Cultured Myoblasts Derived from Rat Soleus Muscle Show Altered Regulation of Proliferation and Myogenesis during the Course of Mechanical Unloading
Abstract
:1. Introduction
2. Results
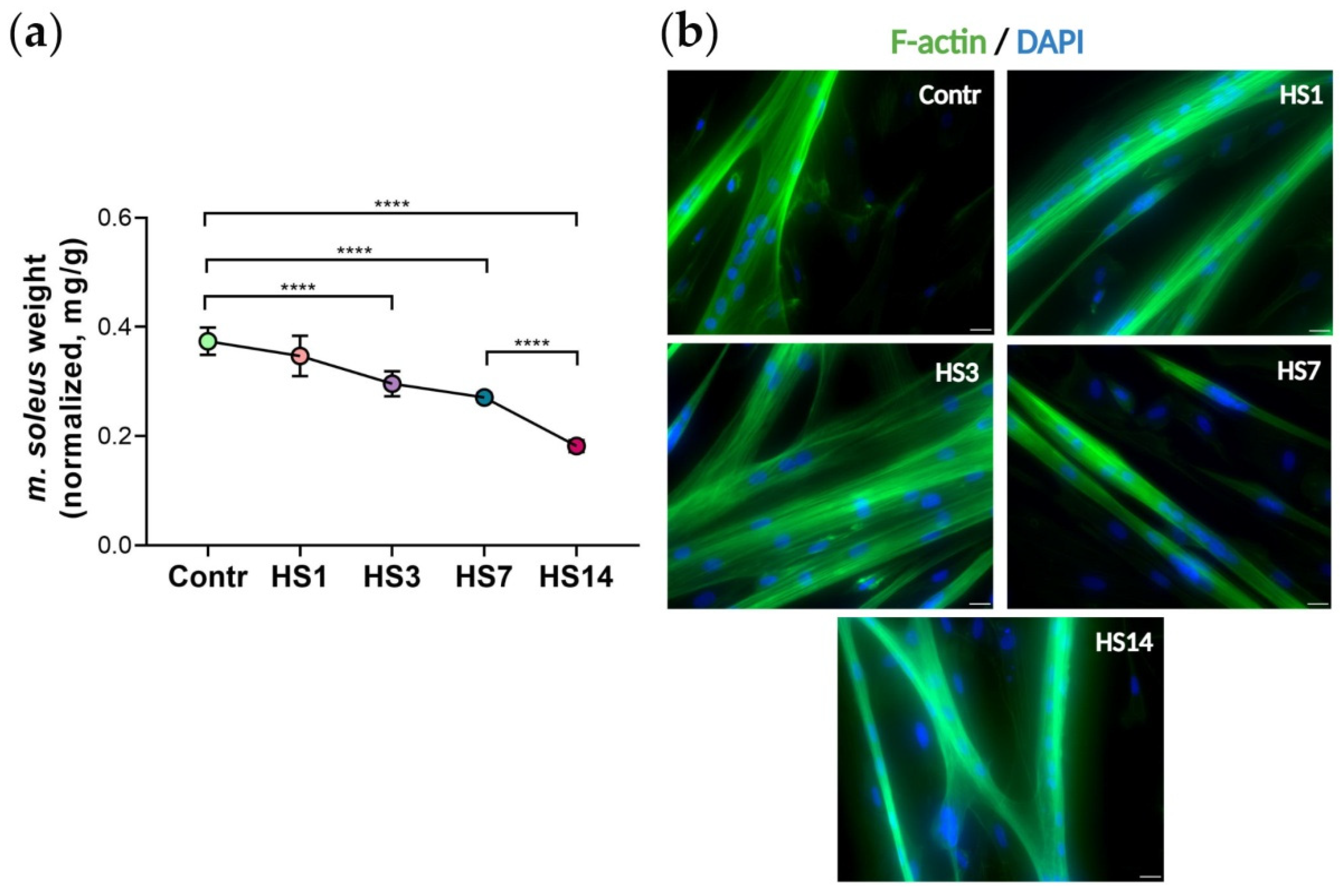
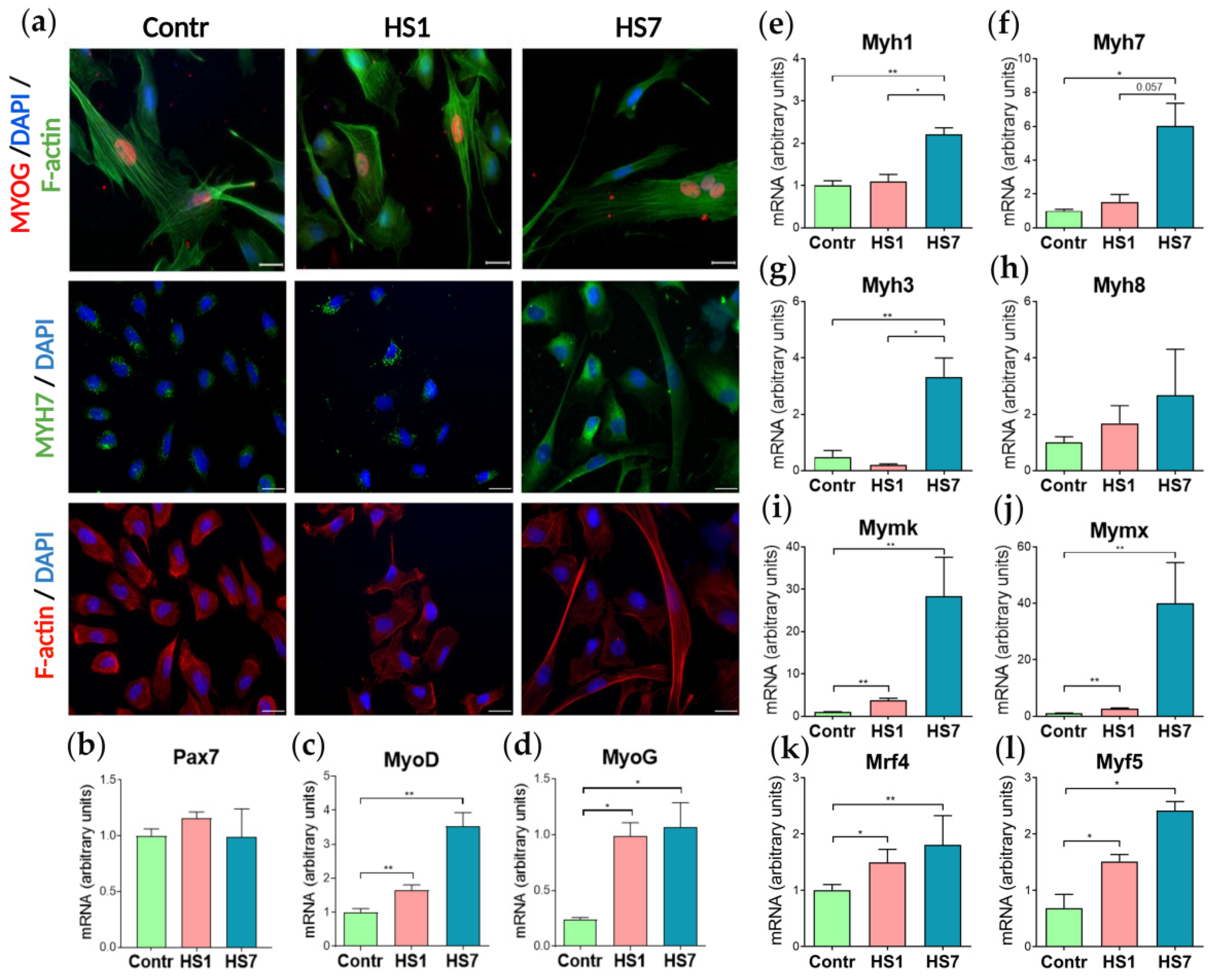
2.1. Soleus Muscle Weight and Myogenic Differentiation of Myoblasts
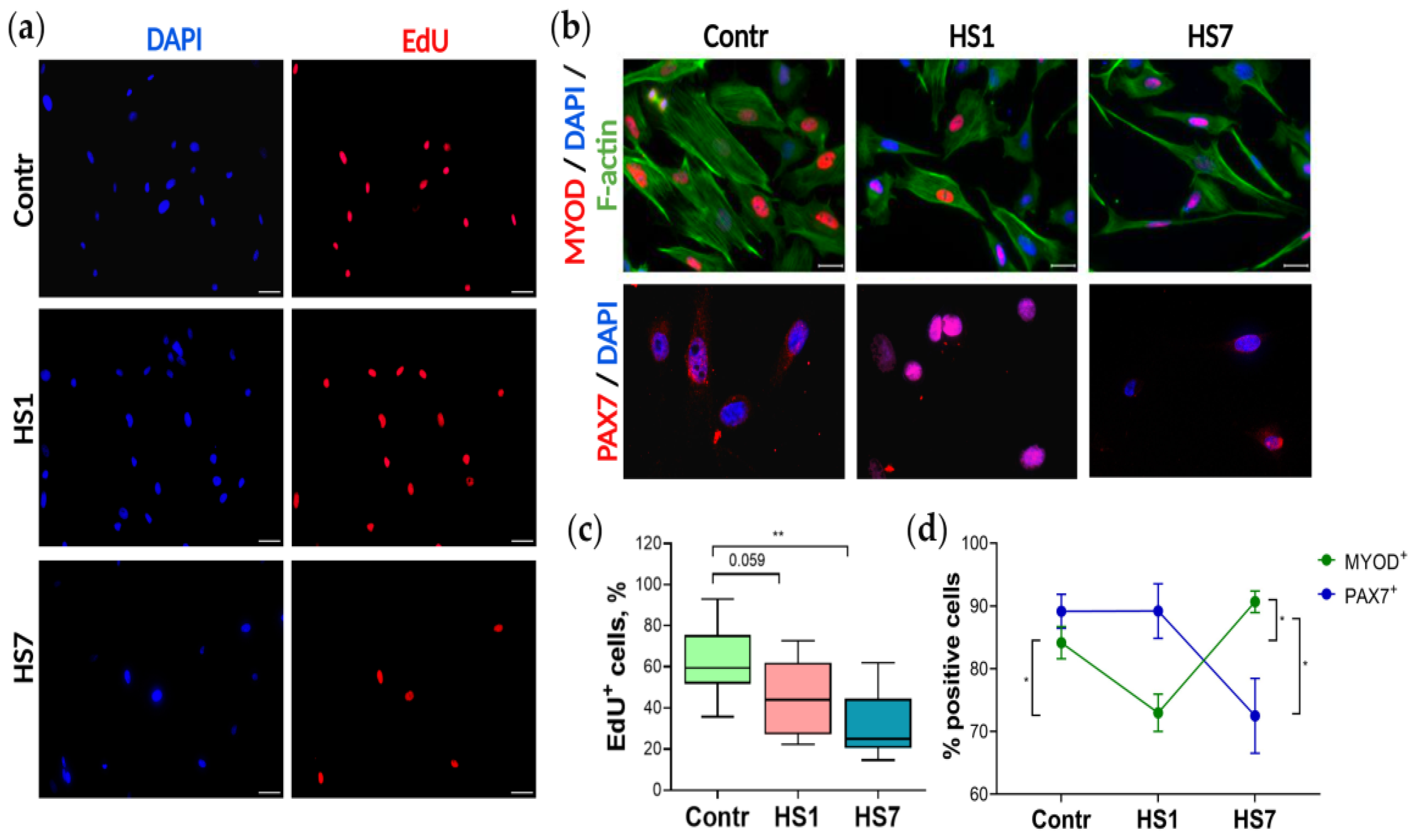
2.2. The Proliferation Activity and Myogenic Commitment of Myoblasts Isolated from Rat Soleus after Mechanical Unloading
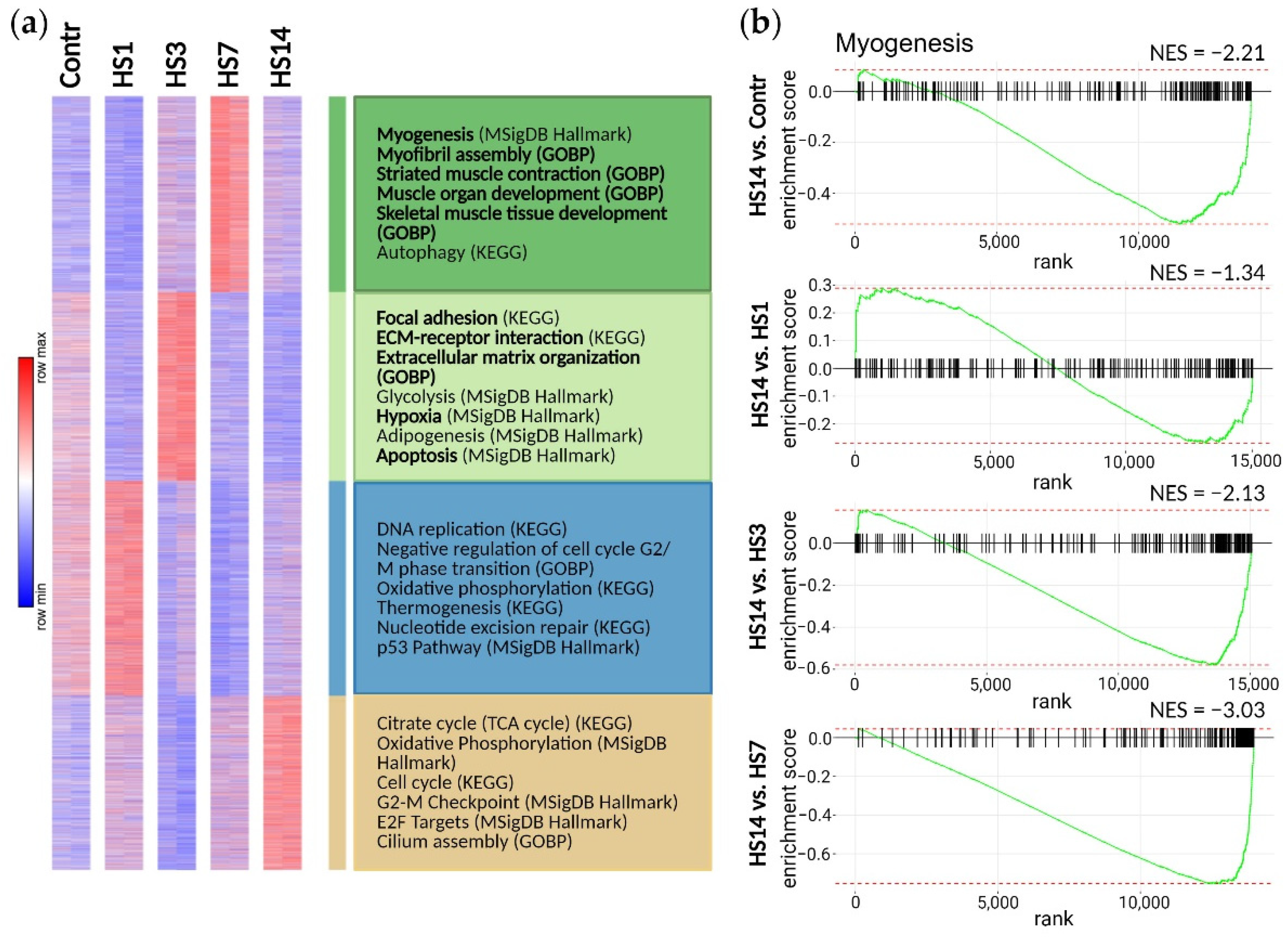
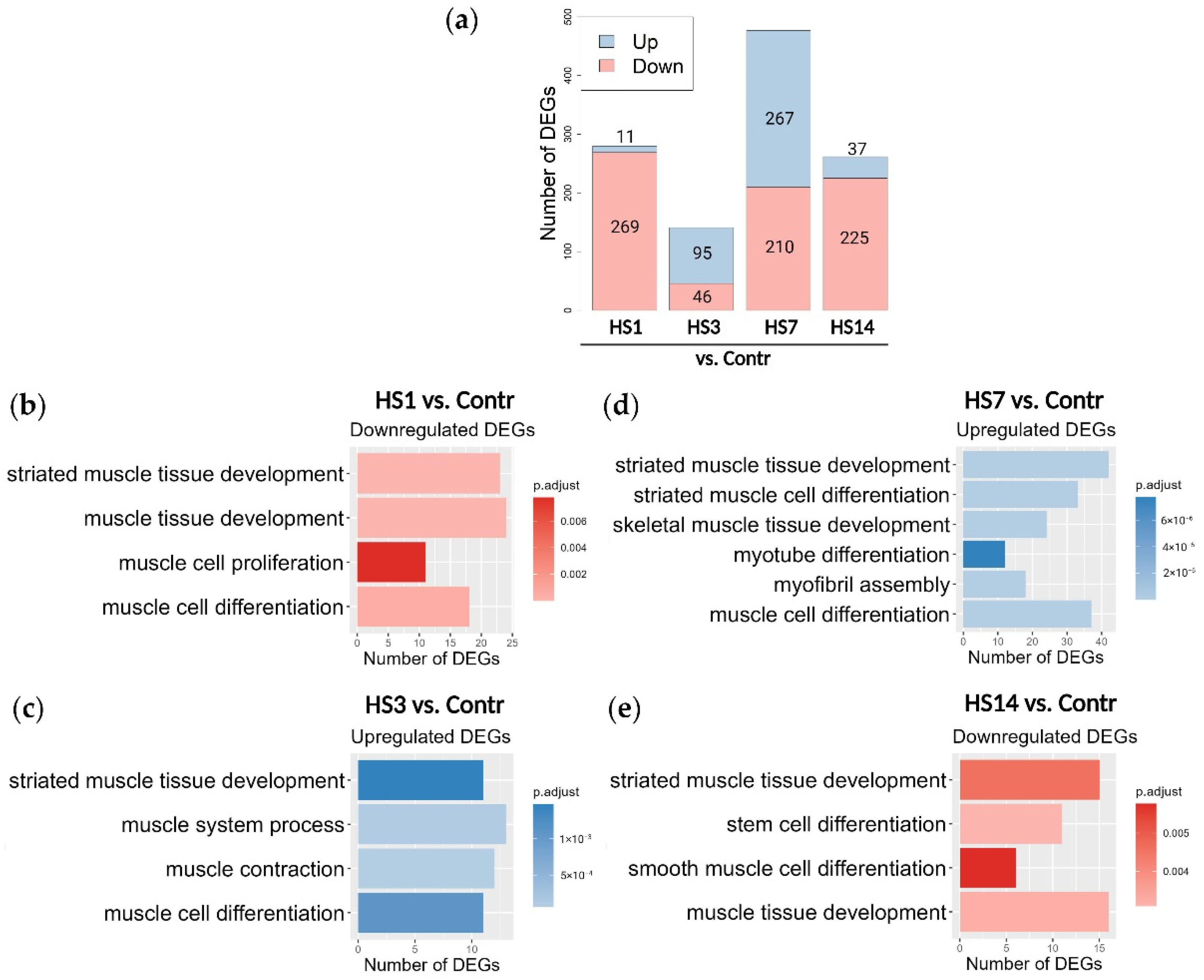
2.3. Transcriptome Analysis Reveals Two Phases in Soleus-Derived Myoblasts’ Response to Mechanical Unloading
3. Discussion
4. Materials and Methods
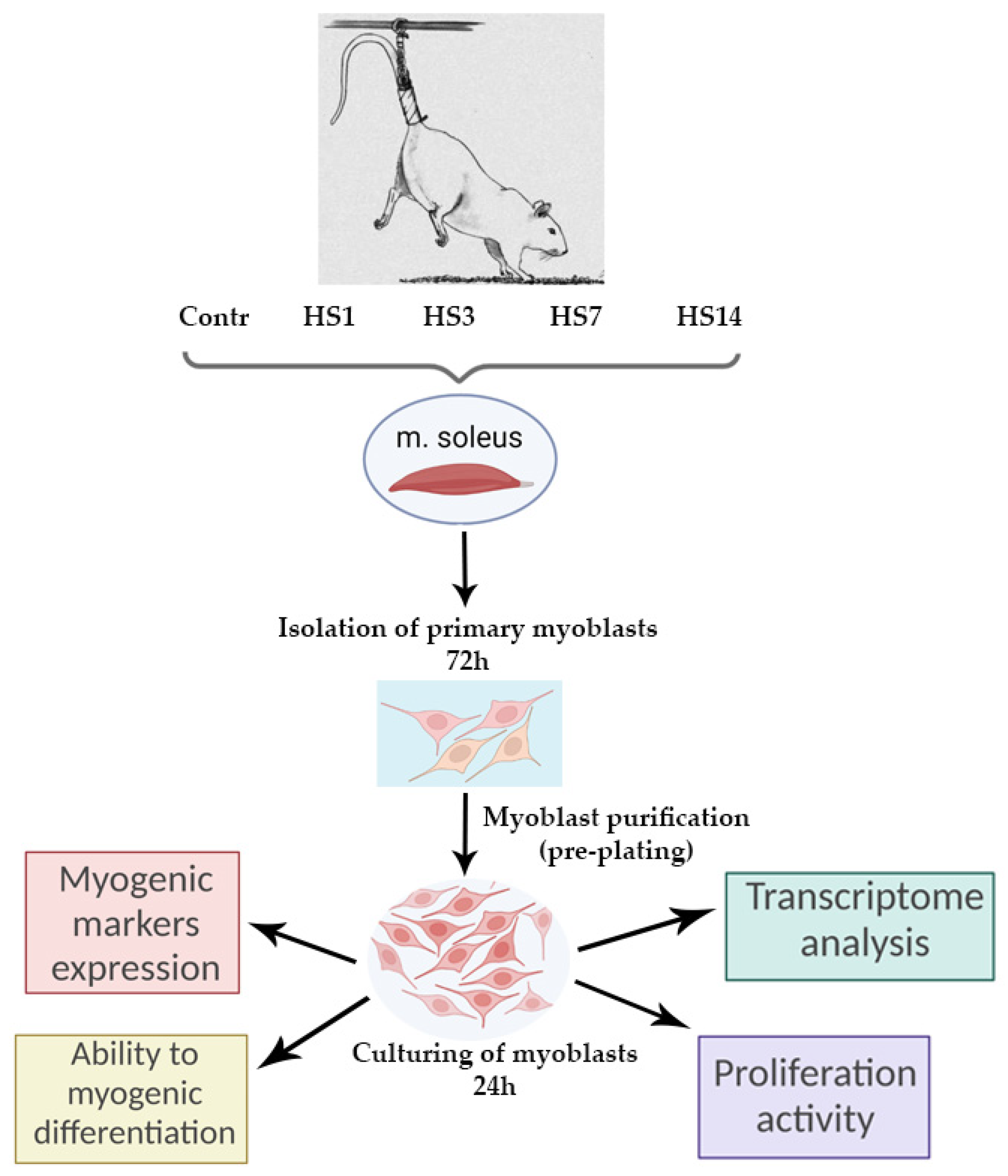
4.1. Experimental Design
4.2. Isolation of SCs
4.3. Proliferation Assay
4.4. RNA Isolation, cDNA Synthesis, and qRT-PCR
4.5. Immunocytochemistry
4.6. RNA-seq Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.C.; Rudnicki, M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar] [CrossRef]
- Brooks, N.E.; Myburgh, K.H. Skeletal muscle wasting with disuse atrophy is multi-dimensional: The response and interaction of myonuclei, satellite cells and signaling pathways. Front. Physiol. 2014, 5, 99. [Google Scholar] [CrossRef] [PubMed]
- Heslop, L.; Morgan, J.E.; Partridge, T.A. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J. Cell Sci. 2000, 113, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Briguet, A.; Courdier-Fruh, I.; Foster, M.; Meier, T.; Magyar, J.P. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. NMD 2004, 14, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Coulton, G.R.; Morgan, J.E.; Partridge, T.A.; Sloper, J.C. The mdx mouse skeletal muscle myopathy: I. A histological, morphometric and biochemical investigation. Neuropathol. Appl. Neurobiol. 1988, 14, 53–70. [Google Scholar] [CrossRef]
- Marshall, P.A.; Williams, P.E.; Goldspink, G. Accumulation of collagen and altered fiber-type ratios as indicators of abnormal muscle gene expression in the mdx dystrophic mouse. Muscle Nerve 1989, 12, 528–537. [Google Scholar] [CrossRef]
- Schiaffino, S.; Rossi, A.C.; Smerdu, V.; Leinwand, L.A.; Reggiani, C. Developmental myosins: Expression patterns and functional significance. Skelet. Muscle 2015, 5, 22. [Google Scholar] [CrossRef]
- Dmitrieva, R.I.; Lelyavina, T.A.; Komarova, M.Y.; Galenko, V.L.; Ivanova, O.A.; Tikanova, P.A.; Khromova, N.V.; Golovkin, A.S.; Bortsova, M.A.; Sergushichev, A.; et al. Skeletal muscle resident progenitor cells coexpress mesenchymal and myogenic markers and are not affected by chronic heart failure-induced dysregulations. Stem Cells Int. 2019, 2019, 5690345. [Google Scholar] [CrossRef] [PubMed]
- Bachman, J.F.; Chakkalakal, J.V. Insights into muscle stem cell dynamics during postnatal development. FEBS J. 2022, 289, 2710–2722. [Google Scholar] [CrossRef] [PubMed]
- Chemello, F.; Wang, Z.; Li, H.; McAnally, J.R.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. USA 2020, 117, 29691–29701. [Google Scholar] [CrossRef]
- Ignatieva, E.V.; Ivanova, O.A.; Komarova, M.Y.; Khromova, N.V.; Polev, D.E.; Kostareva, A.A.; Sergushichev, A.; Dmitrieva, R.I. LMNA mutations G232E and R482L cause dysregulation of skeletal muscle differentiation, bioenergetics, and metabolic gene expression profile. Genes 2020, 11, 1057. [Google Scholar] [CrossRef]
- Chibalin, A.V.; Benziane, B.; Zakyrjanova, G.F.; Kravtsova, V.V.; Krivoi, I.I. Early endplate remodeling and skeletal muscle signaling events following rat hindlimb suspension. J. Cell. Physiol. 2018, 233, 6329–6336. [Google Scholar] [CrossRef] [PubMed]
- Giger, J.M.; Bodell, P.W.; Zeng, M.; Baldwin, K.M.; Haddad, F. Rapid muscle atrophy response to unloading: Pretranslational processes involving MHC and actin. J. Appl. Physiol. 2009, 107, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Kandarian, S.C.; Stevenson, E.J. Molecular events in skeletal muscle during disuse atrophy. Exerc. Sport Sci. Rev. 2002, 30, 111–116. [Google Scholar] [CrossRef]
- Mirzoev, T.; Tyganov, S.; Vilchinskaya, N.; Lomonosova, Y.; Shenkman, B. Key markers of mTORC1-dependent and mTORC1-independent signaling pathways regulating protein synthesis in rat soleus muscle during early stages of hindlimb unloading. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 39, 1011–1020. [Google Scholar] [CrossRef]
- Шенкман, Б.С.; Туртикoва, О.В.; Немирoвская, Т.Л.; Григoрьев, А.И. Сoкратительная активнoсть скелетнoй мышцы и судьба миoядер. Acta Nat. 2010, 2, 62–69. [Google Scholar]
- Nakanishi, R.; Hirayama, Y.; Tanaka, M.; Maeshige, N.; Kondo, H.; Ishihara, A.; Roy, R.R.; Fujino, H. Nucleoprotein supplementation enhances the recovery of rat soleus mass with reloading after hindlimb unloading-induced atrophy via myonuclei accretion and increased protein synthesis. Nutr. Res. 2016, 36, 1335–1344. [Google Scholar] [CrossRef]
- Wang, X.D.; Kawano, F.; Matsuoka, Y.; Fukunaga, K.; Terada, M.; Sudoh, M.; Ishihara, A.; Ohira, Y. Mechanical load-dependent regulation of satellite cell and fiber size in rat soleus muscle. Am. J. Physiol. Cell Physiol. 2006, 290, C981–C989. [Google Scholar] [CrossRef]
- Mitchell, P.O.; Pavlath, G.K. Skeletal muscle atrophy leads to loss and dysfunction of muscle precursor cells. Am. J. Physiol. Cell Physiol. 2004, 287, C1753–C1762. [Google Scholar] [CrossRef]
- Matsuba, Y.; Goto, K.; Morioka, S.; Naito, T.; Akema, T.; Hashimoto, N.; Sugiura, T.; Ohira, Y.; Beppu, M.; Yoshioka, T. Gravitational unloading inhibits the regenerative potential of atrophied soleus muscle in mice. Acta Physiol. 2009, 196, 329–339. [Google Scholar] [CrossRef]
- Radugina, E.A.; Almeida, E.A.C.; Blaber, E.; Poplinskaya, V.A.; Markitantova, Y.V.; Grigoryan, E.N. Exposure to microgravity for 30 days onboard Bion M1 caused muscle atrophy and impaired regeneration in murine femoral Quadriceps. Life Sci. Space Res. 2018, 16, 18–25. [Google Scholar] [CrossRef]
- Ferreira, R.; Neuparth, M.J.; Ascensao, A.; Magalhaes, J.; Vitorino, R.; Duarte, J.A.; Amado, F. Skeletal muscle atrophy increases cell proliferation in mice gastrocnemius during the first week of hindlimb suspension. Eur. J. Appl. Physiol. 2006, 97, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Guitart, M.; Lloreta, J.; Manas-Garcia, L.; Barreiro, E. Muscle regeneration potential and satellite cell activation profile during recovery following hindlimb immobilization in mice. J. Cell. Physiol. 2018, 233, 4360–4372. [Google Scholar] [CrossRef] [PubMed]
- Morey-Holton, E.R.; Globus, R.K. Hindlimb unloading rodent model: Technical aspects. J. Appl. Physiol. 2002, 92, 1367–1377. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Mozdziak, P.E.; Truong, Q.; Macius, A.; Schultz, E. Hindlimb suspension reduces muscle regeneration. Eur. J. Appl. Physiol. Occup. Physiol. 1998, 78, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Barjot, C.; Cotten, M.L.; Goblet, C.; Whalen, R.G.; Bacou, F. Expression of myosin heavy chain and of myogenic regulatory factor genes in fast or slow rabbit muscle satellite cell cultures. J. Muscle Res. Cell Motil. 1995, 16, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Carvajal Monroy, P.L.; Yablonka-Reuveni, Z.; Grefte, S.; Kuijpers-Jagtman, A.M.; Wagener, F.A.; Von den Hoff, J.W. Isolation and characterization of satellite cells from rat head branchiomeric muscles. J. Vis. Exp. JoVE 2015, 101, e52802. [Google Scholar] [CrossRef]
- Charge, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C. Disuse-induced muscle wasting. Int. J. Biochem. Cell Biol. 2013, 45, 2200–2208. [Google Scholar] [CrossRef] [PubMed]
- Eigler, T.; Zarfati, G.; Amzallag, E.; Sinha, S.; Segev, N.; Zabary, Y.; Zaritsky, A.; Shakked, A.; Umansky, K.B.; Schejter, E.D.; et al. ERK1/2 inhibition promotes robust myotube growth via CaMKII activation resulting in myoblast-to-myotube fusion. Dev. Cell 2021, 56, 3349–3363.e3346. [Google Scholar] [CrossRef] [PubMed]
- Friedrichs, M.; Wirsdoerfer, F.; Flohe, S.B.; Schneider, S.; Wuelling, M.; Vortkamp, A. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol. 2011, 12, 26. [Google Scholar] [CrossRef]
- Jones, N.C.; Fedorov, Y.V.; Rosenthal, R.S.; Olwin, B.B. ERK1/2 is required for myoblast proliferation but is dispensable for muscle gene expression and cell fusion. J. Cell. Physiol. 2001, 186, 104–115. [Google Scholar] [CrossRef]
- Delaney, K.; Kasprzycka, P.; Ciemerych, M.A.; Zimowska, M. The role of TGF-β1 during skeletal muscle regeneration. Cell Biol. Int. 2017, 41, 706–715. [Google Scholar] [CrossRef]
- von Maltzahn, J.; Chang, N.C.; Bentzinger, C.F.; Rudnicki, M.A. Wnt signaling in myogenesis. Trends Cell Biol. 2012, 22, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Schmidt, C.; Luke, G.; Allen, S.; Valasek, P.; Muntoni, F.; Lawrence-Watt, D.; Patel, K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J. Cell Sci. 2008, 121, 2939–2950. [Google Scholar] [CrossRef] [PubMed]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 2002, 3, 397–409. [Google Scholar] [CrossRef]
- Qin, L.; Xu, J.; Wu, Z.; Zhang, Z.; Li, J.; Wang, C.; Long, Q. Notch1-mediated signaling regulates proliferation of porcine satellite cells (PSCs). Cell. Signal. 2013, 25, 561–569. [Google Scholar] [CrossRef]
- Kitamoto, T.; Hanaoka, K. Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem Cells 2010, 28, 2205–2216. [Google Scholar] [CrossRef] [PubMed]
- Tsivitse, S. Notch and Wnt signaling, physiological stimuli and postnatal myogenesis. Int. J. Biol. Sci. 2010, 6, 268–281. [Google Scholar] [CrossRef]
- Gerrard, J.C.; Hay, J.P.; Adams, R.N.; Williams, J.C., 3rd; Huot, J.R.; Weathers, K.M.; Marino, J.S.; Arthur, S.T. Current thoughts of notch’s role in myoblast regulation and muscle-associated disease. Int. J. Environ. Res. Public Health 2021, 18, 12558. [Google Scholar] [CrossRef] [PubMed]
- Brack, A.S.; Conboy, M.J.; Roy, S.; Lee, M.; Kuo, C.J.; Keller, C.; Rando, T.A. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 2007, 317, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Wallace, G.Q.; McNally, E.M. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu. Rev. Physiol. 2009, 71, 37–57. [Google Scholar] [CrossRef]
- Shi, X.; Garry, D.J. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006, 20, 1692–1708. [Google Scholar] [CrossRef] [PubMed]
- Filippelli, R.L.; Chang, N.C. Empowering muscle stem cells for the treatment of duchenne muscular dystrophy. Cells Tissues Organs 2022, 211, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Yablonka-Reuveni, Z.; Anderson, J.E. Satellite cells from dystrophic (mdx) mice display accelerated differentiation in primary cultures and in isolated myofibers. Dev. Dyn. 2006, 235, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Ingalls, C.P.; Wenke, J.C.; Armstrong, R.B. Time course changes in [Ca2+]i, force, and protein content in hindlimb-suspended mouse soleus muscles. Aviat. Space Environ. Med. 2001, 72, 471–476. [Google Scholar] [PubMed]
- Smolina, N.; Kostareva, A.; Bruton, J.; Karpushev, A.; Sjoberg, G.; Sejersen, T. Primary Murine Myotubes as a Model for Investigating Muscular Dystrophy. BioMed Res. Int. 2015, 2015, 594751. [Google Scholar] [CrossRef] [PubMed]
- Danoviz, M.E.; Yablonka-Reuveni, Z. Skeletal muscle satellite cells: Background and methods for isolation and analysis in a primary culture system. Methods Mol. Biol. 2012, 798, 21–52. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]

| Pathways | Number of DEGs | Genes |
|---|---|---|
| Extracellular matrix organization GO: 0030198 | 34 | Col1a1, Tgfbi, Pdgfra, Col6a1, Cyp1b1, Has2, Grem1, Adamtsl3, Col16a1, Loxl3, Smoc2, Col8a1, Eng, Itga8, Adamts9, Col14a1, Olfml2a, Adamts2, Lum, Adamts8, Kazald1, Fbln5, Itgb3, Acan, Has1, Csgalnact1, Fmod, Dpt, Foxc2, Mmp3, Adamts16, Ptx3, Il6, Foxc1 |
| Striated muscle tissue development GO: 0014706 | 42 | Pdgfra, Grem1, Dcn, Ifitm3, Wnt5a, Eng, Adamts9, Col14a1, S100b, Meox2, Aldh1a2, Nr2f2, Erbb3, Pdgfrb, Shox2, Sox8, Dll1, Foxc2, Igf1, Eya2, Myh15, Csrp3, Foxc1 |
| Regulation of cytosolic calcium ion concentration GO: 0051480 | 22 | Pdgfra, Thy1, Bdkrb2, Wnt5a, Ramp3, Ptgir, Synpo, Htr2a, Itgb3, Adcy5, Ednrb, Cacna1c, Cmklr1, Avp, Lpar3, Pth1r, Ackr4, Ptgfr, Cacna1g, Ednra, Kdr, C5ar2 |
| Regulation of cellular response to growth factor stimulus GO: 0090287 | 20 | Fbn1, Vegfd, Grem1, Dcn, Smoc2, Acvrl1, Xdh, Wnt5a, Eng, Itga8, Fgf10, Smad6, Fbn2, Dll1, Hjv, Msx1, Vsir, Kdr, Tmem204, Cdkn1c |
| Transmembrane receptor protein serine/threonine kinase signaling pathway GO: 0007178 | 20 | Fbn1, Grem1, Acvrl1, Wnt5a, Lrrc32, Eng, Itga8, Ror2, Fgf10, Cilp, Smad6, Fbn2, Tmem100, Hjv, Msx1, Vsir, Rbpms, Kdr, Lgals9, Cdkn1c |
| Muscle cell differentiation GO:0042692 | 18 | Pdgfra, Grem1, Ifitm3, Enpp3, Nfatc4, Eng, Itga8, Col14a1, Fgf10, Ednrb, Pdgfrb, Shox2, Dll1, Igf1, Msx1, Casp1, Tmem204, Csrp3 |
| ERK1 and ERK2 cascade GO: 0070371 | 15 | Pdgfra, Ramp3, Ece1, Fgf10, Htr2a, Itgb3, Pdgfrb, Avp, Igf1, Ednra, Kdr, Lgals9, Il6, Hand2, C5ar2 |
| Cytokine-mediated signaling pathway GO: 0019221 | 14 | Ifitm3, Il1rn, Wnt5a, Il11ra1, Il33, Ifitm1, Crlf1, Cxcl6, Ackr4, Slit3, Il20rb, Casp1, Il6, Foxc1 |
| Response to transforming growth factor β GO: 0071559 | 13 | Fbn1, Col1a1, Penk, Acvrl1, Wnt5a, Lrrc32, Eng, Itga8, Cilp, Smad6, Fbn2, Star, Cdkn1c |
| BMP signaling pathway GO: 0030509 | 12 | Fbn1, Grem1, Acvrl1, Wnt5a, Eng, Ror2, Smad6, Tmem100, Hjv, Msx1, Vsir, Kdr |
| Regulation of Wnt signaling pathway GO: 0030111 | 12 | Col1a1, Grem1, Wnt5a, Nfatc4, Plpp3, Ror2, Tnn, Fgf10, Nkd2, Cthrc1, Dkk2, Gsc |
| Tumor necrosis factor production GO: 0032640 | 11 | Wnt5a, Nfatc4, Twist2, Il33, Spon2, Ptafr, Igf1, Vsir, Lgals9, Il6, C5ar2 |
| Calcium-mediated signaling GO: 0019722 | 10 | Nfatc4, Erbb3, Ednrb, Cacna1c, Cmklr1, Tmem100, Ackr4, Ptgfr, Igf1, Kdr |
| Cellular response to mechanical stimulus GO: 0071260 | 8 | Col1a1, Aqp1, Eng, Il33, Itgb3, Piezo2, Ednra, Casp1 |
| Phosphatidylinositol 3-kinase signaling GO: 0014065 | 8 | Dcn, Ror2, Hgf, Htr2a, Erbb3, Pdgfrb, Igf1, Kdr |
| Apoptotic process involved in development GO: 1902742 | 5 | Slit3, Foxc2, Robo2, Hand2, Foxc1 |
| Pathways | Number of DEGs | Genes |
|---|---|---|
| Striated muscle tissue development GO: 0014706 | 42 | Tgfb2, Heyl, Sox11, Erbb3, Plagl1, Myo18b, Myog, Srpk3, Msc, Myom1, Tnnt2, Dmd, Actc1, Acta1, Tnnc1, Rgs2, Fhod3, Notch1, Gja5, Dll1, Stac3, Myh7, Cacna1s, Actn2, Tnni1, Mylpf, Lmod3, Chrnd, Tenm4, Myh15, Neb, Myom2, Smyd1, Wt1, Hey2, Myoz2, Nrap, Sox6, Col19a1, Csrp3, Mylk3, Rxrg |
| Muscle cell differentiation GO: 0042692 | 37 | Itga8, Tmod1, Myo18b, Myog, Mymx, Tnnt2, Dmd, Actc1, Acta1, Olfm2, Ldb3, Pdgfb, Rgs2, Mymk, Fhod3, Notch1, Dll1, Stac3, Cacna1s, Actn2, Trim72, Casq2, Tnnt1, Tnnt3, Lmod3, Cfh, Neb, Mypn, Smyd1, Ankrd23, Wt1, Hey2, Myoz2, Nrap, Sox6, Csrp3, Mylk3 |
| Actomyosin structure organization GO: 0031032 | 19 | Tmod1, Tnnt2, Actc1, Acta1, Ldb3, Wnt11, Fhod3, Actn2, Casq2, Tnnt1, Tnnt3, Lmod3, Neb, Mypn, Ankrd23, Myoz2, Nrap, Csrp3, Mylk3 |
| Calcium ion transport GO: 0006816 | 19 | Tgfb2, Vdr, Cemip, Cacng6, Dmd, Srl, Pdgfb, Atp1a2, Stac3, Ryr3, Cacna1s, Casq2, Slc24a3, Hrc, Cacnb1, Camk2b, Nos1, Xcl1, Dhrs7c |
| Myotube differentiation GO: 0014902 | 12 | Myog, Mymx, Dmd, Acta1, Mymk, Notch1, Stac3, Cacna1s, Trim72, Lmod3, Smyd1, Csrp3 |
| Notch signaling pathway GO: 0007219 | 11 | Tgfb2, Heyl, Notch3, Postn, Tp63, Notch1, Dll1, Dtx4, Hey2, Tspan15, Grip2 |
| Potassium ion transport GO:0006813 | 11 | Fxyd1, Atp1a2, Adora1, Gja5, Actn2, Casq2, Slc24a3, Kcnma1, Nos1, Scn4a, Kcna7 |
| Cellular response to transforming growth factor β stimulus GO: 0071560 | 11 | Tgfb2, Sox11, Itga8, Nrep, Itgb6, Postn, Adamtsl2, Cdkn1c, Cdh5, Sox6, Xcl1 |
| Regulation of apoptotic process involved in development GO: 1904748 | 4 | Tgfb2, Vdr, Notch1, Wt1 |
| Pathways | Number of DEGs | Genes |
|---|---|---|
| Wnt signaling pathway GO: 0016055 | 15 | Grem1, Itga3, Cdh3, Dkk3, Fgf10, Hmga2, Bmp2, Klf4, Sulf2, Plpp3, Ror2, Ctnnd2, Nog, Dkk2, Ptpro |
| Transmembrane receptor protein serine/ threonine kinase signaling pathway GO: 0007178 | 14 | Grem1, Itga3, Grem2, Fgf10, Bmp2, Myocd, Jade2, Ror2, Pparg, Msx2, Lgals9, Nog, Tmem100, Gdf10 |
| Canonical Wnt signaling pathway GO: 0060070 | 13 | Grem1, Cdh3, Dkk3, Fgf10, Bmp2, Klf4, Sulf2, Plpp3, Ror2, Ctnnd2, Nog, Dkk2, Ptpro |
| Cytokine-mediated signaling pathway GO: 0019221 | 12 | Grem2, Il1rn, Il11ra1, Il1rl2, Cxcl3, F3, Cxcl6, Pparg, Ereg, Arg1, Il20rb, Foxc1 |
| Stem cell differentiation GO: 0048863 | 9 | Grem1, Hmga2, Myocd, Ednrb, Osr1, Msx2, Sema3e, Hand2, Foxc1 |
| BMP signaling pathway GO: 0030509 | 9 | Grem1, Itga3, Grem2, Bmp2, Ror2, Pparg, Msx2, Nog, Tmem100 |
| Gene Name | Sequence (5′->3′) | GenBank |
|---|---|---|
| Pax7 | 5′-tataagagggagaaccccgga-3′ 5′-gctaatcgaactcactgaggg-3′ | NM_001191984.1 |
| Myf5 | 5′-ccctgatgtatcaaacgcatgt-3′ 5′-cgtgatccgatccactatgct-3′ | NM_001106783.1 |
| Myf6 | 5′-cttgagggtgcggatttcct-3′ 5′-cgcttgctcctccttcctta-3′ | NM_013172.2 |
| MyoG | 5′-ggtcccaacccaggagatca-3′ 5′-acatatcctccaccgtgatgc-3′ | NM_017115.3 |
| MyoD | 5′-tgctctgatggcatgatgga-3′ 5′-ctggacgcctcactgtagta-3′ | NM_176079.2 |
| Gapdh | 5′-cggtgtgaacggatttggc-3′ 5′-ttgaggtcaatgaaggggtcg-3′ | NM_017008.4 |
| Mymx | 5′-gatcgctgctatcacgcct-3′ 5′-ctcacgtcttgggagctcag-3′ | NM_001399466.1 |
| Mymk | 5′-tctttgtggcgttctcccat-3′ 5′-cagggctgtcccatagatgc-3′ | NM_001399315.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komarova, M.Y.; Rozhkov, S.V.; Ivanova, O.A.; Turtikova, O.V.; Mirzoev, T.M.; Dmitrieva, R.I.; Shenkman, B.S.; Vilchinskaya, N.A. Cultured Myoblasts Derived from Rat Soleus Muscle Show Altered Regulation of Proliferation and Myogenesis during the Course of Mechanical Unloading. Int. J. Mol. Sci. 2022, 23, 9150. https://doi.org/10.3390/ijms23169150
Komarova MY, Rozhkov SV, Ivanova OA, Turtikova OV, Mirzoev TM, Dmitrieva RI, Shenkman BS, Vilchinskaya NA. Cultured Myoblasts Derived from Rat Soleus Muscle Show Altered Regulation of Proliferation and Myogenesis during the Course of Mechanical Unloading. International Journal of Molecular Sciences. 2022; 23(16):9150. https://doi.org/10.3390/ijms23169150
Chicago/Turabian StyleKomarova, Margarita Y., Sergey V. Rozhkov, Oksana A. Ivanova, Olga V. Turtikova, Timur M. Mirzoev, Renata I. Dmitrieva, Boris S. Shenkman, and Natalia A. Vilchinskaya. 2022. "Cultured Myoblasts Derived from Rat Soleus Muscle Show Altered Regulation of Proliferation and Myogenesis during the Course of Mechanical Unloading" International Journal of Molecular Sciences 23, no. 16: 9150. https://doi.org/10.3390/ijms23169150






