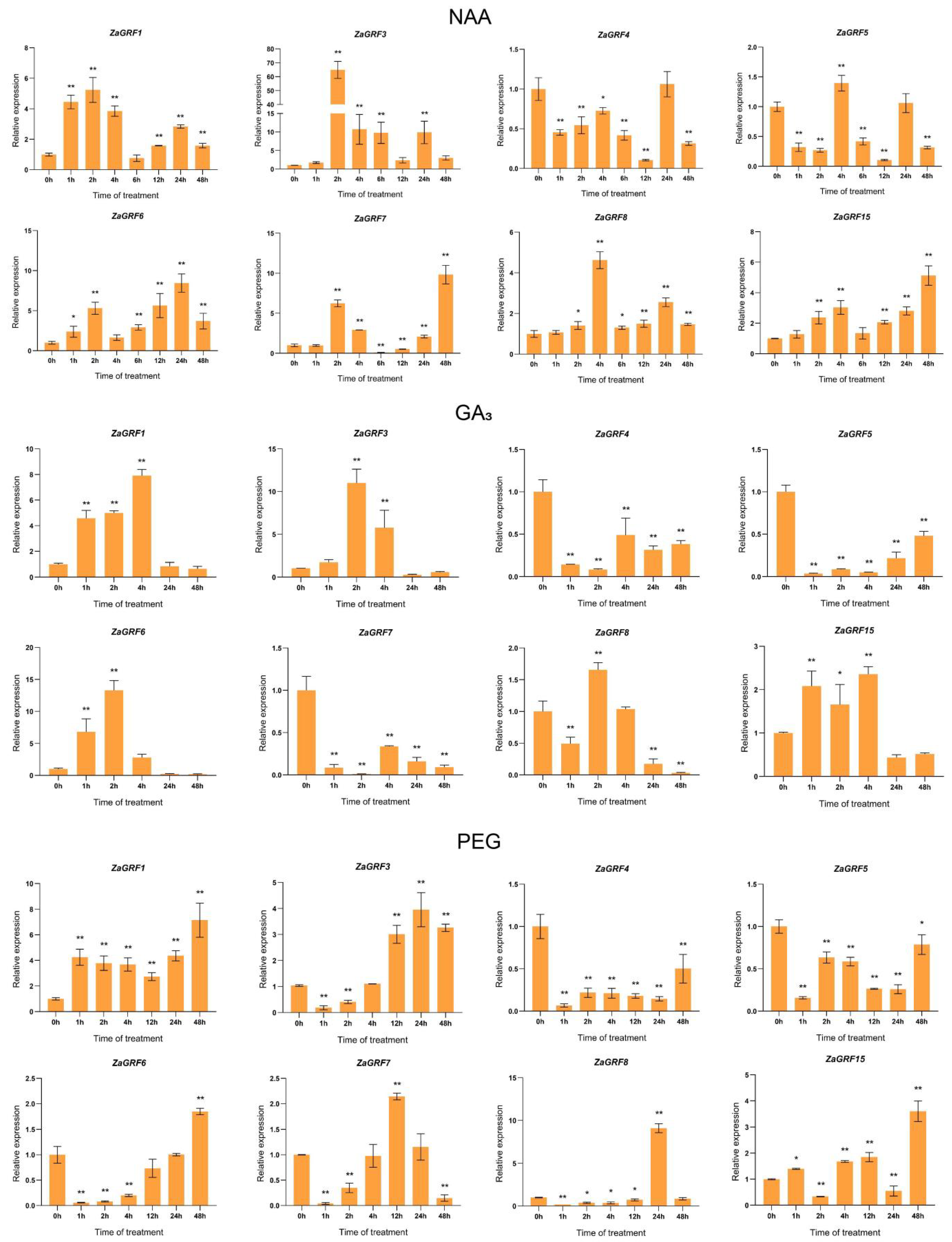
2.8. ZaGRF Gene Expression Profiles in Response to Hormone and Drought Treatment
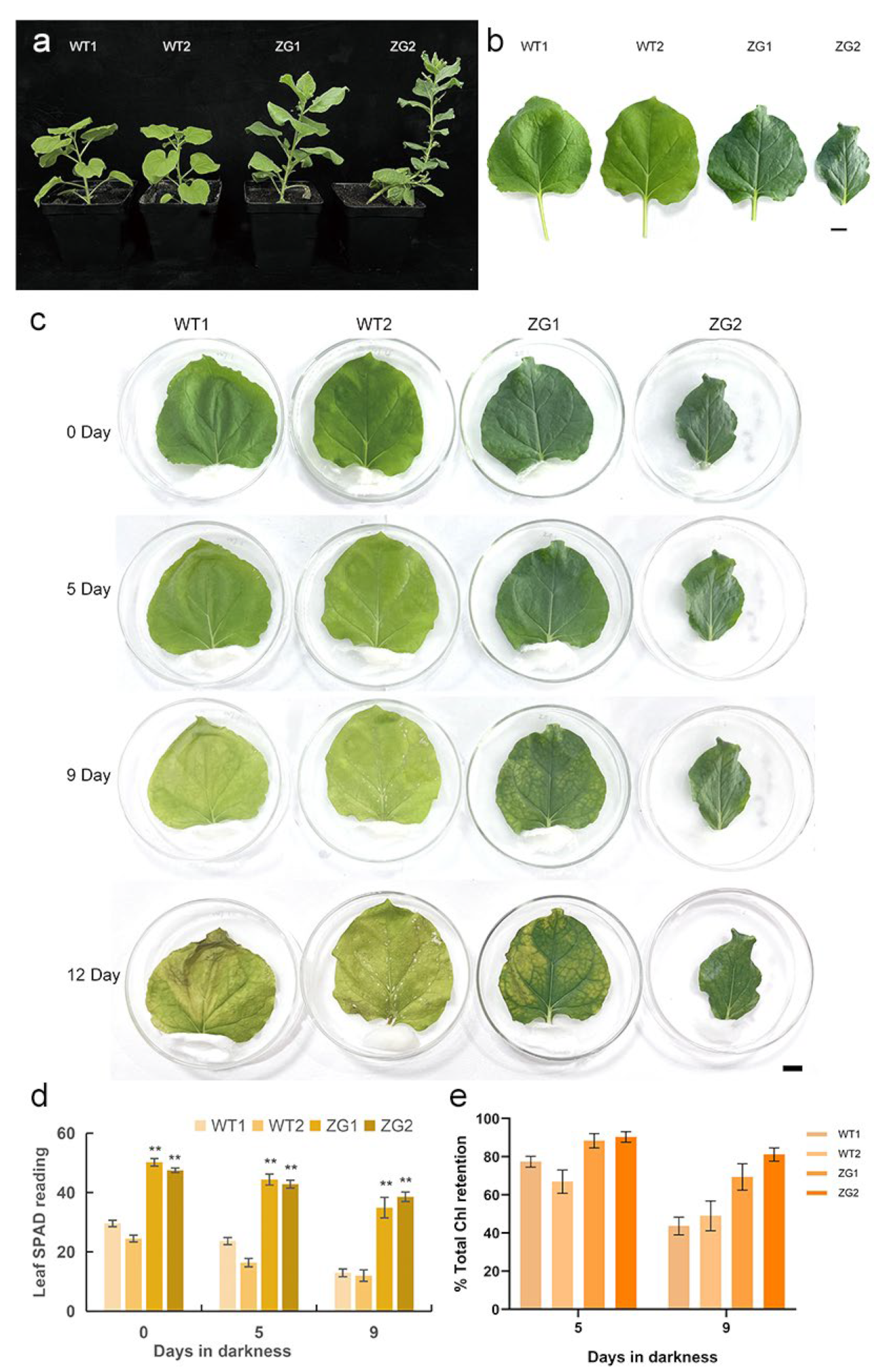
2.9. Overexpression of ZaGRF6 Alters Leaf Size and Longevity in Transgenic Nicotiana Benthamiana
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Protein Identification, Physicochemical Properties, and Mir396 Target Site Analysis
4.3. Prediction of Protein Structure, Subcellular Localization, and Post-Translational Modifications
4.4. Phylogeny, Conserved Motifs, and Gene Structure Analysis
4.5. Chromosome Location and Collinearity Analysis
4.6. Cis-Acting Element and Gene Expression Pattern Analysis
4.7. Plasmid Construction and Plant Transformation
4.8. Chl Measurements after Dark-Induced Senescence
4.9. RNA Isolation and qRT–PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef]
- Zhang, D.-F.; Li, B.; Jia, G.-Q.; Zhang, T.-F.; Dai, J.-R.; Li, J.-S.; Wang, S.-C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, J.H.; Kende, H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in Rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Z.; Jin, J.; Xie, X.; Zhang, H.; Chen, Q.; Luo, Z.; Yang, J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 2018, 639, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, K.; Ning, L.; He, J.; Ma, X.; Li, Z.; Zhang, X.; Yin, D. Genome-wide analysis of the growth-regulating factor family in Peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2019, 20, 4120. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Ma, J.; Song, C.; Zhang, L.; Gao, C.; Zhang, D.; An, N.; Mao, J.; Han, M. Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genet. Genomes 2018, 14, 54. [Google Scholar] [CrossRef]
- Vercruyssen, L.; Tognetti, V.B.; Gonzalez, N.; Van Dingenen, J.; De Milde, L.; Bielach, A.; De Rycke, R.; Van Breusegem, F.; Inzé, D. GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 2015, 167, 817–832. [Google Scholar] [CrossRef]
- Kim, J.-S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y.; et al. Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid– and osmotic stress–responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar] [CrossRef]
- Du, W.; Yang, J.; Li, Q.; Su, Q.; Yi, D.; Pang, Y. Genome-wide identification and characterization of growth regulatory factor family genes in medicago. Int. J. Mol. Sci. 2022, 23, 6905. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; Zhang, L.; Xie, T.; Li, L. Genome-wide identification and analysis of the Growth-Regulating Factor (GRF) gene family and GRF-Interacting Factor family in Triticum aestivum L. Biochem. Genet. 2020, 58, 705–724. [Google Scholar] [CrossRef]
- Wang, M.; Tong, S.; Ma, T.; Xi, Z.; Liu, J. Chromosome-level genome assembly of Sichuan pepper provides insights into apomixis, drought tolerance, and alkaloid biosynthesis. Mol. Ecol. Resour. 2021, 21, 2533–2545. [Google Scholar] [CrossRef]
- Xu, D.; Zhuo, Z.; Wang, R.; Ye, M.; Pu, B. Modeling the distribution of Zanthoxylum armatum in China with MaxEnt modeling. Glob. Ecol. Conserv. 2019, 19, e00691. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, J.; Zhu, L.; Li, T.; Jiang, W.; Zhou, J.; Peng, W.; Wu, C. Zanthoxylum bungeanum Maxim. (Rutaceae): A systematic review of its traditional uses, botany, phytochemistry, pharmacology, pharmacokinetics, and toxicology. Int. J. Mol. Sci. 2017, 18, 2172. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, N.; Liu, X.; Ye, J.; Zhang, J.; Chen, Z.; Xu, F.; Zhang, W.; Liao, Y. Comparative transcriptome analysis identified differentially expressed genes between male and female flowers of Zanthoxylum armatum var novemfolius. Agronomy 2020, 10, 283. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, D.; Xue, M.; Qian, J.; He, Y.; Wang, S. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 2014, 56, 1053–1063. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, W.; Wang, Y.; He, Q.; Shu, F.; Liu, H.; Wang, J.; Wang, J.; Yuan, L.; Deng, H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, B.; Zhang, Z.; Luo, W.; Tang, Y.; Niu, Y.; Chong, K.; Xu, Y. OsGRF6 interacts with SLR1 to regulate OsGA2ox1 expression for coordinating chilling tolerance and growth in rice. J. Plant Physiol. 2021, 260, 153406. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wilmanns, M.; Thornton, J.; Köhn, M. Elucidating human phosphatase-substrate networks. Sci. Signal. 2013, 6, rs10. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Fujikura, U.; Olas, J.J.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018, 14, e1007484. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol. 2017, 176, 946–959. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-L.; Wen, Z.; Yang, K.; Wen, X.-P. Conserved miR396b-GRF regulation is involved in abiotic stress responses in pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2019, 20, 2501. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Wu, C.; Huang, C.; Tie, W.; Yan, Y.; Ding, Z.; Xia, Z.; Wang, W.; Peng, M.; Tian, L.; et al. Genome-wide analysis of the GRF family reveals their involvement in abiotic stress response in cassava. Genes 2018, 9, 110. [Google Scholar] [CrossRef]
- Vercruyssen, L.; Verkest, A.; Gonzalez, N.; Heyndrickx, K.S.; Eeckhout, D.; Han, S.K.; Jégu, T.; Archacki, R.; Van Leene, J.; Andriankaja, M.; et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 2014, 26, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, J.; Wang, Q.; Lv, K.; Du, K.; Zhang, W.; Li, Q.; Kang, X.; Wei, H. Growth-regulating factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 2021, 230, 612–628. [Google Scholar] [CrossRef]
- Zhang, B.; Tong, Y.; Luo, K.; Zhai, Z.; Liu, X.; Shi, Z.; Zhang, D.; Li, D. Identification of GROWTH-REGULATING FACTOR transcription factors in lettuce (Lactuca sativa) genome and functional analysis of LsaGRF5 in leaf size regulation. BMC Plant Biol. 2021, 21, 485. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dan, Z.; Gao, F.; Chen, P.; Fan, F.; Li, S. Rice GROWTH-REGULATING FACTOR7 modulates plant architecture through regulating GA and indole-3-acetic acid metabolism. Plant Physiol. 2020, 184, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, S.; Åstot, C.; Sitbon, F.; Moritz, T.; Olsson, O.; Sandberg, G. Transgenic tobacco plants co-expressing Agrobacterium iaa and ipt genes have wild-type hormone levels but display both auxin- and cytokinin-overproducing phenotypes. Plant J. 2000, 23, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.-F.; Zhao, D.-G. Expression of IPT in Asakura-sanshoo (Zanthoxylum piperitum (L.) DC. f. inerme Makino) alters tree architecture, delays leaf senescence, and changes leaf essential oil composition. Plant Mol. Biol. Report. 2016, 34, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed]
- Márquez-López, R.E.; Quintana-Escobar, A.O.; Loyola-Vargas, V.M. Cytokinins, the Cinderella of plant growth regulators. Phytochem. Rev. 2019, 18, 1387–1408. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; The Babraham Institute: Babraham, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene | Gene ID | Gene Locus | Start/Stop Codon | No. of Amino Acids | MW/kD | PI | Instability Index |
|---|---|---|---|---|---|---|---|
| ZaGRF1 | Zardc16696.t1 | Chr11 | 13,736,257~13,739,667 | 355 | 39.93 | 9.35 | 53.17 |
| ZaGRF2 | Zardc21503.t1 | Chr15 | 3,863,606~3,866,118 | 508 | 55.39 | 9.15 | 40.83 |
| ZaGRF3 | Zardc26531.t1 | Chr18 | 73,130,360~73,133,975 | 498 | 53.65 | 9.39 | 57.36 |
| ZaGRF4 | Zardc29111.t1 | Chr20 | 29,291,029~29,292,832 | 384 | 42.22 | 8.30 | 59.43 |
| ZaGRF5 | Zardc30654.t1 | Chr21 | 65,349,679~65,351,091 | 284 | 31.46 | 9.15 | 54.91 |
| ZaGRF6 | Zardc37188.t1 | Chr26 | 32,600,857~32,604,386 | 333 | 36.81 | 8.66 | 63.71 |
| ZaGRF7 | Zardc37898.t1 | Chr27 | 5,546,773~5,549,425 | 293 | 32.88 | 9.90 | 50.78 |
| ZaGRF8 | Zardc42395.t1 | Chr31 | 51,746,517~51,750,356 | 652 | 70.82 | 6.24 | 47.54 |
| ZaGRF9 | Zardc45509.t1 | Unanchored 456 | 284,498~286,320 | 384 | 42.28 | 8.60 | 59.09 |
| ZaGRF10 | Zardc49103.t1 | Unanchored 5139 | 19,631~21,787 | 381 | 41.88 | 8.23 | 57.88 |
| ZaGRF11 | Zardc50859.t1 | Unanchored 7776 | 8700~10,505 | 391 | 43.07 | 7.76 | 59.18 |
| ZaGRF12 | Zardc52135.t1 | Unanchored 9706 | 24,661~26,568 | 474 | 51.70 | 7.24 | 58.53 |
| ZaGRF13 | Zardc52753.t1 | Unanchored 10725 | 65,943~67,423 | 272 | 31.14 | 8.75 | 70.16 |
| ZaGRF14 | Zardc53033.t1 | Unanchored 11223 | 3049~5043 | 503 | 55.15 | 6.98 | 57.08 |
| ZaGRF15 | Zardc54515.t1 | Unanchored 13411 | 6765~8221 | 327 | 36.87 | 8.79 | 59.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Chen, J.; Li, J.; Li, Y.; Zeng, X. Genome-Wide Identification and Analysis of the Growth-Regulating Factor Family in Zanthoxylum armatum DC and Functional Analysis of ZaGRF6 in Leaf Size and Longevity Regulation. Int. J. Mol. Sci. 2022, 23, 9043. https://doi.org/10.3390/ijms23169043
Huang Y, Chen J, Li J, Li Y, Zeng X. Genome-Wide Identification and Analysis of the Growth-Regulating Factor Family in Zanthoxylum armatum DC and Functional Analysis of ZaGRF6 in Leaf Size and Longevity Regulation. International Journal of Molecular Sciences. 2022; 23(16):9043. https://doi.org/10.3390/ijms23169043
Chicago/Turabian StyleHuang, Yanhui, Jiajia Chen, Jianrong Li, Yan Li, and Xiaofang Zeng. 2022. "Genome-Wide Identification and Analysis of the Growth-Regulating Factor Family in Zanthoxylum armatum DC and Functional Analysis of ZaGRF6 in Leaf Size and Longevity Regulation" International Journal of Molecular Sciences 23, no. 16: 9043. https://doi.org/10.3390/ijms23169043
APA StyleHuang, Y., Chen, J., Li, J., Li, Y., & Zeng, X. (2022). Genome-Wide Identification and Analysis of the Growth-Regulating Factor Family in Zanthoxylum armatum DC and Functional Analysis of ZaGRF6 in Leaf Size and Longevity Regulation. International Journal of Molecular Sciences, 23(16), 9043. https://doi.org/10.3390/ijms23169043






