Profiling of the Candidate Interacting Proteins of SELF-PRUNING 6A (SP6A) in Solanum tuberosum
Abstract
:1. Introduction
2. Results
2.1. Yeast Two-Hybrid Library Construction
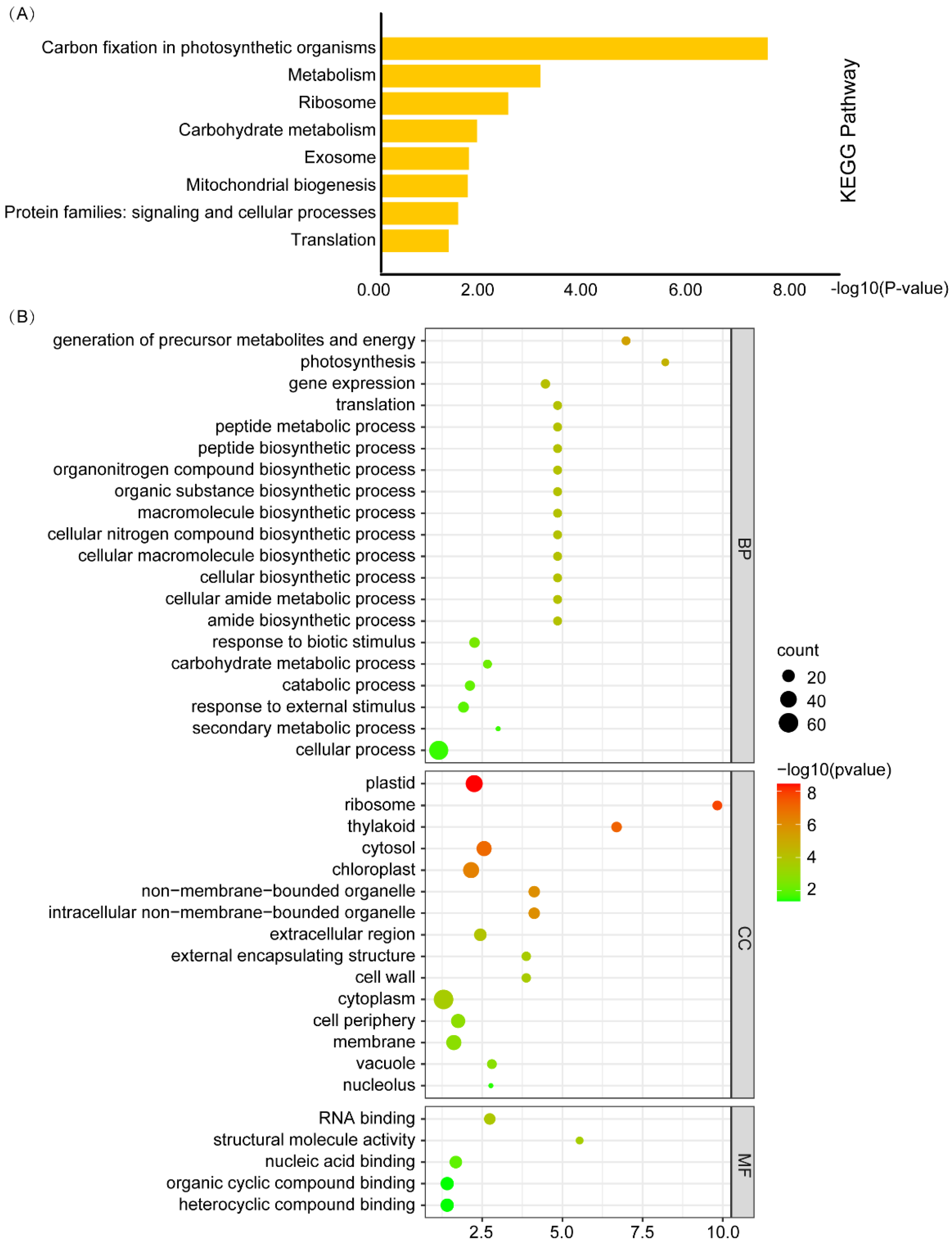
2.2. Screening of StSP6A Interactors
2.3. Yeast Two-Hybrid and Bimolecular Fluorescence Complementation Analysis of the Interactors of StSP6A In Vitro and In Vivo
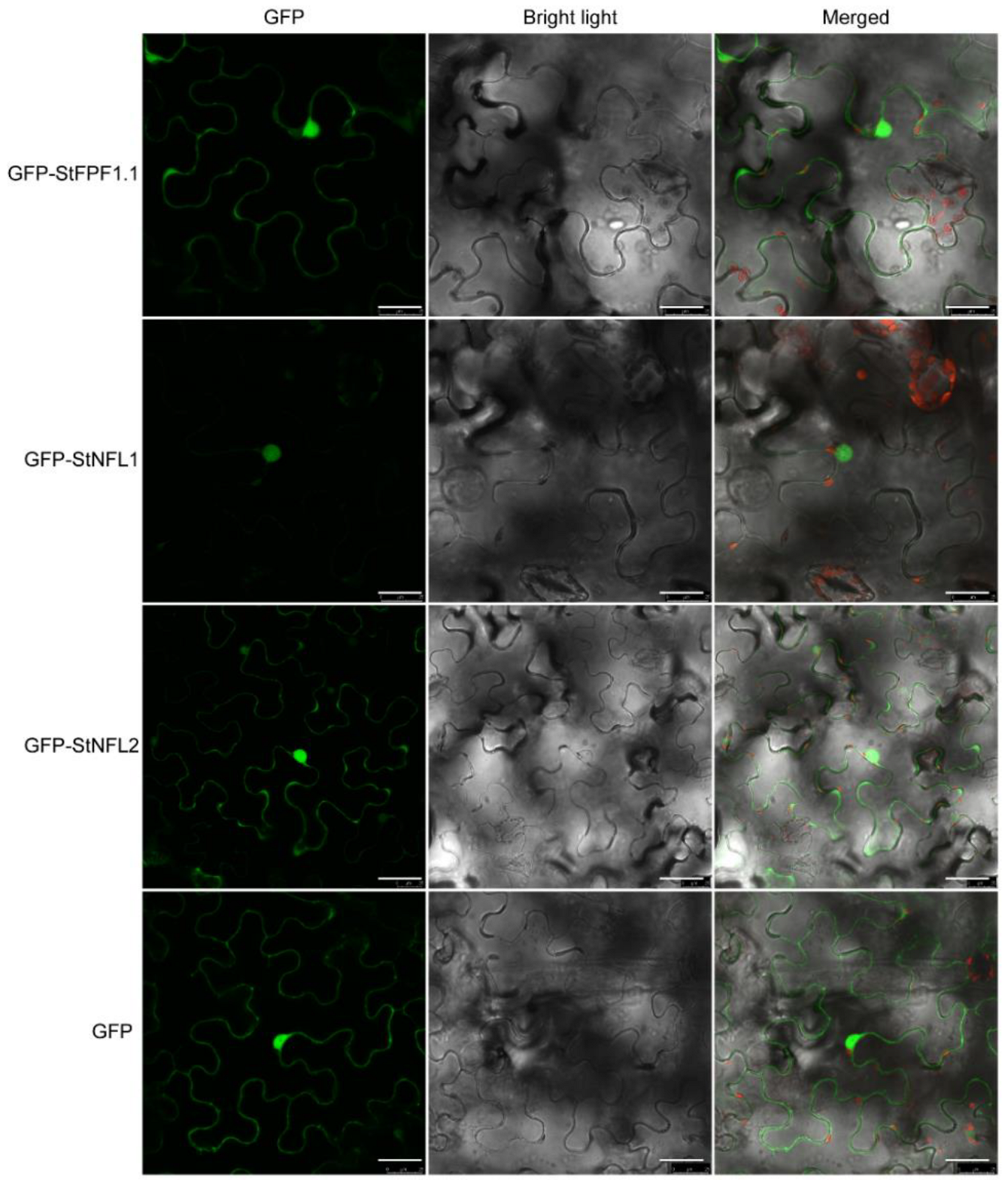
2.4. Subcellular Localization of the StFPF1.1, StNFL1, and StNFL2
2.5. Expression Patterns of StFPF1.1, StNFL1, StNFL2, and StSP6A in Response to Tuberization or Flowering
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and Quantitative Reverse Transcription PCR Analysis
4.3. Yeast Library Construction
4.4. BK-StSP6A Vector Construction
4.5. Y2H Library Screening and Y2H Assays
4.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis and Visualization
4.7. Bimolecular Fluorescence Complementation (BiFC) Assays
4.8. Subcellular Localization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by flowering locust. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Cruz-Oró, E.; Prat, S. Conserved function of flowering locust (FT) homologues as signals for storage organ differentiation. Curr. Opin. Plant Biol. 2015, 23, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Hastilestari, B.R.; Lorenz, J.; Reid, S.; Hofmann, J.; Pscheidt, D.; Sonnewald, U.; Sonnewald, S. Deciphering source and sink responses of potato plants (Solanum tuberosum L.) to elevated temperatures. Plant Cell Environ. 2018, 41, 2600–2616. [Google Scholar] [CrossRef] [PubMed]
- Lehretz, G.G.; Sonnewald, S.; Hornyik, C.; Corral, J.M.; Sonnewald, U. Post-transcriptional Regulation of FLOWERING LOCUS T Modulates Heat-Dependent Source-Sink Development in Potato. Curr. Biol. 2019, 29, 1614–1624.e3. [Google Scholar] [CrossRef]
- Zhou, T.; Song, B.; Liu, T.; Shen, Y.; Dong, L.; Jing, S.; Xie, C.; Liu, J. Phytochrome F plays critical roles in potato photoperiodic tuberization. Plant J. 2019, 98, 42–54. [Google Scholar] [CrossRef]
- Teo, C.-J.; Takahashi, K.; Shimizu, K.; Shimamoto, K.; Taoka, K.-I. Potato Tuber Induction is Regulated by Interactions Between Components of a Tuberigen Complex. Plant Cell Physiol. 2017, 58, 365–374. [Google Scholar] [CrossRef]
- Jing, S.; Sun, X.; Yu, L.; Wang, E.; Cheng, Z.; Liu, H.; Jiang, P.; Qin, J.; Begum, S.; Song, B. Transcription factor StABI5-like 1 binding to the flowering locust homologs promotes early maturity in potato. Plant Physiol. 2022, 189, 1677–1693. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.G.F.; Sonnewald, U.; Bachem, C.W. Source-Sink Regulation Is Mediated by Interaction of an FT Homolog with a SWEET Protein in Potato. Curr. Biol. 2019, 29, 1178–1186.e6. [Google Scholar] [CrossRef]
- Nicolas, M.; Torres-Perez, R.; Wahl, V.; Cruz-Oró, E.; Rodríguez-Buey, M.L.; Zamarreño, A.M.; Martín-Jouve, B.; García-Mina, J.M.; Oliveros, J.C.; Prat, S. Spatial control of potato tuberization by the TCP transcription factor branched1b. Nat. Plants 2022, 8, 281–294. [Google Scholar] [CrossRef]
- Plantenga, F.D.M.; Bergonzi, S.; Abelenda, J.A.; Bachem, C.W.; Visser, R.G.F.; Heuvelink, E.; Marcelis, L.F.M. The tuberization signal StSP6A represses flower bud development in potato. J. Exp. Bot. 2019, 70, 937–948. [Google Scholar] [CrossRef]
- Tang, D.; Jia, Y.; Zhang, J.; Li, H.; Cheng, L.; Wang, P.; Bao, Z.; Liu, Z.; Feng, S.; Zhu, X.; et al. Genome evolution and diversity of wild and cultivated potatoes. Nature 2022, 606, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, C.; Hou, X.; Xi, W.; Shen, L.; Tao, Z.; Wang, Y.; Yu, H. FTIP1 Is an Essential Regulator Required for Florigen Transport. PLoS Biol. 2012, 10, e1001313. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, C.; Teo, Z.W.N.; Zhang, B.; Yu, H. The MCTP-SNARE Complex Regulates Florigen Transport in Arabidopsis. Plant Cell 2019, 31, 2475–2490. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, L.; Shen, L.; Yu, H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2016, 2, 16075. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, Z.; Li, S.; Hou, M.; Zhou, Y.; Zhao, Y.; Li, G.; Zhao, H.; Ma, H. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018, 19, 726. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, X.; Li, K.; Song, S.; Jing, Y.; Lu, S. Screening and Identification of Candidate GUN1-Interacting Proteins in Arab. Thaliana. Int. J. Mol. Sci. 2021, 22, 11364. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Yang, L.; Zhuang, X.; Zhang, L.; Qi, H. Yeast Two-Hybrid Screen Identifies PKA-Rialpha Interacting Proteins during Mouse Spermiogenesis. Genes 2021, 12, 1941. [Google Scholar] [CrossRef]
- Guo, J.; Hu, Y.; Zhou, Y.; Zhu, Z.; Sun, Y.; Li, J.; Wu, R.; Miao, Y.; Sun, X. Profiling of the Receptor for Activated C Kinase 1a (RACK1a) interaction network in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2019, 520, 366–372. [Google Scholar] [CrossRef]
- Lakatos, L.; Klein, M.; Höfgen, R.; Bánfalvi, Z. Potato StubSNF1 interacts with StubGAL83: A plant protein kinase complex with yeast and mammalian counterparts. Plant J. 1999, 17, 569–574. [Google Scholar] [CrossRef]
- Begum, S.; Jing, S.; Yu, L.; Sun, X.; Wang, E.; Kawochar, A.; Qin, J.; Liu, J.; Song, B. Modulation of JA signalling reveals the influence of StJAZ1-like on tuber initiation and tuber bulking in potato. Plant J. 2022, 109, 952–964. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Xin, R.; Kim, D.-H.; Sung, S.; Lange, T.; Huq, E. NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 2016, 143, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Poirier, B.C.; Feldman, M.J.; Lange, B.M. bHLH093/NFL and bHLH061 are required for apical meristem function in Arabidopsis thaliana. Plant Signal. Behav. 2018, 13, e1486146. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, B.; Navarro, C.; Bijsterbosch, G.; Lange, T.; Prat, S.; Visser, R.G.F.; Bachem, C.W.B. StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. Plant J. 2007, 52, 362–373. [Google Scholar] [CrossRef]
- Xu, M.-L.; Jiang, J.-F.; Ge, L.; Xu, Y.-Y.; Chen, H.; Zhao, Y.; Bi, Y.-R.; Wen, J.-Q.; Chong, K. FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep. 2005, 24, 79–85. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, Q.; Xie, Z.; Yu, B.; Zeng, R.; Min, Q.; Huang, J. OsFPFL4 is Involved in the Root and Flower Development by Affecting Auxin Levels and ROS Accumulation in Rice (Oryza sativa). Rice 2020, 13, 2–15. [Google Scholar] [CrossRef]
- Kania, T.; Russenberger, D.; Peng, S.; Apel, K.; Melzer, S. FPF1 promotes flowering in Arabidopsis. Plant Cell 1997, 9, 1327–1338. [Google Scholar] [CrossRef]
- Ge, L.; Chen, H.; Jiang, J.-F.; Zhao, Y.; Xu, M.-L.; Xu, Y.-Y.; Tan, K.-H.; Xu, Z.-H.; Chong, K. Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol. 2004, 135, 1502–1513. [Google Scholar] [CrossRef]
- Sharma, P.; Lin, T.; Hannapel, D.J. Targets of the StBEL5 Transcription Factor Include the FT Ortholog StSP6A. Plant Physiol. 2016, 170, 310–324. [Google Scholar] [CrossRef]
- Lin, T.; Sharma, P.; Gonzalez, D.H.; Viola, I.L.; Hannapel, D.J. The Impact of the Long-Distance Transport of a BEL1-Like Messenger RNA on Development. Plant Physiol. 2013, 161, 760–772. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhang, X.; Zhang, J.; Li, F.; Xiang, J. Envelope Proteins of White Spot Syndrome Virus (WSSV) Interact with Litopenaeus vannamei Peritrophin-Like Protein (LvPT). PLoS ONE 2015, 10, e0144922. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zong, Y.; Liu, C.; Lu, R.; Huang, J.; Xu, J.; Wang, Y. Construction and identification of barley yeast two-hybrid library under salt stress. Acta Agric. Shanghai 2019, 35, 1–5. [Google Scholar]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schütze, K.; Batistic, O.; Weckermann, K.; Näke, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, E.; Liu, T.; Sun, X.; Jing, S.; Zhou, T.; Liu, T.; Song, B. Profiling of the Candidate Interacting Proteins of SELF-PRUNING 6A (SP6A) in Solanum tuberosum. Int. J. Mol. Sci. 2022, 23, 9126. https://doi.org/10.3390/ijms23169126
Wang E, Liu T, Sun X, Jing S, Zhou T, Liu T, Song B. Profiling of the Candidate Interacting Proteins of SELF-PRUNING 6A (SP6A) in Solanum tuberosum. International Journal of Molecular Sciences. 2022; 23(16):9126. https://doi.org/10.3390/ijms23169126
Chicago/Turabian StyleWang, Enshuang, Tengfei Liu, Xiaomeng Sun, Shenglin Jing, Tingting Zhou, Tiantian Liu, and Botao Song. 2022. "Profiling of the Candidate Interacting Proteins of SELF-PRUNING 6A (SP6A) in Solanum tuberosum" International Journal of Molecular Sciences 23, no. 16: 9126. https://doi.org/10.3390/ijms23169126





