Essential Acidovorax citrulli Virulence Gene hrpE Activates Host Immune Response against Pathogen
Abstract
:1. Introduction
2. Results
2.1. Confirmation of the Mutant and Complementary Strains
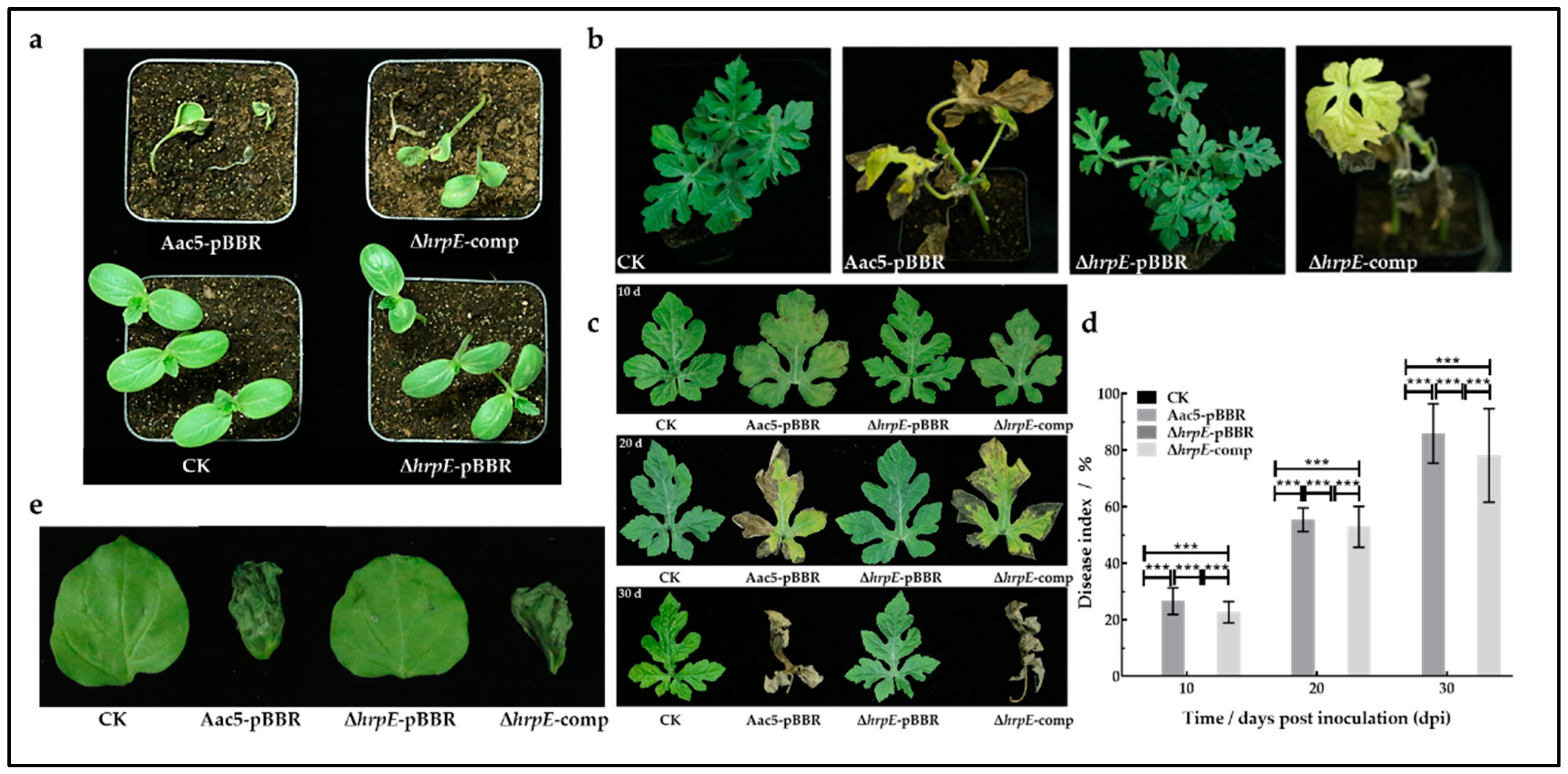
2.2. HrpE Is Essential for A. citrulli Pathogenicity
2.3. A. citrulli HrpE Is Essential for HR on Non-Host Plants
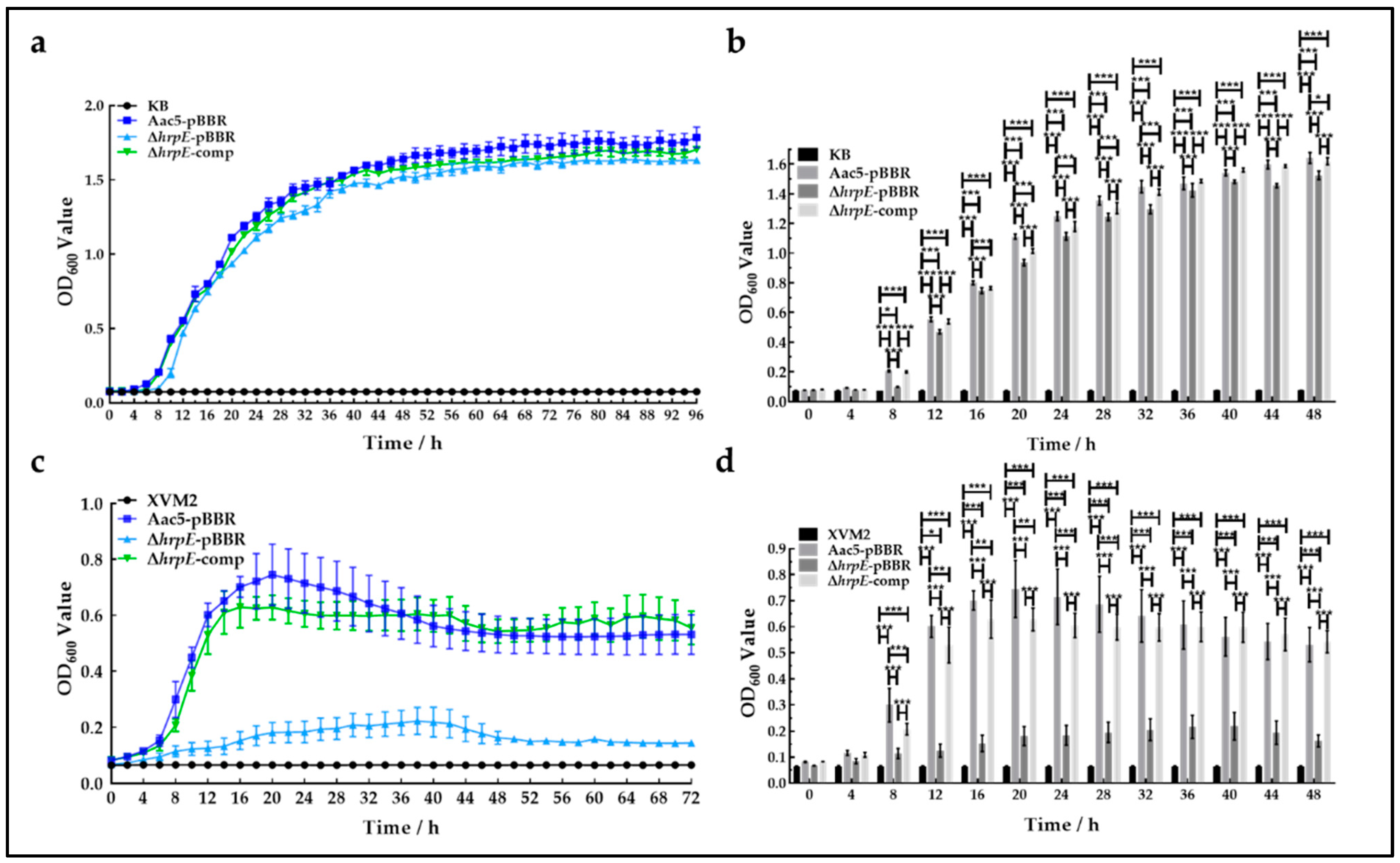
2.4. HrpE Reduces the Growth Ability of A. citrulli In Vitro and In Vivo
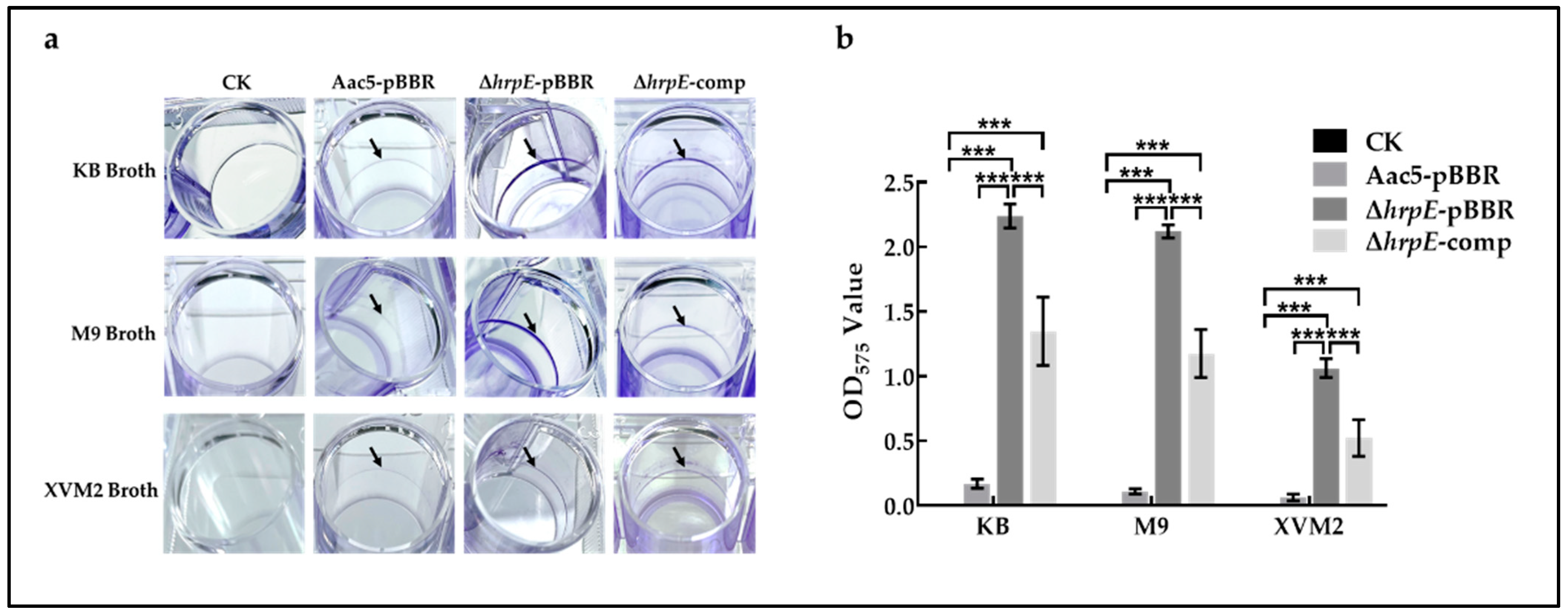
2.5. Deletion of hrpE Enhanced the Biofilm Formation Ability of A. citrulli
2.6. Deletion of hrpE Reduced the Swimming Motility and Twitching Motility of A. citrulli
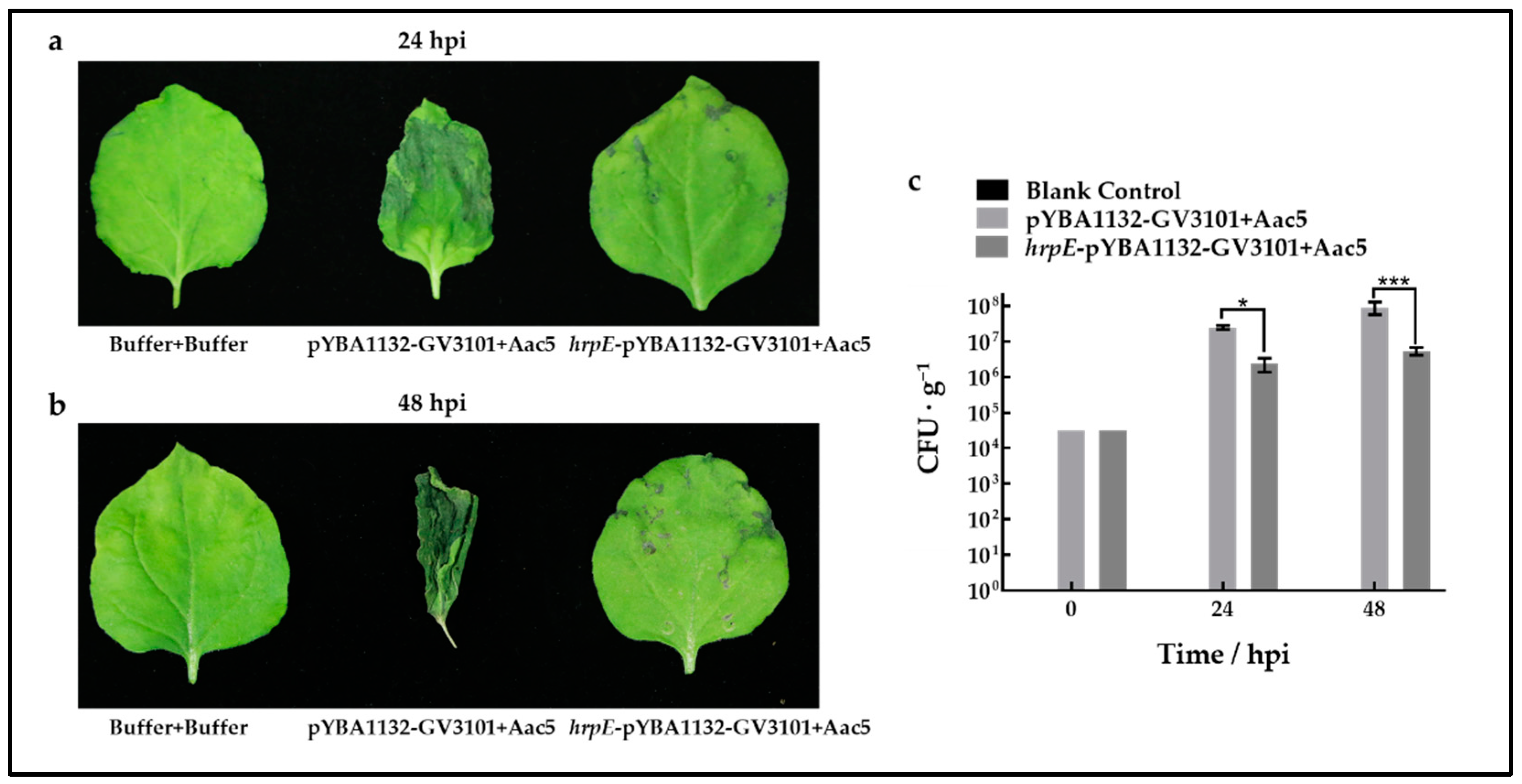
2.7. Pre-Treatment with HrpE Helps Host Plants Resist A. citrulli Infection
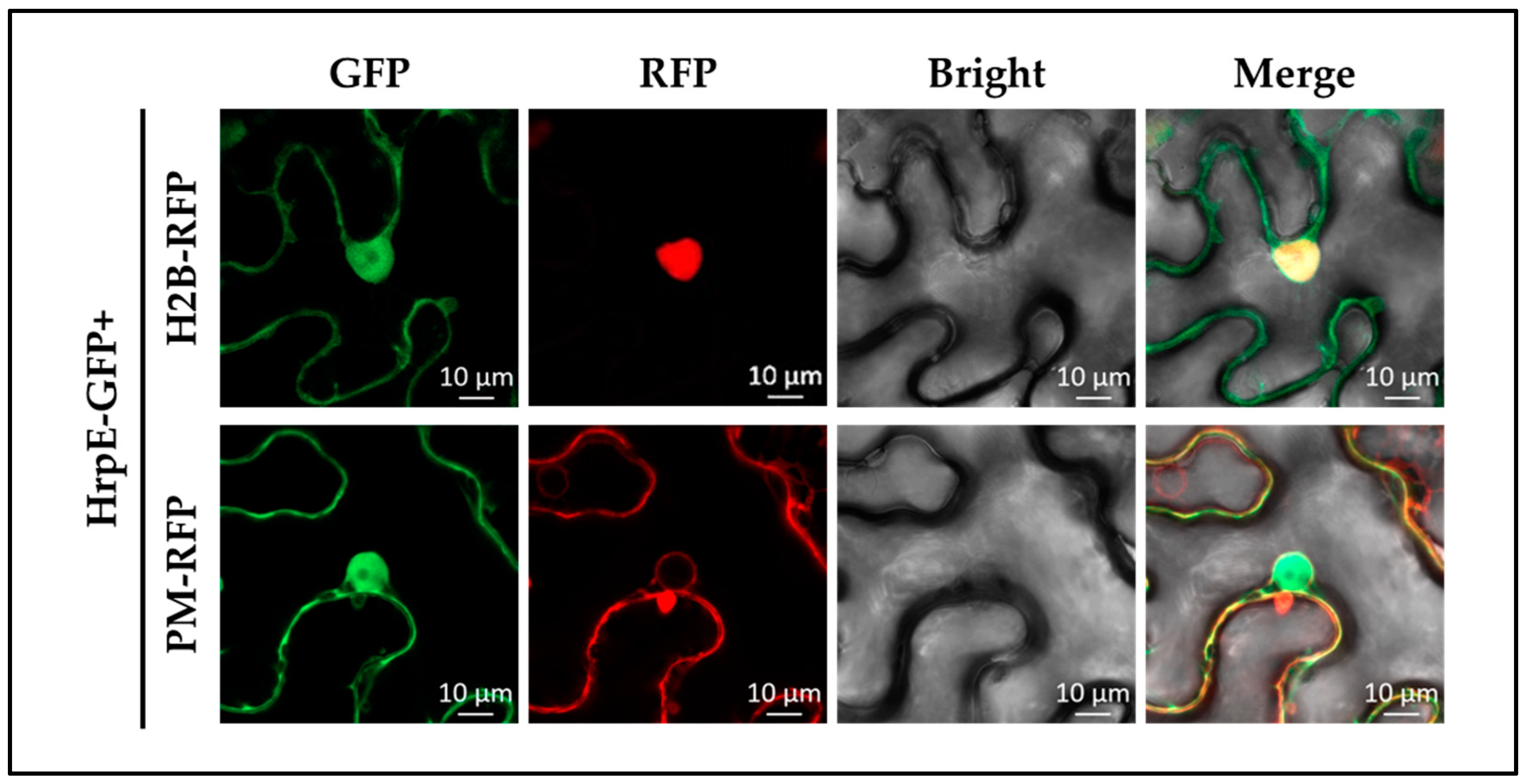
2.8. HrpE Localized at Cytomembrane and Nuclear in Hosts
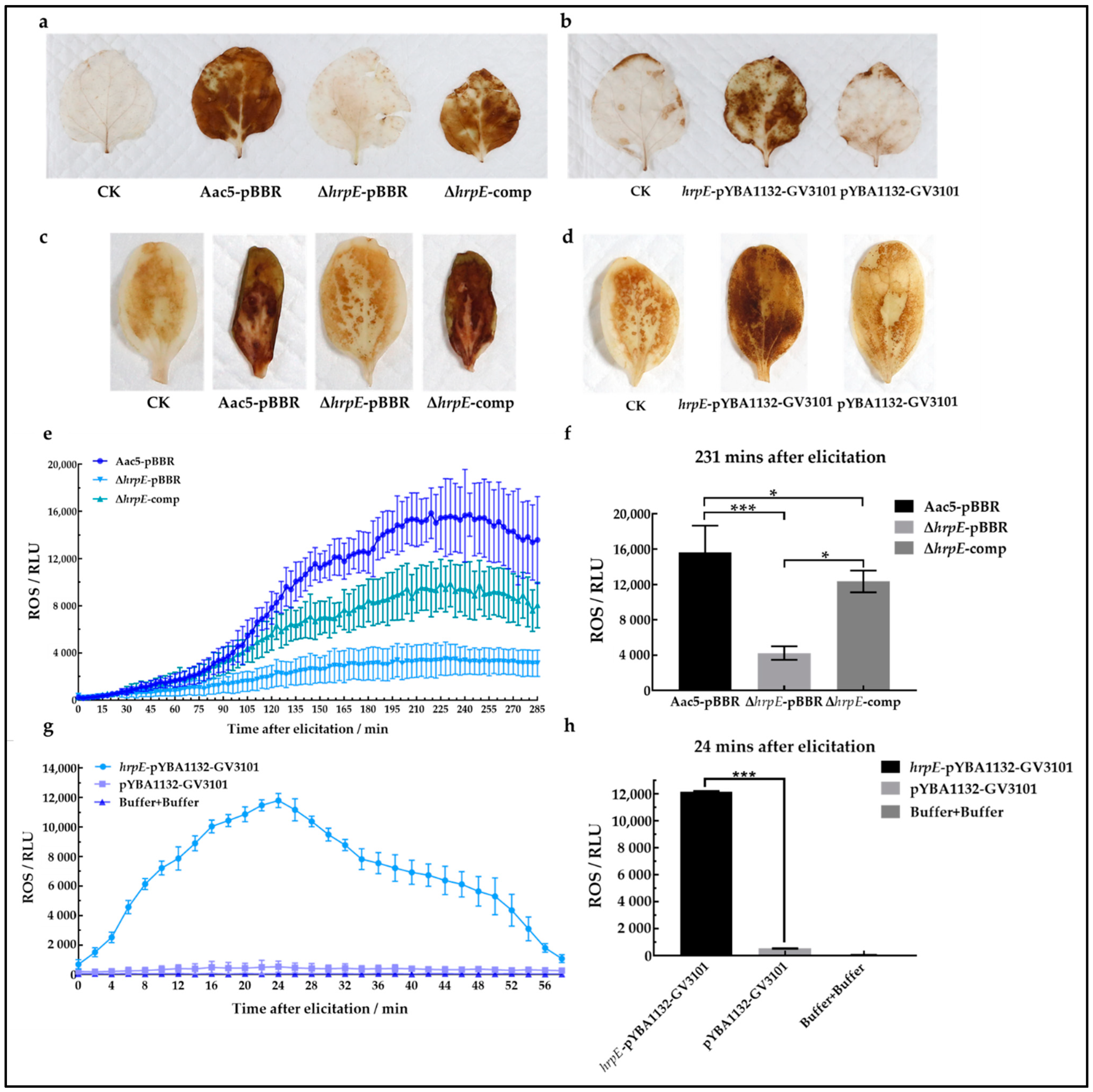
2.9. HrpE Induces ROS Burst in Hosts
2.10. HrpE Induced Callose Deposition in Hosts
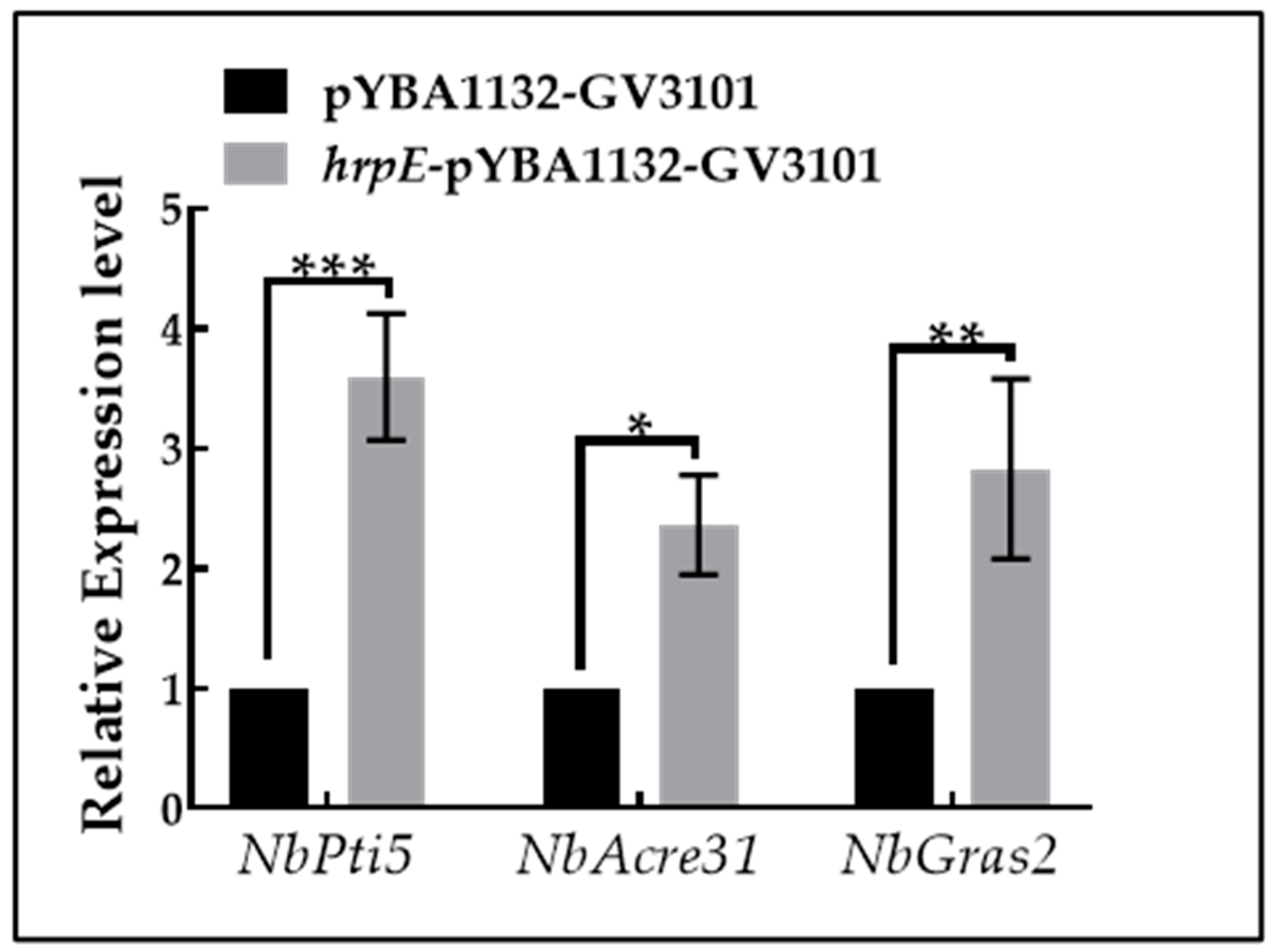
2.11. Expression of PTI-Marker Genes Induced by HrpE in Tobacco
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, Growth Conditions, and Primer Design
4.2. Plant Materials and Culture Conditions
4.3. Construction of the hrpE Mutant and Its Complement Strain
4.4. Pathogenicity Assays
4.4.1. Seed-to Seedling Transmission Assays
4.4.2. Watermelon Spray Inoculation Assays
4.4.3. N. benthamiana Inoculation Assays
4.5. HR on Non-Host Tobacco
4.5.1. Qualitative Determination
4.5.2. Quantitative Determination
4.6. Growth Ability In Vivo and In Vitro
4.6.1. Growth Ability In Vitro
4.6.2. Growth Ability In Vivo
4.7. Biofilm Formation Assay
4.8. Swimming and Twitching Motility Assays
4.8.1. Swimming Motility Assay
4.8.2. Twitching Motility Assay
4.9. Analysis of Aac5 Growth in Watermelon and Tobacco Leaves Pre-Treated with HrpE
4.9.1. Construction of Transient Expression Strains
4.9.2. Assays on N. benthamiana
4.9.3. Watermelon Seedling Assays
4.10. Subcellular Localization of HrpE in N. benthamiana
4.11. ROS Production Assays
4.11.1. ROS Production in N. benthamiana Leaves
- Qualitative determination of ROS induced by protein
- Qualitative determination of ROS induced by strains
- Quantitative determination of ROS induced by protein
- Quantitative determination of ROS induced by strains
4.11.2. ROS Production in Watermelon Cotyledons
- Induced by protein
- Induced by strains
4.12. Callose Staining
4.12.1. Strain Induction
4.12.2. Protein Induction
4.13. DNA/RNA Extraction and RT-qPCR
4.14. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webb, R.E.; Goth, R.W. A seedborne bacterium isolated from watermelon. Plant Dis. Report. 1965, 49, 818–821. [Google Scholar]
- Schaad, N.W.; Postnikova, E.; Sechler, A.; Claflin, L.E.; Vidaver, A.K.; Jones, J.B.; Agarkova, I.; Ignatov, A.; Dickstein, E.; Ramundo, B.A. Reclassification of subspecies of Acidovorax avenae as A. avenae (Manns 1905) emend., A. cattleyae (Pavarino, 1911) comb. nov., A. citrulli (Schaad et al., 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 2008, 31, 434–446. [Google Scholar] [CrossRef]
- Bahar, O.; Kritzman, G.; Burdman, S. Bacterial fruit blotch of melon: Screens for disease tolerance and role of seed transmission in pathogenicity. Eur. J. Plant Pathol. 2009, 123, 71–83. [Google Scholar] [CrossRef]
- Burdman, S.; Walcott, R.R. Acidovorax citrulli: Generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 2012, 13, 805–815. [Google Scholar] [CrossRef]
- Tian, Y.L.; Zhao, Y.Q.; Zhou, J.J.; Sun, T.; Luo, X.; Kurowski, C.; Gong, W.R.; Hu, B.S.; Walcott, R.R. Prevalence of Acidovorax citrulli in commercial Cucurbit seedlots during 2010-2018 in China. Plant Dis. 2020, 104, 255–259. [Google Scholar] [CrossRef]
- Walcott, R.R.; Langston, J.D.B.; Sanders, F.H.; Gitaitis, R.D. Investigating intraspecific variation of Acidovorax avenae subsp. citrulli using DNA fingerprinting and whole cell fatty acid analysis. Phytopathology 2000, 90, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Walcott, R.R.; Fessehaie, A.; Castro, A.C. Differences in Pathogenicity between two Genetically Distinct Groups of Acidovorax avenae subsp. citrulli on Cucurbit Hosts. J. Phytopathol. 2004, 152, 277–285. [Google Scholar] [CrossRef]
- Yan, S.S.; Yang, Y.W.; Wang, T.L.; Zhao, T.C.; Schaad, N.W. Genetic diversity analysis of Acidovorax citrulli in China. Eur. J. Plant Pathol. 2013, 136, 171–181. [Google Scholar] [CrossRef]
- Zivanovic, M.; Walcott, R.R. Further characterization of genetically distinct groups of Acidovorax citrulli Strains. Phytopathology 2017, 107, 29–35. [Google Scholar] [CrossRef]
- Islam, M.R.; Hossain, M.R.; Jesse, M.I.; Jung, H.; Kim, H.; Park, J.I.; Nou, I.S. Development of molecular marker linked with bacterial fruit blotch resistance in melon (Cucumis melo L.). Genes 2020, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Hossain, M.R.; Jesse, M.I.; Jung, H.; Kim, H.; Park, J.I.; Nou, I.S. Characterization, identification, and expression profiling of genome-wide R-genes in melon and their putative roles in bacterial fruit blotch resistance. BMC Genet. 2020, 21, 11872–11877. [Google Scholar] [CrossRef]
- Emanuel, F.A.; Claudeana, S.C.; Elizabeth, R.A.; Marco, A.S.G.; Glauber, H.S.N.; Elineide, B.S. New sources of melon accessions with resistance to bacterial fruit blotch at different phenological stages of melon growth and to multiple strains of Acidovorax citrulli. Euphytica 2021, 217, 79. [Google Scholar] [CrossRef]
- Gu, X.F.; Zeng, Q.C.; Wang, Y.; Li, J.S.; Wang, Q. Comprehensive genomic analysis of Bacillus subtilis 9407 reveals its biocontrol potential against bacterial fruit blotch. Phytopathol. Res. 2021, 3, 4. [Google Scholar] [CrossRef]
- Shin, H.J.; Kim, H.; Beuchat, L.R.; Ryu, J.H. Antimicrobial activities of organic acid vapors against Acidovorax citrulli, Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes on Cucurbitaceae seeds. Food Microbiol. 2020, 92, 103569. [Google Scholar] [CrossRef]
- Perrino, E.V.; Ladisa, G.; Calabrese, G. Flora and plant genetic resources of ancient olive groves of Apulia (Southern Italy). Genet. Resour. Crop Evol. 2014, 61, 23–53. [Google Scholar] [CrossRef]
- Jones, J.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Wang, Y.; Rory, N.P.; Thorsten, N.; Wang, Y.C. Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 2022, 20, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.H.; Jiang, Z.Y.; Bi, G.Z.; Nomura, K.; Liu, M.H.; Wang, Y.P.; Cai, B.Y.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2020, 592, 105–109. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Chakravarthy, S.; Velásquez, A.C.; McLane, H.L.; Zeng, L.; Nakayashiki, H.; Duck, H.P.; Alan, C.; Gregory, B.M. Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact. 2010, 23, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.T.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2020, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Portaliou, A.G.; Tsolis, K.C.; Loos, M.S.; Zorzini, V.; Economou, A. Type III secretion: Building and operating a remarkable nanomachine. Trends Biochem. Sci. 2016, 41, 175–189. [Google Scholar] [CrossRef]
- Lindgren, P.B. The role of hrp genes during plant-bacterial interactions. Annu. Rev. Phytopathol. 1997, 35, 129–152. [Google Scholar] [CrossRef] [PubMed]
- Tampakaki, A.P.; Skandalis, N.; Gazi, A.D.; Bastaki, M.N.; Sarris, P.F.; Charova, S.N.; Kokkinidis, M.; Panopoulos, N.J. Playing the “Harp”: Evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 2010, 48, 347–370. [Google Scholar] [CrossRef]
- Weber, E.; Koebnik, R. Domain structure of HrpE, the Hrp pilus subunit of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 2005, 187, 6175–6186. [Google Scholar] [CrossRef]
- Weber, E.; Tuula, O.R.; Huguet, E.; Hause, G.; Romantschuk, M.; Korhonen, T.K.; Bonas, U.; Koebnik, R.; Martin, L.U.; Institut, F.G. The type III-dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 2005, 187, 2458–2468. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.L.; Hu, W.Q.; Brown, I.; McGhee, G.; Hart, P.; Jones, A.L.; He, S.Y. Visualization of secreted Hrp and Avr proteins along the Hrp pilus during type III secretion in Erwinia amylovora and Pseudomonas syringae. Mol. Microbiol. 2001, 40, 1129–1139. [Google Scholar] [CrossRef]
- Cui, Y.P.; Zou, L.F.; Zou, H.S.; Li, Y.R.; Muhammad, Z.; Chen, G.Y. HrpE3 is a type III effector protein required for full virulence of Xanthomonas oryzae pv. oryzicola in rice. Mol. Plant Pathol. 2013, 14, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Natalia, G.; Cecilia, V.V.; Germán, G.S.; Ainelén, P.; Jorgelina, O. HrpE, the major component of the Xanthomonas type three protein secretion pilus, elicits plant immunity responses. Sci. Rep. 2018, 8, 9842–9855. [Google Scholar] [CrossRef]
- Sheikh, T.M.M.; Zhang, L.Y.; Zubair, M.; Hanif, A.; Li, P.; Farzand, A.; Ali, H.; Bilal, M.S.; Hu, Y.Q.; Chen, X.C.; et al. The type III accessory protein HrpE of Xanthomonas oryzae pv. oryzae surpasses the secretion role, and enhances plant resistance and photosynthesis. Microorganisms 2019, 7, 572. [Google Scholar] [CrossRef]
- Sheikh, T.M.M.; Haider, M.S.; Hanif, A.; Ali, H.; Khan, A.R.; Li, P.; Zubair, M.; Farzand, A.; Tariq, L.; Ouyang, X.; et al. N-terminal HrpE from Xanthomonas oryzae pv. oryzae mediates the regulation of growth and photosynthesis in rice. Plant Growth Regul. 2022, 96, 383–396. [Google Scholar] [CrossRef]
- Weber, E.; Koebnik, R. Positive selection of the Hrp pilin HrpE of the plant pathogen Xanthomonas. J. Bacterial. 2006, 188, 1405–1410. [Google Scholar] [CrossRef]
- Wei, Z.M.; Laby, R.J.; Zumoff, C.H.; Bauer, D.W.; He, S.Y.; Collmer, A.; Beer, S.V. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 1992, 257, 85–87. [Google Scholar] [CrossRef]
- Dong, H.S.; Delaney, T.P.; Bauer, D.W.; Beer, S.V. Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J. 1999, 20, 207–215. [Google Scholar] [CrossRef]
- Krause, M.; Durner, J. Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol. Plant Microbe Interact. 2004, 17, 131–139. [Google Scholar] [CrossRef]
- Peng, J.L.; Dong, H.S.; Dong, H.P.; Delaney, T.P.; Bonasera, J.M.; Beer, S.V. Harpin-elicited hypersensitive cell death and pathogen resistance require the NDR1 and EDS1 genes. Physiol. Mol. Plant Pathol. 2003, 62, 317–326. [Google Scholar] [CrossRef]
- Choi, M.S.; Kim, W.; Lee, C.; Oh, C. Harpins, multi-functional proteins secreted by gram-negative plant pathogenic bacteria. Mol. Plant Microbe Interact. 2013, 26, 1115–1122. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zhao, M.; Yan, J.P.; Yang, L.L.; Yang, Y.W.; Guan, W.; Walcott, R.R.; Zhao, T.C. Involvement of hrpX and hrpG in the virulence of Acidovorax citrulli strain Aac5, causal agent of bacterial fruit blotch in cucurbits. Front. Microbiol. 2018, 9, 507–519. [Google Scholar] [CrossRef]
- Yan, W.R.; Wang, T.L.; Yang, Y.W.; Dai, L.Y.; Zhao, T.C. Biological function analysis of hrcN gene of Acidovorax citrulli. Acta Phytopathol. Sinica 2015, 45, 33–40. [Google Scholar] [CrossRef]
- Yu, C.; Wu, L.N.; Zhao, L.P.; Zhang, X.X.; Yang, Y.W.; Guan, W.; Zhao, T.C. Functional analysis of hrcQ in type III secretion system of Acidovorax citrulli. Plant Protect. 2019, 45, 48–56. [Google Scholar] [CrossRef]
- Eckshtain-Levi, N.; Munitz, T.; Živanović, M.; Traore, S.M.; Spröer, C.; Zhao, B.Y.; Welbaum, G.; Walcott, R.R.; Sikorski, J.; Burdman, S. Comparative analysis of type III secreted effector genes reflects divergence of Acidovorax citrulli strains into three distinct lineages. Phytopathology 2014, 104, 1152–1162. [Google Scholar] [CrossRef]
- Jimenez-Guerrero, I.; Perez-Montano, F.; Mateus, D.S.G.; Wagner, N.; Shkedy, D.; Zhao, M.; Pizarro, L.; Bar, M.; Walcott, R.R.; Sessa, G.; et al. Show me your secret(ed) weapons: A multifaceted approach reveals a wide arsenal of type III-secreted effectors in the cucurbit pathogenic bacterium Acidovorax citrulli and novel effectors in the Acidovorax genus. Mol. Plant Pathol. 2020, 21, 17–37. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zhao, M.; Jiang, J.; Yang, L.L.; Yang, Y.W.; Yang, S.S.; Walcott, R.R.; Qiu, D.W.; Zhao, T.C. Identification and functional analysis of AopN, an Acidovorax citrulli effector that induces programmed cell death in plants. Int. J. Mol. Sci. 2020, 21, 6050. [Google Scholar] [CrossRef]
- Zhang, X.X.; Yang, Y.W.; Zhao, M.; Yang, L.L.; Jiang, J.; Walcott, R.R.; Yang, S.S.; Zhao, T.C. Acidovorax citrulli type III effector AopP suppresses plant immunity by targeting the watermelon transcription factor WRKY6. Front. Plant Sci. 2020, 11, 579218. [Google Scholar] [CrossRef]
- Traore, S.M.; Eckshtain-Levi, N.; Miao, J.M.; Sparks, A.C.; Wang, Z.B.; Wang, K.R.; Li, Q.; Burdman, S.; Walcott, R.R.; Welbaum, G.E.; et al. Nicotiana species as surrogate host for studying the pathogenicity of Acidovorax citrulli, the causal agent of bacterial fruit blotch of cucurbits. Mol. Plant Pathol. 2019, 20, 800–814. [Google Scholar] [CrossRef]
- Bluhm, B.H.; Zhao, X.; Flaherty, J.E.; Xu, J.R.; Dunkle, L.D. Ras2 regulates growth and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interac. 2007, 20, 627–636. [Google Scholar] [CrossRef]
- Wengelnik, K.; Marie, C.; Marjorie, R.; Ulla, B. Expression and localization of hrpa1, a protein of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 1996, 178, 1061. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef]
- Broek, A.V.; Vanderleyden, J. The role of bacterial motility, chemotaxis, and attachment in bacteria-plant interactions. Mol. Plant Microbe Interact. 1995, 8, 800–810. [Google Scholar] [CrossRef]
- Torres, M.A. ROS in biotic interactions. Physiol. Plantarum 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Estrella, L.; Victoria, P.; Jérôme, R.; Victor, F.; Brigitte, M.M.; Jurriaan, T. Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef]
- Wang, Y.P.; Zou, L.f.; Zhou, D.; Chen, G.Y. Key roles of hrpE gene of Xanthomonas oryzae pv. oryzicola in formation of Hrp pilus and pathogenicity in rice. Acta Phytopathol. Sinica 2009, 39, 392–398. [Google Scholar]
- Wei, W.; Plovanich-Jones, A.; Deng, W.L.; Jin, Q.L.; Collmer, A.; Huang, H.C.; He, S.Y. The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 2247–2252. [Google Scholar] [CrossRef]
- Martin, R.; Elina, R.; Suvi, T. Hrp pilus—Reaching through the plant cell wall. Eur. J. Plant Pathol. 2001, 107, 153–160. [Google Scholar] [CrossRef]
- Jin, Q.L.; He, S.Y. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 2001, 294, 2556–2558. [Google Scholar] [CrossRef]
- He, S.Y.; Jin, Q.L. The Hrp pilus: Learning from flagella. Curr. Opin. Microbiol. 2003, 6, 15–19. [Google Scholar] [CrossRef]
- Block, A.; Li, G.Y.; Fu, Z.Q.; Alfano, J.R. Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 2008, 11, 396–403. [Google Scholar] [CrossRef]
- Galan, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 2014, 68, 415–438. [Google Scholar] [CrossRef]
- Hu, Y.Z.; Ding, Y.X.; Cai, B.Y.; Qin, X.H.; Wu, J.N.; Yuan, M.H.; Wan, S.W.; Zhao, Y.; Xin, X.F. Bacterial effectors manipulate plant abscisic acid signaling for creation of an aqueous apoplast. Cell Host Microbe 2022, 30, 518–529.e6. [Google Scholar] [CrossRef]
- Guan, W.; Wang, T.L.; Huang, Q.; Zhao, M.; Tian, E.Y.; Liu, Y.F.; Liu, B.; Yang, Y.W.; Zhao, T.C. Transcriptomic and functional analyses reveal roles of AclR, a LuxR-type global regular, in regulating motility and virulence of Acidovorax citrulli. Mol. Plant Microbe Interact. 2021, 34, 952–961. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379. [Google Scholar] [CrossRef]
- Zhou, J.M.; Zhang, Y.L. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Jose, O.A.B.; Daudi, A.; Butt, V.S.; Bolwell, G.P. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 2012, 236, 765–779. [Google Scholar] [CrossRef]
- Li, P.; Zhao, L.L.; Qi, F.; Htwe, N.M.P.S.; Li, Q.Y.; Zhang, D.W.; Lin, F.C.; Shangguan, K.K.; Liang, Y. The receptor-like cytoplasmic kinase PIPK regulates broad-spectrum ROS signaling in multiple layers of plant immune system. Mol. Plant 2021, 14, 1652–1667. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Kvitko, B.H.; Collmer, A. Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. Methods Mol. Biol. 2011, 712, 109–128. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Wang, T.L.; Guan, W.; Huang, Q.; Yang, Y.W.; Yan, W.R.; Sun, B.X.; Zhao, T.C. Quorum-sensing contributes to virulence, twitching motility, seed attachment and biofilm formation in the wild type strain Aac-5 of Acidovorax citrulli. Microb. Pathogenesis 2016, 100, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.G.; Jiang, W.J.; Ni, X.Y.; Lin, M.; Zhang, W.; Tian, G.Z. Multiplication of Acidovorax citrulli in planta during infection of melon seedlings requires the ability to synthesize leucine. Plant Pathol. 2014, 63, 784–791. [Google Scholar] [CrossRef]
- Tang, D.J.; Chen, X.L.; Jia, Y.; Liang, Y.W.; He, Y.P.; Lu, T.T.; Zhu, C.R.; Han, B.; An, S.Q.; Tang, J.L. Genome-wide screen and functional analysis in Xanthomonas reveal a large number of mRNA-derived sRNAs, including the novel RsmA-sequester RsmU. Mol. Plant Pathol. 2020, 21, 1573–1590. [Google Scholar] [CrossRef]
- Guan, W.; Wang, T.L.; Huang, Q.; Tian, E.Y.; Liu, B.; Yang, Y.W.; Zhao, T.C. A LuxR-type regulator, AcrR, regulates flagellar assembly and contributes to virulence, motility, biofilm formation, and growth ability of Acidovorax citrulli. Mol. Plant Pathol. 2020, 21, 489–501. [Google Scholar] [CrossRef]
- Zhao, Y.D.; Zhu, X.Y.; Xu, G.J.; Wang, X.L. OsEIL6 is involved in regulating rice resistance to Magnaporthe oryzae. Plant Protect. 2020, 4, 12–18. [Google Scholar] [CrossRef]
- Gelvin, S.B. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, V.V.; Kapoor, A.; Zhang, K.L.; Zhu, J.J.; Zhou, T.; Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 2007, 447, 735–738. [Google Scholar] [CrossRef] [PubMed]
- Priller, J.P.; Reid, S.; Konein, P.; Dietrich, P.; Sonnewald, S. The Xanthomonas campestris pv. vesicatoria type-3 effector XopB inhibits plant defence responses by interfering with ROS production. PLoS ONE 2016, 11, e0159107. [Google Scholar] [CrossRef]
- Sang, Y.; Macho, A.P. Analysis of PAMP-triggered ROS burst in plant immunity. Methods Mol. Biol. 2017, 1578, 143–153. [Google Scholar] [CrossRef]
- Hirai, H.; Furukawa, T.; Katsuragi, Y.; Che, F.S. Purification of flagellin from Acidovorax avenae and analysis of plant immune responses induced by the purified flagellin. Bio-Protocol 2016, 6. [Google Scholar] [CrossRef]
- Kim, H.S.; Thammarat, P.; Lommel, S.A.; Hogan, C.S.; Charkowski, A.O. Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant-microbe interactions. Mol. Plant Microbe Interact. 2011, 24, 773–786. [Google Scholar] [CrossRef]
- Chen, C.; Liu, S.; Liu, Q.; Niu, J.; Liu, P.; Zhao, J.; Jian, H. An ANNEXIN-Like protein from the cereal cyst nematode Heterodera avenae suppresses plant defense. PLoS ONE 2015, 10, e0122256. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, W.; Zhao, M.; Fei, N.; Yang, L.; Qiao, P.; Walcott, R.; Yang, Y.; Zhao, T. Essential Acidovorax citrulli Virulence Gene hrpE Activates Host Immune Response against Pathogen. Int. J. Mol. Sci. 2022, 23, 9144. https://doi.org/10.3390/ijms23169144
Ji W, Zhao M, Fei N, Yang L, Qiao P, Walcott R, Yang Y, Zhao T. Essential Acidovorax citrulli Virulence Gene hrpE Activates Host Immune Response against Pathogen. International Journal of Molecular Sciences. 2022; 23(16):9144. https://doi.org/10.3390/ijms23169144
Chicago/Turabian StyleJi, Weiqin, Mei Zhao, Nuoya Fei, Linlin Yang, Pei Qiao, Ron Walcott, Yuwen Yang, and Tingchang Zhao. 2022. "Essential Acidovorax citrulli Virulence Gene hrpE Activates Host Immune Response against Pathogen" International Journal of Molecular Sciences 23, no. 16: 9144. https://doi.org/10.3390/ijms23169144
APA StyleJi, W., Zhao, M., Fei, N., Yang, L., Qiao, P., Walcott, R., Yang, Y., & Zhao, T. (2022). Essential Acidovorax citrulli Virulence Gene hrpE Activates Host Immune Response against Pathogen. International Journal of Molecular Sciences, 23(16), 9144. https://doi.org/10.3390/ijms23169144





