Abstract
Ecdysteroids are widely investigated for their role during the molting cascade in insects; however, they are also involved in the development of the female reproductive system. Ecdysteroids are synthesized from cholesterol, which is further converted via a series of enzymatic steps into the main molting hormone, 20-hydoxyecdysone. Most of these biosynthetic conversion steps involve the activity of cytochrome P450 (CYP) hydroxylases, which are encoded by the Halloween genes. Three of these genes, spook (spo), phantom (phm) and shade (shd), were previously characterized in the desert locust, Schistocerca gregaria. Based on recent sequencing data, we have now identified the sequences of disembodied (dib) and shadow (sad), for which we also analyzed spatiotemporal expression profiles using qRT-PCR. Furthermore, we investigated the possible role(s) of five different Halloween genes in the oogenesis process by means of RNA interference mediated knockdown experiments. Our results showed that depleting the expression of SchgrSpo, SchgrSad and SchgrShd had a significant impact on oocyte development, oviposition and hatching of the eggs. Moreover, the shape of the growing oocytes, as well as the deposited eggs, was very drastically altered by the experimental treatments. Consequently, it can be proposed that these three enzymes play an important role in oogenesis.
Keywords:
biosynthesis; ecdysone; ecdysteriod; hemimetabola; hormone; insect; oogenesis; reproduction 1. Introduction
Ecdysteroids are lipophilic polyhydroxylated hormones that are widely investigated for their regulatory role in the molting of insects. They initiate the process of replacing the old cuticle by a newer and larger one, which allows the insects to grow and to pass from one instar to the next. In larval insects, ecdysone (E) is produced in the glandular cells of the prothoracic glands (PG) and released in the hemolymph. In peripheral tissues, such as the Malpighian tubules, E will be converted into the active molting hormone, 20-hydroxyecdysone (20E). In adult insects, the prothoracic glands are known to degenerate and ecdysteroidogenesis is taken over by the reproductive organs. Furthermore, in addition to the prothoracic glands and the reproductive organs, some other sites, such as oenocytes and epidermis, may also function as sources of ecdysteroids in insects, as reviewed by Delbecque et al. (1990), Gilbert et al. (2002) and Lafont and Koolman (2009) [1,2,3].
The response to 20E is mediated at the site of the cell nucleus by a heterodimeric receptor complex composed of the ecdysone receptor (EcR) and the ultraspiracle/retinoid-X-receptor (USP/RXR) [4]. In general, the 20E bound complex interacts with ecdysone response elements (EcRE) and as such, may induce transcription of 20E target genes. First, the early genes, including the ecdysone-induced protein 74 and 75 (E74 and E75), as well as Broad-Complex (Br-C), will be transcribed [5,6,7,8]. With a slight delay, transcription of the early-late genes, including E78, hormone receptor 3 and 39 (HR3 and HR39), will take place [9,10,11] and, together with the early genes, these will induce the transcription of the late genes, such as fushi tarazu transcription factor 1 (FTZ-F1) [12]. However, besides this 20E-dependent activation, the ligand-independent activity of these nuclear receptors has recently been described in Drosophila [13].
In contrast to vertebrates, insects are not able to synthesize cholesterol, the biosynthetic precursor of ecdysteroids, from single carbon molecules, but need to obtain sterols from their diet. Already in the intestine of the insects, plant-derived phytosterols will be dealkylated to cholesterol [2]. In the microsomes of a cell, cholesterol will be further converted into 7-dehydrocholesterol (7dC), which is then released into the cytosol and converted into diketol. The exact sequence of enzymatic reactions involved in this conversion is unknown and described as the ‘Black Box’ in ecdysteroidogenesis. One rate-limiting cytochrome P450 (CYP) enzyme in this ‘Black Box’ is encoded by the Halloween gene spook (spo) [14,15]. When returned into the microsomes, diketol will be hydroxylated into ketodiol, which, at its turn, is hydroxylated to ketotriol by a 25-hydroxylase [16,17]. In the mitochondria of the cell, ketotriol will be hydroxylated to 2-deoxyecdysone (2dE) by a 22-hydroxylase, and 2dE will be further hydroxylated to E by a 2-hydroxylase [18,19]. These hydroxylation steps are catalyzed by cytochrome P450 (CYP) enzymes that are encoded by the Halloween genes phantom (phm), disembodied (dib) and shadow (sad), respectively [16,17,18,19]. Finally, E will be released in the hemolymph and in peripheral tissues hydroxylated to 20E by a 20-hydroxylase which is encoded by the Halloween gene shade (shd) [20]. For an extensive overview of the ecdysteroid biosynthesis pathway, the reader is referred to the following reviews: Gilbert and Warren (2005), Lafont (2005) and Rewitz et al. (2006) [21,22,23].
Together with juvenile hormones (JHs), ecdysteroids are considered as the classic insect hormones involved in oogenesis of insects; however, their function and importance differ between distinct insect orders [24]. The dominant role of ecdysteroids in the oogenesis process in higher Diptera is widely investigated in the fruit fly, Drosophila melanogaster. Ecdysteroids are needed to establish and maintain the stem cell niche [25], to stimulate follicle cell formation and border cell migration [26], to coordinate the onset of vitellogenesis with the availability of nutrients [27], to stimulate yolk polypeptide synthesis in the fat body [28,29], and finally, to induce choriogenesis [30] (for an extensive overview of these different roles of ecdysteroids, see Belles and Piulachs (2014) and Swevers (2018)) [31,32]. In contrast to this extensive knowledge on the role of ecdysteroids in higher Diptera, their function in insects that use JH as the primary regulator of ovarian maturation (such as cockroaches and locusts) is less documented. Reports have shown that the ovaries of the migratory locust, Locusta migratoria, produce large amounts of ecdysteroids at the end of the gonadotrophic cycle. These ecdysteroids are mainly incorporated in the terminal oocytes as a maternal source for embryogenesis [33]. Ecdysteroids have also been described to induce meiotic re-initiation in this same species, thereby stimulating ovum maturation [34]. In the German cockroach, Blattella germanica, 20E was found to induce the start of the choriogenesis process when vitellogenesis is completed [35,36]. More recently, we have silenced EcR and RXR in the desert locust, Schistocerca gregaria, and have also shown that these ecdysteroid receptor components are essential for normal choriogenesis [37]. In this species, transcript levels of SchgrSpo and SchgrPhm were found to rise at the end of the first gonadotrophic cycle, in accordance with the accumulation of ecdysteroids in oocytes and their role in choriogenesis [38,39]. Our current study provides further transcript profiling of SchgrSpo and SchgrPhm and three additional Halloween genes (SchgrDib, SchgrSad and SchgrShd) in S. gregaria oogenesis. RNAi-mediated knockdown of SchgrSpo, SchgrSad and SchgrShd genes significantly affected oocyte development, oviposition and hatching of the eggs, suggesting that they play an important role in the female reproductive physiology of the desert locust, an insect species in which JH is known to be the main regulator.
2. Results
2.1. Sequence Analysis of SchgrDib and SchgrSad
The complete SchgrDib and SchgrSad open reading frame (ORF) sequences were found in in-house S. gregaria transcriptome and recently published S. gregaria genome databases (SCHGR_00006992 and SCHGR_00008835, respectively) (Supplementary Figures S1 and S2) [40]. The SchgrDib ORF (GenBank acc. no. KY404120) comprises 1578 nucleotides encoding a predicted protein of 526 amino acids, whereas the SchgrSad ORF (GenBank acc. No. MZ780957) comprises 1443 nucleotides encoding a predicted protein of 481 amino acids. These amino acid sequences are included in a multiple sequence alignment with previously identified insect DIB (Supplementary Figure S3) or SAD proteins (Supplementary Figure S4) [19,41,42,43,44]. SchgrDib and SchgrSad displayed 47.5% and 40.7% identity with their respective functionally characterized orthologs in D. melanogaster. Furthermore, several key motifs for CYP450 enzymes, namely, the mitochondrial signal sequence, the P/G rich domain, helix-C, helix-I, helix-K, the PERF-motif, and the heme-binding domain, could be identified (Supplementary Figures S3 and S4) [45].
2.2. Tissue Distribution and Developmental Transcript Profile during the First Gonadotrophic Cycle
Tissue and temporal distribution profiles of SchgrDib, SchgrSad and SchgrShd transcripts are shown in Figure 1, whereas transcript profiles of SchgrSpo and SchgrPhm were previously published by Marchal et al. (2011) [39]. Transcript levels of the Halloween genes SchgrDib, SchgrSad and SchgrShd were determined in the brain, corpora cardiaca, suboesophageal ganglion, prothoracic glands, thoracic ganglia, fat body, midgut and ovaries of virgin females and the testes and accessory glands of virgin males, both 10 days after the final molt, using qRT-PCR (Figure 1A,C,E). Transcript levels of SchgrDib were found to be significantly higher in the ovaries, while SchgrSad transcript levels were found to be significantly higher in the prothoracic glands of female adult locusts when compared to other tissues (Figure 1A,C). SchgrShd on the other hand, showed a wide distribution profile with high transcript levels in the ovaries, suboesophageal ganglion and brains of female adult locusts and testes and accessory glands of male adult locusts (Figure 1E). Furthermore, the temporal distribution profiles of SchgrDib, SchgrSad and SchgrShd were determined in the ovaries throughout the first reproductive cycle (Figure 1B,D,F). SchgrDib transcript levels were the highest on the day of molting to the adult stage; throughout the rest of the first reproductive cycle, the transcript levels were lower and remained relatively stable, showing no correlation with the ecdysteroid titer (Figure 1B). In contrast to SchgrDib transcript levels, the temporal expression of SchgrSad and SchgrShd was increased towards the end of reproductive cycle, correlating with the observed peak in ecdysteroid titer (Figure 1D,F).
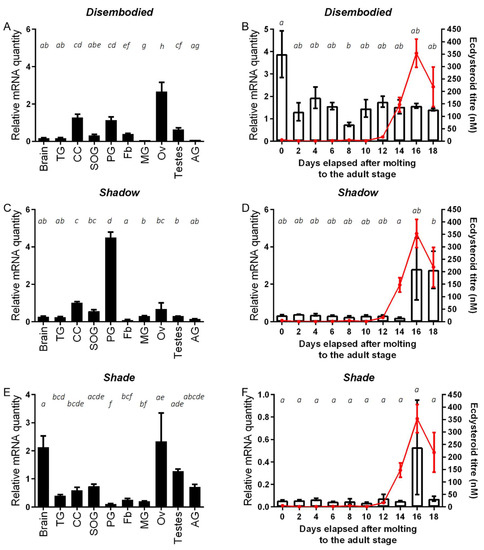
Figure 1.
Tissue and temporal distribution of SchgrDib, SchgrSad and SchgrShd transcripts. Relative transcript levels of (A) SchgrDib, (C) SchgrSad and (E) SchgrShd measured in different tissues of adult locusts, using qRT-PCR. All tissues were dissected from adult female locusts 10 days after the final molt, except for the male accessory glands (AG) and testes. The data represent mean ± S.E.M. of three independent pools (10 animals/pool), run in duplicate and normalized to ribosomal protein 49 (RP49) and elongation factor 1α (EF1α) transcript levels. Other abbreviations on the X-axis: TG: thoracic ganglia; CC: corpora cardiaca; SOG: suboesophageal ganglion; PG: prothoracic glands; Fb: fat body; MG: midgut; Ov: ovaries. Temporal distribution profile of (B) SchgrDib, (D) SchgrSad and (F) SchgrShd in the ovaries during the first reproductive cycle. Using qRT-PCR, relative transcript levels of SchgrDib, SchgrSad and SchgrShd were measured every other day in the ovaries, starting on the day of molting to the adult stage (AdD0). Data represent mean ± S.E.M. (bars) of three independent pools of ten animals each, run in duplicate and normalized to β-actin and EF1α transcript levels. The ecdysteroid titer (red line), expressed in nM, throughout the first reproductive cycle was measured with an enzyme immunoassay (EIA). The data represent mean ± S.E.M. of 6–18 hemolymph samples taken from different animals per time point. Significant differences are indicated using letters as described by Piepho (2018), meaning that conditions having a letter in common do not significantly differ from each other (One-way ANOVA with Tukey’s multiple comparison test on log-transformed data) [46].
2.3. RNA Interference of the Individual Halloween Genes
To investigate the role of ecdysteroids in the female reproductive physiology of S. gregaria, the locust orthologs of five ecdysteroid biosynthesis genes were silenced using RNAi. Female locusts were injected with dsRNA targeting SchgrSpo (dsSpo), SchgrPhm (dsPhm), SchgrDib (dsDib), SchgrSad (dsSad) or SchgrShd (dsShd) on day 6 of the 5th nymphal stage (N5D6), day 1 (AdD1), day 5 (AdD5) and day 9 (AdD9) of the adult stage. Control locusts were injected with dsGFP following the same injection scheme. First, one group of locusts was sacrificed twelve days after molting to the adult stage (AdD12) to investigate the possible effects of the RNAi-mediated knockdown on oocyte development and on the transcript levels of several genes of interest. Second, a group of locusts was kept alive to investigate the possible effects of the RNAi treatment on mating, fecundity and fertility (Supplementary Figure S5).
2.3.1. Knockdown Efficiency and Effect on Transcript Levels of Target Genes
Transcript levels of the Halloween genes were investigated in the dissected ovaries of female locusts twelve days after the final molt using qRT-PCR. Analysis of the normalized Ct-values indicated that the levels of SchgrSpo, SchgrPhm, SchgrDib, SchgrSad and SchgrShd transcripts were significantly reduced in their respective knockdown conditions (dsSpo, dsPhm, dsDib, dsSad and dsShd) with 93%, 88%, 46%, 98% and 98%, respectively (p = 0.0234, p = 0.0036, p = 0.0237, p = 0.0130, p = 0.0043, respectively; two-sided unpaired t-test on log-transformed data including Welch’s correction for comparison of SchgrSad transcript levels between dsGFP and dsSad) (Supplementary Figure S6). Furthermore, SchgrPhm transcript levels were significantly lower in the dsSpo- and dsSad- treated females (69%; p = 0.0413 and 74%; p = 0.0299, respectively; two-sided Unpaired t-test on log-transformed data) and SchgrDib transcript levels in dsShd- treated females (42%; p = 0.0253; two-sided Unpaired t-test on log-transformed data) when results were compared to the control (dsGFP) condition (Supplementary Figure S6).
The effect of knocking down the ecdysteroid biosynthesis gene orthologs on JH signalling and vitellogenin synthesis was investigated by measuring the transcript levels of the JH receptor gene methoprene-tolerant (SchgrMet) and the JH response gene Krüppel-homolog 1 (SchgrKr-h1) in the fat body and ovaries, as well as the transcript levels of vitellogenin 1 and vitellogenin 2 (SchgrVg1 and SchgrVg2) in the fat body, of all experimental animals. However, no significant differences in expression of these genes were observed when compared with dsGFP-injected (control) females (Supplementary Figure S7).
2.3.2. Observations of Oocyte Size, Mating, Oviposition and Hatching
Analysis of the average length and width of basal oocytes in fifteen female locusts, dissected on AdD12, per condition showed that the oocytes of dsSpo-, dsSad-, dsShd-treated females had an abnormal, more spherical shape when compared to the typical ovoid shape of the oocytes of dsGFP-treated females (Figure 2A–C + Supplementary Figure S8). Moreover, basal oocytes in these three conditions were significantly shorter than oocytes derived from control animals (dsSpo p < 0.0001; dsSad p = 0.0008; dsShd p = 0.0059; One-way ANOVA with Dunnett’s Multiple Comparisons Test; Figure 2D), whereas their width did not significantly differ between any of the tested conditions (One-way ANOVA with Dunnett’s Multiple Comparisons Test; Figure 2E). Furthermore, plotting the average length in function of the average width showed that oocytes of dsSpo-, dsSad-, dsShd-treated females were significantly shorter than control oocytes at a given width, whereas estimations of the oocyte volume did not reveal any significant differences between the knockdown and control conditions (Kruskal–Wallis test with Dunn’s multiple comparison test; Supplementary Figure S9A). Knockdown of SchgrPhm and SchgrDib did not result in this phenotype (Figure 2F). Confocal imaging of DAPI-stained follicles did not reveal any obvious structural differences in the follicular epithelium between the various knockdown (dsSpo, dsPhm, dsDib, dsSad and dsShd-injected) conditions and the dsGFP-injected control (Supplementary Figure S10).
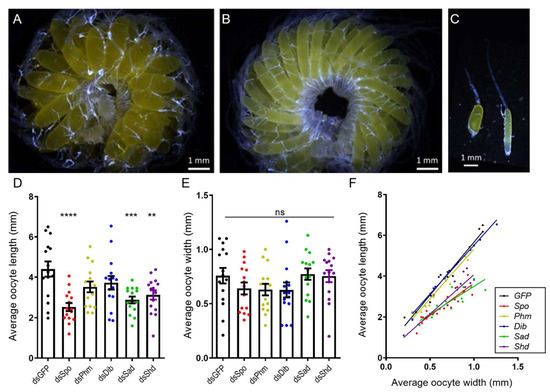
Figure 2.
Oocyte development in dsRNA-injected adult females twelve days after the final molt. For each female locust, the width and length of five individual basal oocytes were measured twelve days after the final molt (AdD12). (A) Representative ovary for the observed phenotype upon RNAi-mediated knockdown of SchgrSpo, SchgrSad or SchgrShd (average size of basal oocytes: length = 2.84 ± 0.12 mm, width = 0.72 ± 0.03 mm). (B) Ovary representative for locusts in which GFP, SchgrPhm and SchgrDib were targeted (average size of basal oocytes: length = 3.88 ± 0.19 mm, width = 0.67 ± 0.04 mm). (C) Representative ovariole for the observed phenotype upon RNAi-mediated knockdown of SchgrSpo, SchgrSad or SchgrShd (left, basal oocyte: length = 2.4 mm, width = 0.9 mm) and the control (right, basal oocyte: length = 3.5 mm, width = 0.6 mm). Scale bars A-C = 1 mm. (D,E) The average length (D) or width (E) of five basal oocytes per locust and per condition, represented as mean ± S.E.M.; each dot represents one data point (n = 15). Significant differences are indicated by (an) asterisk(s) (**** p < 0.0001; *** p < 0.001 and ** p < 0.01; ns = ‘not significant’; One-way ANOVA with Dunnett’s Multiple Comparisons Test). (F) The data represent the average length (Y-axis) and width (X-axis) of five basal oocytes collected from each individual female treated with dsSpo, dsPhm, dsDib, dsSad, dsShd or dsGFP (coloured dots), as well as the linear regression (coloured lines) (n = 15 per condition). The Pearson‘s correlation coefficients for the dsSpo, dsPhm, dsDib, dsSad, dsShd or dsGFP knockdown conditions are 0.8954, 0.9771, 0.9888, 0.8587, 0.9205 and 0.9840, respectively (p < 0.0001 in all conditions). The linear regression curves of dsSpo-, dsSad- and dsShd- treated females are significantly different from the control condition (p = 0.00029 for dsSpo-treated females and p < 0.0001 for dsSad- and dsShd-treated females; ANCOVA).
Although no significant differences in the occurrence of mating were observed between any of the tested conditions (Supplementary Figure S11A), the cumulative percentage of ovipositing females over time significantly differed between dsGFP- (control) (83.33%) and dsSpo- (65%) or dsSad- (50%) injected females (p = 0.0335 and p = 0.0448, respectively; Mantel–Cox test; Figure 3A). Nevertheless, the average number of days between copulation and egg laying, as well as the number of laid eggs, did not differ significantly between any of the tested conditions (Figure 3B,C). However, in accordance with the oocyte measurements, the eggs deposited by dsSpo-, dsSad-, dsShd-treated females were significantly shorter (p = 0.0028, p = 0.0004 and p = 0.0003, respectively; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test) and eggs from dsSpo-treated females were also significantly wider than eggs derived from dsGFP-treated females (p = 0.0364; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 3D–F). Furthermore, the estimated volume of the eggs was not significantly affected in the knockdown conditions when compared to the control condition (One-way ANOVA with Dunnett’s multiple comparison test; Supplementary Figure S9C). Observational analysis of hatching showed that the average number of days between egg laying and hatching did not significantly differ between any of the tested conditions (Figure 3G), that the average number of hatchlings derived from dsSpo-, dsSad- and dsShd-treated females was significantly lower (p = 0.0012, p = 0.0079 and p = 0.0286, respectively; One-way ANOVA with Dunnett’s Multiple Comparisons Test; Figure 3H) and that dsSpo- and dsSad-treatment conditions had a correspondingly lower percentage of hatching success (=100 × number of hatchlings/number of eggs laid; p = 0.0021 and p = 0.0103, respectively; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 3I), when compared to dsGFP-treated females.
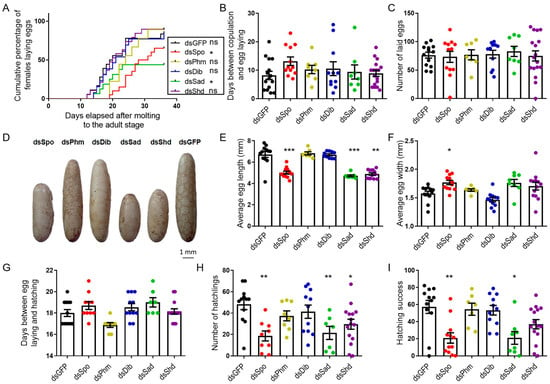
Figure 3.
Effects of RNAi-mediated knockdown of the individual Halloween genes (SchgrSpo, SchgrPhm, SchgrDib, SchgrSad and SchgrShd) on egg size, egg laying and hatching of adult female S. gregaria. (A) Cumulative percentage of egg laying by dsRNA-injected female locusts: 65.00% (n = 12) for dsSpo-, 88.89% (n = 7) for dsPhm-, 84.62% (n = 11) for dsDib-, 50.00% (n = 8) for dsSad-, 89.47% (n = 13) for dsShd- and 83.33% (n = 12) for dsGFP-treated females. The curves of dsSpo- and dsSad-treated females significantly differ from the curve of dsGFP-treated females (p = 0.0335 and p = 0.0448, respectively; Mantel–Cox test) (* p < 0.05, ns = ‘not significant’). (B) The number of days between copulation and egg laying is presented as the mean ± S.E.M.; each dot represents one data point. (C) The number of eggs laid per condition is presented as the mean ± S.E.M.; each dot represents one data point. (D) Representative examples of eggs for each dsRNA treatment (dsSpo, dsPhm, dsDib, dsSad, dsShd or dsGFP). Scale bar = 1 mm. (E,F) The average length (E) or width (F) of 20 eggs per locust and per condition is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (*** p < 0.001, ** p < 0.01, * p < 0.05; Kruskal–Wallis test with Dunn’s Multiple Comparison Test). (G) The number of days between egg laying and hatching is presented as the mean ± S.E.M.; each dot represents one data point. (H) The number of hatchlings per condition is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (** p < 0.01, * p < 0.05; One-way ANOVA with Dunnett’s Multiple Comparison Test). (I) The hatching success (percentage of hatching eggs) is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (** p < 0.01, * p < 0.05; Kruskal–Wallis test with Dunn’s Multiple Comparison Test).
2.4. RNA Interference of Halloween Gene Combinations
Since injections of dsSpo, dsSad and dsShd resulted in females containing significantly shorter, more spherical, basal oocytes than dsGFP-injected (control) females, while neither dsPhm nor dsDib injections revealed this phenotype, two combinations of Halloween genes were targeted, i.e., SchgrSpo, SchgrSad and SchgrShd (dsSpo/Sad/Shd) and SchgrPhm and SchgrDib (dsPhm/Dib) in a follow-up experiment. The dsSpo/Sad/Shd knockdown condition was selected to verify whether an even more pronounced effect would be generated, whereas the dsPhm/Dib condition was included to find out whether the spherical oocyte/egg phenotype would perhaps appear upon a combined knockdown. Similarly, as for the separate gene knockdown experiment, a first group of injected locusts was used to investigate possible effects on oocyte development and on the transcript levels of several genes of interest, whereas a second group of locusts was kept alive to enable us to observe mating, egg laying and hatching.
2.4.1. Knockdown Efficiency and Effect on Transcript Levels of Target Genes
qRT-PCR analysis of the Halloween gene transcript levels in 12-day adult females revealed that SchgrPhm (66%) and SchgrShd (81%) were significantly downregulated in the dsSpo/Sad/Shd knockdown condition (p < 0.0001 for both transcripts; two-sided Unpaired t-test on log-transformed data including the Welch’s correction for the comparison of the SchgrShd transcript levels) and SchgrPhm (90%) and SchgrDib (60%) were significantly downregulated in the dsPhm/Dib knockdown condition (p < 0.0001 for both transcripts; two-sided Unpaired t-test on log-transformed data; Supplementary Figure S12). Additionally, no significant differences were observed for SchgrMet and SchgrKr-h1 transcript levels in fat body and ovaries, as well as SchgrVg1 and SchgrVg2 transcript levels in fat body of dsSpo/Sad/Shd- and dsPhm/Dib-treated females, when compared with dsGFP-injected control females (Supplementary Figure S13).
2.4.2. Observations of Oocyte Size, Mating, Oviposition and Hatching
The analysis of oocyte length and width indicated that basal oocytes of dsSpo/Sad/Shd-treated females were significantly shorter than oocytes of the dsGFP-treated females (p < 0.0001; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 4A). However, the average oocyte width of dsSpo/Sad/Shd-treated females did not significantly differ from the control condition, meaning that, in line with the knockdown of the individual Halloween genes, oocytes of the combined knockdown condition were significantly shorter than control oocytes at a certain width (Figure 4B,C). Furthermore, their estimated oocyte volume did not significantly differ from the estimated oocyte volume of the control condition (Kruskal–Wallis test with Dunn’s multiple comparison test; Supplementary Figure S9B). Consequently, oocytes from dsSpo/Sad/Shd-treated females had a more spherical shape when compared to the ovoid shape of oocytes from control females (Supplementary Figure S8). In contrast to the findings for dsPhm- and dsDib-treated females, basal oocytes of females treated with dsPhm/Dib were significantly shorter and thinner than basal oocytes from dsGFP-injected (control) females (p = 0.0072 and p = 0.0155, respectively; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 4A–C; Supplementary Figure S8). As a consequence, these oocytes were also found to have a significantly smaller estimated volume than in the control condition (p = 0.0126; Kruskal–Wallis test with Dunn’s multiple comparison test; Supplementary Figure S9B). Furthermore, no major differences were observed in DAPI-stained follicular epithelia between knockdown (dsSpo/Sad/Shd and dsPhm/Dib) and control (dsGFP) conditions (Supplementary Figure S10).
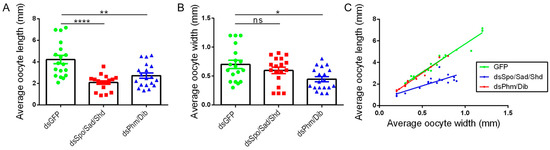
Figure 4.
Oocyte development in adult females that were injected with a combination of dsSpo, dsSad and dsShd or with a combination of dsPhm and dsDib. (A,B) The average length (A) or the average width (B) of five basal oocytes per female locust twelve days after the final molt and per condition are represented as mean ± S.E.M.; each dot represents one data point (n = 15). Significant differences are indicated by (an) asterisk(s) (**** p < 0.0001; ** p < 0.01, * p < 0.05 and ns = ‘not significant’; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test). (C) The data represent the average length (Y-axis) and width (X-axis) of five basal oocytes collected from dsSpo/Sad/Shd-, dsPhm/Dib-, or dsGFP-treated females twelve days after the final molt (coloured dots), as well as the linear regression (coloured lines) (n = 15 per condition). The Pearson‘s correlation coefficients for the dsSpo/Sad/Shd, dsPhm/Dib and dsGFP knockdown conditions are 0.8276, 0.9824 and 0.9778, respectively (p < 0.0001 in all conditions). The linear regression curve of dsSpo/Sad/Shd is significantly different from the control condition (p < 0.0001; ANCOVA).
Despite the observed effects on oocyte size, observations of mating revealed no significant differences between any of the tested conditions (Supplementary Figure S11B). However, only 19% of the dsSpo/Sad/Shd-treated females (4 out of 21) were able to lay eggs over a period of 45 days (Figure 5A). This result is in huge contrast with the 87.5% and 96% which were observed for the dsPhm/Dib- and dsGFP-treated females, respectively. Although most females of the dsPhm/Dib condition were able to lay their eggs, it took them significantly more time in comparison to dsGFP-injected (control) females (p = 0.0187; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 5B). Moreover, females from the dsPhm/Dib knockdown condition were found to lay significantly more eggs than females from the control condition (p = 0.0134; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 5C). Measurements of the egg length and width indicated that eggs derived from both knockdown conditions, dsSpo/Sad/Shd and dsPhm/Dib, were significantly shorter than these derived from dsGFP-injected (control) females (p = 0.0027 and p = 0.0019, respectively; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test), whereas their width was not significantly affected (Kruskal–Wallis test with Dunn’s multiple comparisons test; Figure 5D–F). Furthermore, the estimated volume of the eggs did not significantly differ between all conditions (Kruskal–Wallis test with Dunn’s multiple comparison test; Supplementary Figure S9D). Besides the significant effect observed on egg laying, eggs of the dsPhm/Dib knockdown condition took significantly more time to hatch and, although the number of hatchlings was not significantly different, the hatching success was significantly lower when compared to the dsGFP control condition (p = 0.0028, p = 0.8076 and p = 0.0188, respectively; Kruskal–Wallis test with Dunn’s Multiple Comparisons Test; Figure 5G–I). The average number of days between egg laying and hatching, the average number of hatchlings and the average hatching success (=100 × number of hatchlings/number of eggs laid) were not affected significantly after simultaneously targeting SchgrSpo, SchgrSad and SchgrShd.
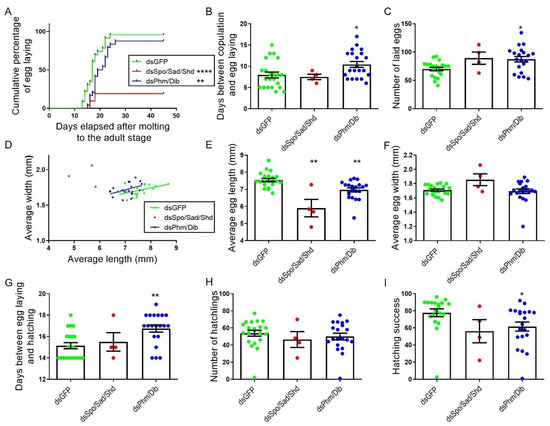
Figure 5.
Effects of RNAi-mediated knockdown of SchgrSpo/SchgrSad/SchgrShd and SchgrPhm/SchgrDib on egg size, egg laying and hatching of female S. gregaria. (A) Cumulative percentage of egg laying by dsRNA-injected female locusts: 19.05% for dsSpo/Sad/Shd- (n = 4), 87.5% for dsPhm/Dib- (n = 24) and 95.65% for dsGFP-treated (n = 23) females. The curves of dsSpo/Sad/Shd and dsPhm/Dib-treated females significantly differ from the curve of dsGFP-treated females (**** p < 0.0001 for dsSpo/Sad/Shd and ** p < 0.01 for dsPhm/Dib; Mantel–Cox test). (B) The number of days between copulation and egg laying is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (* p < 0.05; Kruskal–Wallis test with Dunn’s Multiple Comparisons test). (C) The number of eggs laid per condition is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (* p < 0.05; Kruskal–Wallis test with Dunn’s Multiple Comparisons test). (D) The data represent the average length (Y-axis) and width (X-axis) of eggs laid by dsSpo/Sad/Shd-, dsPhm/Dib- or dsGFP-treated females (coloured dots), as well as the linear regression (coloured lines) (dsSpo/Sad/Shd n = 4, dsPhm/Dib n = 20 and dsGFP n = 21). The Pearson‘s correlation coefficients for the dsSpo/Sad/Shd, dsPhm/Dib and dsGFP knockdown conditions are −0.6416 (p = 0.3584), 0.4309 (p = 0.0655) and 0.4624 (p = 0.0348), respectively. (E) The average length of seven eggs per locust is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (** p < 0.01; Kruskal–Wallis test with Dunn’s Multiple Comparisons test). (F) The average width of seven eggs per locust is presented as the mean ± S.E.M.; each dot represents one data point. (G) The number of days between egg laying and hatching is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (** p < 0.01; Kruskal–Wallis test with Dunn’s Multiple Comparisons test). (H) The number of hatchlings per condition is presented as the mean ± S.E.M.; each dot represents one data point. (I) The hatching success percentage is presented as the mean ± S.E.M.; each dot represents one data point. Significant differences are indicated by (an) asterisk(s) (* p < 0.05; Kruskal–Wallis test with Dunn’s Multiple Comparisons test).
3. Discussion
3.1. Characteristics and Expression Patterns of SchgrDib, SchgrSad and SchgrShd
In the current study, the nucleotide sequences of the Halloween genes Disembodied (CYP302A1, SchgrDib) and Shadow (CYP315a1, SchgrSad) in the desert locust, S. gregaria, were described (Supplementary Figures S2 and S3). Based on their amino acid sequence similarities with other known DIB and SAD proteins, several key motifs of cytochrome P450 (CYP450) enzymes were identified (Supplementary Figures S4 and S5) [19,41,42,43,44]. Earlier phylogenetic studies have shown that DIB and SAD belong to a monophyletic group of mitochondrial P450s [47], exhibiting common characteristics, such as charged residues in the N-terminal target sequence and two positively charged residues near the heme-binding domain that facilitate redox partner interactions [45,48]. DIB was identified in D. melanogaster and B. mori as a C22-hydroxylase, converting 2,22-dideoxyecdysone to 2-deoxyedysone, whereas SAD was identified as a C2-hydroxylase, converting 2-deoxyecdysone to ecdysone [19,42]. Recently, Dib and Sad orthologs were identified in L. migratoria, and based on their conserved phylogenetic relationship with representative insects, it was suggested that both enzymes are involved in the synthesis of the molting hormone [41]. Since SchgrDib and SchgrSad have 89% and 81% sequence identity with their respective orthologs in L migratoria and since they show 48% and 41% sequence identity with the functionally conserved orthologs in D. melanogaster [19], it is likely that both enzymes are also part of the ecdysteroidogenesis pathway in the desert locust.
Tissue and temporal distribution profiles of the Halloween genes SchgrSpo and SchgrPhm in adult locusts were previously published by Marchal et al. [39]. SchgrSpo and SchgrPhm relative transcript levels were highest in the prothoracic glands and in the ovaries of adult female locusts. Similarly, transcript levels of SchgrSad were highest in the prothoracic glands and high relative transcript levels of SchgrDib and SchgrShd were found in the ovaries (Figure 1A,C,E). Contrary to most insect species, the prothoracic glands still persist in adult locusts after the final molt, but they do not release ecdysteroids anymore [49]. Therefore, the functional importance of the Halloween gene expression in the adult prothoracic glands is still unclear. The expression of the Halloween gene transcript levels in the ovaries is consistent with ecdysteroid synthesis in the adult ovaries as has been described in Diptera and Lepidoptera [50,51,52]. More recently, research in a hemipteran species, Nilaparvata lugens, and an orthopteran species, L. migratoria, also showed the relatively high transcript levels of Dib and Shd in the ovaries of female adults [41,53]. Contrary to temporal distributions of the Halloween genes in A. aegypti and the previously published distributions of SchgrSpo and SchgrPhm [39,52], no correlation was observed between SchgrDib transcript levels and the circulating ecdysteroid titer in the adult female S. gregaria. On the other hand, transcript levels of SchgrSad and SchgrShd increased during the female reproductive cycle, which coincided with the observed peak in ecdysteroid titer (Figure 1B,D,F). However, a high variation could be observed at these time points, suggesting that transcription of these genes may be tightly regulated, showing dynamic pulses of transcript levels when induced. A similar pattern was seen for the transcript levels of BlageShd in the ovaries of the German cockroach B. germanica [54]. In the brown planthopper, N. lugens, it was suggested that both NilluCYP307B1 (Spo) and NilluCYP314A (Shd) are rate-limiting enzymes for ecdysteroidogenesis [53]. Therefore, it is evident that the expression profile of these genes correlates well with the circulating ecdysteroid titer [14,45]. Since DIB does not act as rate-limiting enzyme, its transcript levels are probably less strictly regulated.
3.2. Halloween Gene Expression Is Crucial in Female S. gregaria Reproductive Physiology Affecting Egg Shape, Oviposition and Hatching
Both experimental conditions, knocking down the Halloween genes SchgrSpo, SchgrSad and SchgrShd, separately (dsSpo, dsSad and dsShd) or in combination (dsSpo/Sad/Shd), led to the observation of abnormally shaped, more spherical oocytes that were significantly shorter in length than the control condition, while their width and estimated volume were not reduced (Figure 2 and Figure 4; Supplementary Figures S8 and S9). A very similar phenotype was seen in B. germanica and T. castaneum after depletion of Notch [55,56]. The Notch pathway is an essential regulator of cell proliferation and differentiation and, as such, is involved in determining cell fate [57,58]. In the panoistic ovary of B. germanica, Notch was found to play a role in inducing ovarian follicle elongation. Furthermore, also in three holometabolan insect species, D. melanogaster, A. mellifera and T. castaneum, the Notch pathway was found to play an important role in oogenesis [56,58,59,60]. Interestingly, in 2018, Yatsenko and Shcherbata identified a coordinated role between ecdysone signalling and Notch signaling in germline niche formation in Drosophila [61]. Furtermore, Ramos et al. (2020) suggested that the Notch downstream component Eyes Absent affects the ecdysteroidogenic pathway in B. germanica [54]. Therefore, it is possible that a depletion of Halloween gene expression in S. gregaria might also functionally interact with the Notch signaling pathway and, consequently, disturb the oogenesis process.
To verify if the more spherical oocyte phenotype observed after injections of female locusts with dsSpo, dsSad, dsShd or dsSpo/Sad/Shd might be correlated with disruption of the follicular epithelium, we have visualized DAPI-stained ovarian follicles by confocal microscopy. However, comparison with follicles derived from dsGFP-injected control females did not reveal any obvious structural differences in the follicular epithelial layer, which retained its normal appearance. Therefore, although differences at submicroscopic or molecular levels cannot be excluded, the more spherical oocyte shape does not appear to be associated with any obvious abnormalities in the surrounding epithelium.
Although the observed effect on egg laying in the dsSpo/Sad/Shd-treated females was more severe than the effect after treatment with dsSpo, dsSad or dsShd separately, qPCR-based transcript analysis revealed that only the SchgrShd transcript levels were significantly reduced (Supplementary Figure S12). This may be due to compensatory mechanisms, similarly as previously reported for other Halloween gene knockdown experiments in S. gregaria [39] and in other organisms [62,63,64]. Furthermore, this qPCR analysis only represents one single timepoint at the end of the experiment when the locusts were sacrificed.
In contrast to the observed phenotypes in dsSpo-, dsSad, dsShd- and dsSpo/Sad/Shd-treated females, knockdown of SchgrPhm or SchgrDib separately did not reveal any effect on oocyte development, even though their transcript levels were significantly downregulated (Figure 2, Supplementary Figures S6 and S8). However, downregulating both transcripts at the same time (dsPhm/Dib) resulted in a delayed oocyte development which was represented by oocytes that were significantly smaller in length and width than the ones from the control condition (Figure 4). Additionally, a significantly increased number of days between copulation and egg laying, as well as a significantly increased number of laid eggs, was observed for these dsPhm/Dib-treated females in comparison to dsGFP-treated females (Figure 5). It is generally known that female insects often exhibit a negative correlation between the number and the size of their eggs, constituting a trade-off between the offspring size and the available reproductive resources [65]. More specifically, in the desert locust, it was recently demonstrated that egg size and egg number may vary depending on their density-dependent phase state, having smaller but more eggs in isolated locusts when compared to crowded ones [66]. Moreover, already in 1999, it was shown that ovaries of solitarious S. gregaria females contain less ecdysteroids than gregarious ones [67]. Taken together, it might be proposed that the reproductive trade-off between solitarious and gregarious ovaries is correlated with the ecdysteroid content of these ovaries. Consequently, it can be suggested that knockdown of the Halloween genes, which is expected to lead to reduced ecdysteroid levels in the ovaries, may trigger changes in oocyte/egg size and number that are in line with differences that were previously reported between population density dependent locust phases.
Besides their effect on oocyte development, treatment with dsSpo, dsSad and dsSpo/Sad/Shd also affected egg laying, having significantly fewer females that successfully deposited their eggs than in the control condition (Figure 3 and Figure 5). These results are in accordance with the results obtained after knocking down the nuclear receptor complex (SchgrECR/SchgrRXR) in the same insect species which resulted in severely impaired oviposition [37]. Furthermore, significantly fewer eggs from the dsSpo-, dsSad- and dsPhm/Dib-treated females were hatching, in comparison to the eggs of the dsGFP-treated females (hatching success; Figure 3 and Figure 5). Since ecdysteroids are known to be incorporated into the developing oocytes to support embryogenesis [33], it could be suggested that knockdown of the Halloween genes may have resulted in insufficient ecdysteroid accumulation, hence disturbing the development of the embryo.
Knockdown of different S. gregaria Halloween genes did not always result in a very similar phenotypic outcome. Such variations were also seen in T. castaneum females, where knockdown of TricaShd resulted in arrested oocyte maturation, whereas knockdown of TricaPhm did not [68]. These discrepancies in the results for different genes acting in the same biosynthetic pathway could possibly be explained by the fact that Spo and Shd are known as important rate-limiting enzymes in the ecdysteroid biosynthesis pathway in different insect species [14,53]. Therefore, it could be suggested that the partial and transient depletion of SchgrPhm and SchgrDib (which is inherent to knockdown strategies) may have allowed for the synthesis of a level of ecdysteroids that was still sufficient for normal oocyte development. Another possible explanation could be that, in addition to E or 20E, one or more other ecdysteroids might also be capable of mediating this process. Interestingly, in some arthropod species, such as the spider mite, Tetranychus urticae, the Phm enzyme is lacking, and as a consequence, Ponasterone A is acting as the main ecdysteroid hormone [69]. In addition, since it is well known that multiple ecdysteroids are incorporated in the developing oocytes of different insect species in a conjugated form [70,71,72], it is possible that some of these conjugates, and hence, some hydroxylation steps, could be more important than others. Finally, it can also not be fully excluded that some enzymes encoded by Halloween genes might still exert as yet unknown activities in addition to their canonical role in the ecdysteroidogenesis process. However, since the knockdowns of three different enzymes, each responsible for a distinct step in the ecdysteroid biosynthesis pathway, have resulted in an identical phenotype, the latter option seems less likely.
3.3. Cross-Talk between Ecdysteroids and Other Hormonal Pathways
Multiple reports in different insect species proposed that cross-communication between JH and ecdysteroid signaling is taking place during the oogenesis process [36,70,73,74,75]. To further investigate this, the effect of knocking down the Halloween genes on transcript levels of SchgrMet and SchgrKr-h1 was investigated using qRT-PCR. Both genes were previously indicated to be a good measure for the activity of JH [76,77,78,79]. Furthermore, transcript levels of SchgrVg1 and SchgrVg2 after knockdown of the different Halloween genes, separately and in combination, were investigated to evaluate the effect on vitellogenin synthesis. Other than expected, none of the tested knockdown conditions has induced any significant effect on SchgrVg1 and SchgrVg2 in the fat body and on SchgrMet or SchgrKr-h1 expression levels in fat body or ovaries of females twelve days after the final molt (Supplementary Figures S7 and S13). Nevertheless, previously published results from our lab showed that downregulating the nuclear receptor (SchgrECR/SchgrRXR) complex resulted in reduced SchgrMet transcript levels in the fat body [37]. These results were in accordance with results obtained in the cockroaches Diploptera punctata and B. germanica, suggesting that ecdysteroid signaling is involved in terminating the reproductive cycle [36,74]. However, why knockdown of Halloween genes did not result in a similar effect and how the interaction between ecdysteroid and JH signaling is regulated remains elusive. Perhaps, the effect of ECR/RXR on the JH signaling pathway might be ligand-independent, as this receptor complex was also found to prevent cell death in the PG of Drosophila in a ligand-independent manner [13].
Similarly to the results obtained after knockdown of SchgrECR/SchgrRXR, vitellogenin transcript levels were not significantly influenced by the Halloween gene RNAi treatments. These results indicate that, contrary to what had been suggested previously by J. Girardie and A. Girardie (1998), vitellogenin synthesis in locusts is most probably not directly regulated by ecdysteroids, while it is clearly dependent on JH [24].
The present study has shown that RNAi-mediated knockdown of the Halloween genes SchgrSpo, SchgrSad and SchgrShd, which code for ecdysteroid biosynthesis enzymes, had a clear effect on the development and shape of growing oocytes in adult female locusts, as well as on the shape (length/width ratio) of the deposited eggs. Moreover, oviposition and hatching success were negatively affected by this knockdown. Future research may shed more light on the functional interactions between ecdysteroid and JH signaling in the oogenesis process of female S. gregaria locusts.
4. Materials and Methods
4.1. Rearing of Animals
Desert locusts (S. gregaria) were reared under crowded conditions at a temperature of 30 ± 1 °C, 40–60% relative humidity and a 14/10 light/dark cycle. They were fed ad libitum with fresh cabbage leaves and rolled oats. Mature females in breeding cages were allowed to deposit their eggs in pots filled with a moistened turf/sand (2:1) mixture, which were collected and replaced on a weekly basis. Collected egg pots were placed in new empty cages, resulting in groups of hatchlings that differed no more than seven days in age. The adult locusts that were used for investigating the tissue distribution and temporal expression profiles of SchgrDib, SchgrSad and SchgrShd were derived from cages in which locusts were collected that all had molted to adulthood on the same day.
4.2. Sequence Analysis of SchgrDib and SchgrSad
The nucleotide sequences coding for the full-length S. gregaria orthologs of dib (KY404120) and sad (MZ780957) were retrieved from the recently published S. gregaria genome database [40], and also confirmed by an in-house transcriptome database (unpublished data). Using the M-Coffee web server in the default settings (http://tcoffee.crg.cat/apps/tcoffee/do:mcoffee; page accessed on 7 July 2020) [80], both sequences were aligned to orthologous sequences from four representative insect species (Locusta migratoria, Drosophila melanogaster, Bombyx mori and Anopheles gambiae) and conserved residues were highlighted using Boxshade (https://embnet.vital-it.ch/software/BOX_form.html; page accessed on 7 July 2020) [19,41,42,43,44].
4.3. Tissue Collection and RNA Extraction
To study the tissue distribution of SchgrDib, SchgrSad and SchgrShd, adult locusts were collected on their day of molting from the 5th nymphal stage to the adult stage, and ten days after this final molt (AdD10), the following tissues were dissected under a binocular microscope and rinsed in locust Ringer solution (150 mM NaCl, 1.7 mM CaCl2, 10 mM KCl, 4.3 mM MgCl2, 4 mM NaHCO3, 90 mM Sucrose, 5 mM Trehalose, pH 7.2): brain, corpora cardiaca, suboesophageal ganglion, prothoracic glands, thoracic ganglia, fat body, midgut and ovaries of virgin females, as well as testes and accessory glands of virgin males. These tissues were collected in three independent pools, each containing samples derived from 10 individual locusts, and were transferred to MagNA Lyser Green Beads containing Tubes (Roche) or RNase-free Screw Cap Microcentrifuge tubes.
To study the temporal transcript expression profiles of SchgrDib, SchgrSad and SchgrShd, groups of locusts that molted to adulthood on the same day were collected and ovaries of female locusts were dissected every other day, until day 18 of the adult stage. Samples were rinsed in locust Ringer solution and collected in MagNA Lyser Green Beads containing Tubes (Roche) in three independent pools, each containing six samples of individual locusts.
During the described RNAi experiments (see Section 4.6), virgin female locusts were dissected under a binocular microscope twelve days after the final molt. Tissues of interest (Ovaries and Fat body) were rinsed in locust Ringer solution and transferred to MagNA Lyser Green Beads containing Tubes (Roche). Samples derived from three individual locusts were pooled, generating five pools from a total of 15 female locusts. All samples were snap-frozen in liquid nitrogen and stored at −80 °C until further processing.
According to the different tissue collection strategies, different RNA extraction methods were used for larger, fat containing tissues and relatively small tissues containing low RNA quantities. Tissues collected in MagNA Lyser Green Bead Tubes (Roche) were homogenized in Qiazol (Qiagen) using the MagNa Lyser instrument according to the manufacturer’s instructions (1 min, 6500 rpm; Roche) and total RNA was extracted using the Qiagen® RNeasy Lipid Tissue Kit, performing the optional DNase digestion protocol (RNase-Free DNase set, Qiagen, Hilden Germany). From smaller tissues collected in RNase-free Screw Cap Microcentrifuge tubes, total RNA was extracted using the RNAqueous-Micro Kit (Ambion) according to the manufacturer’s protocol, including the recommended DNase step. The purity and concentration of the RNA samples was analyzed using a Nanodrop spectrophotometer (Nanodrop ND-1000, Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Extracted RNA was reverse transcribed to equal amounts of cDNA using the PrimeScriptTM reagent kit (Perfect Real Time) (Takara Bio Inc., Saint-Germain-en-Laye, France) according to the manufacturer’s protocol. The resulting cDNA was diluted ten-fold in Milli-Q water (Millipore).
4.4. Quantitative Real-Time PCR
The tissue and temporal distributions of SchgrDib, SchgrSad and SchgrShd were analyzed using quantitative (real-time) reverse transcription PCR (qRT-PCR) and the QuantStudio® 3 Real-Time PCR Instrument (Applied Biosystems); 4 µL of cDNA was mixed with 5 µL Fast SYBR® Green Master Mix (Applied Biosystems), 0.5 µL forward and 0.5 µL reverse primer (10 µM; primer sequences are listed in Table A1) and genes of interest were amplified under the following temperature conditions: 20 s at 95.0 °C and 40 cycles of 95 °C for 1 s and 60 °C for 20 s. Primers were validated and amplification specificity was determined as described by Lenaerts et al. (2017) [81]. Raw data were normalized according to the ddCt-method using a calibrator sample and two most stably expressed housekeeping genes, ribosomal protein 49 (RP49) and elongation factor 1α (EF1α) for the tissue distribution and β-actin and EF1α for the temporal distribution. Each sample was measured in duplicate.
Knockdown efficiencies after RNAi-mediated knockdown of the ecdysteroid biosynthesis genes and transcript levels of genes of interest were analyzed in a similar way. In the experiment wherein Halloween genes were knocked down separately (See 2.6 RNA interference experiments), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and CG13220 were selected as stably expressed housekeeping genes for all tissue samples. In the experiment wherein a combined knockdown of Halloween genes was performed, EF1α and RP49 were selected.
4.5. Ecdysteroid Measurements Using an Enzyme Immunoassay
During the first reproductive cycle, hemolymph samples were collected every other day until day 18 of the adult stage and pooled in five groups of three locusts each. Locusts were pierced at their cuticle behind the hind leg and 10 µL of hemolymph was collected using a capillary and transferred to 270 µL of cold methanol (100%). Samples were stored at −20 °C until further processing. Ecdysteroid titers were measured in hemolymph samples using the enzyme immunoassay (EIA) as previously described by Marchal et al. (2011) [39]. In this assay, a rabbit L2 polyclonal antiserum with high affinity for ecdysone, 3-deoxyecdysone, 2-deoxyecdysone and a lower affinity (6- to 8-fold) for 20-hydroxyecdysone was used.
4.6. RNA Interference Experiments
Production of dsRNA. dsRNA constructs targeting SchgrSpo (dsSpo), SchgrPhm (dsPhm), SchgrDib (dsDib), SchgrSad (dsSad), SchgrShd (dsShd) and GFP (dsGFP) were synthesized using the MEGAscript® RNAi Kit (Ambion) according to the manufacturer’s protocol. Since this kit makes use of a T7 RNA polymerase, genes of interest were amplified using T7 promoter flanked primers (see Table A2). Results were analyzed by horizontal 1% agarose gel electrophoresis and visualized with GelRedTM (Biotum) under UV light. The band of the expected size was excised and further purified using the GenElute™ Gel extraction Kit (Sigma-Aldrich Co., St. Louis, MO, USA). The resulting DNA fragment was cloned into a pCR4-TOPO vector using the TOPO® TA Cloning Kit (Invitrogen), and after transformation of DH5α competent bacteria (Escherichia coli), and subsequent cloning, the plasmid DNA was purified using the GenEluteTM HP plasmid miniprep kit (Sigma Aldrich). The sequence of the insert was determined by Sanger sequencing to confirm its target specificity. When sequences were confirmed, dsRNA was produced using the T7 RNA polymerase and purified using a two-step purification protocol including a nuclease digestion and a column-based purification step. The purified dsRNA constructs were verified by horizontal 1% agarose gel electrophoresis and their concentrations were measured with a NanoDropTM 1000 Spectrophotometer.
Knockdown of different Halloween genes separately. Female locusts were collected on the day of molting to the fifth nymphal stage (N5D0) and injected with 5 µL of 80 ng/µL dsRNA, diluted in S. gregaria Ringer solution, targeting SchgrSpo, SchgrPhm, SchgrDib, SchgrSad, SchgrShd or GFP at day 6 (N5D6) of this last nymphal stage using a Hamilton syringe. Boost injections were given in the adult stage on day 1 (AdD1), day 5 (AdD5), day 9 (AdD9) and, in case of locusts destined for copulation behavior analysis, on day 13 (AdD13). On day 8 of the adult stage, female locusts were separated from male locusts and each knockdown condition was further divided into two different groups. One group of female locusts was dissected on day 12 of the adult stage (AdD12) to collect the ovaries and fat body for qRT-PCR analysis (n = 15 for all conditions) and to collect single ovarioles for staining the nuclei of the follicle epithelium cells (n = 4 for all conditions), while another group (n = 17, 20, 9, 13, 16, 19 for dsGFP-, dsSpo-, dsPhm-, dsDib-, dsSad- and dsShd-injected females, respectively) was kept alive for an observational analysis of their copulation behavior, as well as oviposition and hatching.
Combined Halloween gene knockdown experiment. In accordance with the previous knockdown experiment, female locusts were again collected on N5D0 and injected with dsRNA on N5D6. Boost injections were given on AdD1, AdD5, AdD9 and, in case of locusts destined for copulation behavior analysis, AdD13. In this experiment, Halloween genes SchgrSpo, SchgrSad and SchgrShd were targeted simultaneously by injecting 400 ng of each dsRNA construct (dsSpo/Sad/Shd) in each animal. Besides the control condition (dsGFP), a third condition targeting both SchgrPhm and SchgrDib, by injecting 400 ng of each dsRNA construct (dsPhm/Dib), was included. Female locusts were separated from male locusts on day 8 of the adult stage and divided into two different groups, one for tissue collection on AdD12, transcript analysis by qRT-PCR and visualization of the follicle epithelium cell nuclei (n = 15 and n = 4 for all three conditions, respectively) and one for observational analysis of copulation behavior, oviposition and hatching (n = 23, 21, 24 for dsGFP-, dsSpo/Sad/Shd- and dsPhm/Dib-injected females, respectively).
4.7. Oocyte Size, Copulation Behavior, Oviposition and Hatching
To determine the impact of RNAi-mediated knockdown of ecdysteroid biosynthesis enzymes on the female reproductive physiology, five individual oocytes were dissected from each female (fifteen females per condition) at an age of 12 days after the finale molt, of which the length and width were measured under a binocular microscope using a micrometer scale. Images of the ovaries and ovarioles were obtained using a light microscope (Zeiss SteREO Discovery. V8; Imaging sofware program Zen 2012, accessed from Leuven, Belgium) equipped with an AxioCam ICc3 camera using the AxioVision 4.7 (Carl Zeiss-Benelux, Oberkochen, Germany). Furthermore, using the calculated average length and width of the oocyte size measurements, an estimation of the oocyte’s volume was made by using the formula for calculating the volume of an ellipsoid body:
The processes of mating, oviposition and hatching were monitored and quantitatively analyzed. Locusts, which had molted to the adult stage on the same day, were kept in a gender-mixed group until day 8 of the adult stage. Males were then removed from the cages and females were transferred to individual containers. From day 9 of the adult stage on, each female locust was combined with a mature virgin male, which was at least one week older, for two hours. Mating was considered successful when there was a direct connection between the female and the male genitals. In this case, the male locust was allowed to stay with the female for another 22 h. In case no mating was observed, the male locust was removed from the container after two hours and the experiment was repeated the following day. After 24 h of mating, the male locusts were removed, and the female locusts received a pot filled with a moistened turf/sand (2:1) mixture, in which they could lay their eggs. These pots were controlled daily for presence of deposited egg batches and were replaced every four days. In case eggs were observed, the total number of eggs was counted for each individual female and the average egg length/width was measured as described above. In addition, an estimated volume of the eggs was calculated using Equation (1). All egg-containing pots were controlled daily for appearance of hatchlings. Finally, the number of hatchlings was counted and the percentage of hatching success (=100 × the number of hatchlings/number of eggs laid) was determined per condition. Supplementary Figure S5 shows a schematic overview of the experimental strategy that was followed in this study.
4.8. Visualisation of the Follicle Epithelium Cell Nuclei
On day 12 of the adult stage, single ovarioles were collected from the ovaries of dsSpo-, dsPhm-, dsDib-, dsSad-, dsShd-, dsSpo/Sad/Shd-, dsPhm/Dib- or dsGFP-treated females and freed from their surrounding membrane. The obtained ovarioles were fixated in 4% paraformaldehyde dissolved in PBS for 30 min at room temperature. After the fixation, ovarioles were washed for 2 times 5 min in PBS and incubated with 15 µg/mL 4′,6-diamidino-2-phenylindole (DAPI), which is known to bind to A-T rich regions of the DNA, for 10 min. Subsequently, ovarioles were washed in 0.2% Tween PBT and mounted on a glass slide using Mowiol solution (4 mM Mowiol® 4-88, 65 M glycerol, finally diluted 1:3 in 0.2 M Tris pH = 8.5). Samples were stored in the dark at 4 °C and imaged using a confocal scanning microscope (FV1000-IX81, Olympus, Hamburg, Germany) and FluoViewer 4.2 software (Olympus accessed from Leuven, Belgium).
4.9. Statistical Analysis
Depending on the performed experiment, different statistical approaches were used. In the case of qRT-PCR analysis, transcript expression levels were normalized in Excel according to the ddCT-method using a calibrator sample and two selected most stably expressed housekeeping genes (see Section 2.4), which were selected using the geNorm software and the NormqPCR package in RStudio [82,83,84]. Afterwards, relative expression levels were converted to Graphpad Prism 6 (Graphpad Software Inc., San Diego, CA, USA) and log-transformed in order to create normalized data. Log-transformed data of the tissue and temporal distributions of SchgrDib, SchgrSad and SchgrShd were further analyzed using one-way ANOVA including the Tukey multiple comparisons test. Log-transformed, normalized expression levels obtained during the described knockdown experiments were always compared against the dsGFP conditions using a two-sided unpaired t-test, including the Welch’s correction when variances were significantly different from each other.
In the case of oocyte/egg length/width measurements and oocyte/egg volume estimations, the calculations were made in Excel and results were plotted using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA) software. Normally distribution of the data was tested using the Shapiro–Wilk test. If the results were normally distributed, significant differences were determined using one-way ANOVA analysis with Dunnett’s multiple comparisons correction, if the results did not constitute a normal distribution, significant differences were determined using Kruskal–Wallis test with Dunn’s multiple comparisons correction. Furthermore, using the same software, the average length was plotted in function of the average width and simple linear regression was performed. Significant differences between the slopes and intercepts of the linear regression curves were determined using ANCOVA analysis.
Lastly, in the case of analyzing the effect of RNAi mediated knockdown on copulation behavior, oviposition and hatching, two different strategies were used. First, the cumulative percentages of females copulating and females laying eggs were represented by a survival curve. Differences between the different curves of the different knockdown conditions were determined using the Mantel–Cox test. Second, the number of days between egg copulation and egg laying, the number of laid eggs, the number of days between egg laying and hatching, the number of hatchlings and the hatchling success were represented as the mean ± S.E.M. The data were analyzed for a normal distribution using the Shapiro–Wilk test and in case of normally distributed data, significant differences were determined using one-way ANOVA analysis with Dunnett’s multiple comparisons correction. In case the results did not constitute a normal distribution, significant differences were determined using Kruskal–Wallis analysis with Dunn’s multiple comparisons correction.
Supplementary Materials
These are available online at https://www.mdpi.com/article/10.3390/ijms23169232/s1.
Author Contributions
Experiments were designed by S.S., C.L., E.M. and J.V.B.; S.S, C.L., M.d.R.P.B., D.C. and P.P. performed the experiments and S.S. and C.L. analyzed the obtained data. E.M. and J.V.B. supervised the project. The first draft of the manuscript was written by S.S., C.L. and J.V.B. and further reviewed and edited by all authors. J.V.B. was the principle investigator responsible for funding acquisition and project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Special Research Fund of KU Leuven (GOA/11/02; C14/15/050; C14/19/069), the European Union’s Horizon 2020 Research and Innovation program (No. 634361), and the Research Foundation of Flanders (FWO) (G0F2417N; G090919N) to J.V.B. S.S. obtained a PhD fellowship from FWO (1S42119N). C.L. received a PhD fellowship from the Agency for Innovation by Science and Technology (IWT; 121547) and a postdoctoral mandate from the KU Leuven internal funds (PDM 18/111).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data incorporated in the figures can be made available upon request to the corresponding author.
Acknowledgments
The authors would like to thank Evert Bruyninckx and Evelien Herinckx for their technical support and Rik Verdonck, Lina Verbakel and Rania Abou El Asrar for their work on the S. gregaria transcriptome databases. They are also very grateful to Lina Verbakel and Joachim Van Lommel for their help during the dissections and to Jean-Paul Delbecque (Université de Bordeaux, France) for his kind gifts of antiserum and tracer.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Oligonucleotide sequences of primers used in qRT-PCR.
Table A1.
Oligonucleotide sequences of primers used in qRT-PCR.
| Reference genes | Forward primer | Reverse primer |
|---|---|---|
| α-tubulin1A | 5′-TGACAATGAGGCCATCTATG-3′ | 5′-TGCTTCCATACCCAGGAATGA-3′ |
| CG13220 | 5′-TGTTCAGTTTTGGCTCTGTTCTGA-3′ | 5′-ACTGTTCTCCGGCAGAATGC-3′ |
| Ubi | 5′-GACTTTGAGGTGTGGCGTAG-3′ | 5′-GGATCACAAACACAGAACGA-3′ |
| RP49 | 5′-CGCTACAAGAAGCTTAAGAGGTCAT-3′ | 5′-CCTACGGCGCACTCTGTTG-3′ |
| β-actin | 5′-AATTACCATTGGTAACGAGCGATT-3′ | 5′-TGCTTCCATACCCAGGAATGA-3′ |
| EF1α | 5′-GATGCTCCAGGCCACAGAGA-3′ | 5′-TGCACAGTCGGCCTGTGAT-3′ |
| GAPDH | 5′-GTCTGATGACAACAGTGCAT-3′ | 5′-GTCCATCACGCCACAACTTTC-3′ |
| Target genes | Forward primer | Reverse primer |
| SchgrSpo | 5′-CAACATCTTCACCAGCTACATGTG-3′ | 5′-GGGTCGTCGTAGTCGAAGGA-3′ |
| SchgrPhm | 5′-CGCAGAGCCCGGACAAC-3′ | 5′-CGAACATGTCGGCCATGA-3′ |
| SchgrDib | 5′-CCCAGGCTGCTATCGAGACT-3′ | 5′-CGACGACCAGGCCTATGTAGTT-3′ |
| SchgrSad | 5′-ATCGTGGCCGAGATTACGAA-3′ | 5′-AGCACCATCTCCGGATCCT-3′ |
| SchgrShd | 5′-CCGCCGTCATGACTTCATA-3′ | 5′-GTGAGCTCCCAAGCGTGG-3′ |
| SchgrEcR | 5′-AAGGTTGATAATGCGGAATATGC-3′ | 5′-GTGATGGGCGCTCTGAAAAT-3′ |
| SchgrRXR | 5′-AATGCCTCGCTATGGGAATG-3′ | 5′-TCCTTTGTTCGCTGCCTTTC-3′ |
| SchgrVg1 | 5′-CCGCTGAACATCACTGCAAT-3′ | 5′-ACTTGGGCCAAATGGATGAG-3′ |
| SchgrVg2 | 5′-GCTACCCGCAATCTGTAAAATACA-3′ | 5′-CGACTGTGAAAGGGCATTGA-3′ |
| SchgrKr-h1 | 5′-CTCCAAGACGTTCATCCAGAG-3′ | 5′-TGCTTGGAGCAGGTGAAG-3′ |
| SchgrMet | 5′-GGTGCCTGAAGAGGAAGAAA-3′ | 5′-ATGGAGGTGATGAAGGAGAAAG-3′ |
Abbreviations: Ubi = ubiquitin conjugating enzyme 10, RP49 = ribosomal protein 49, EF1α = elongation factor 1 alpha, GAPDH = glyceraldehyde 3-phosphate dehydrogenase, Spo = spook, Phm = phantom, Dib = disembodied, Sad = shadow, Shd = shade, EcR = ecdysone receptor, RXR = retinoid-X-receptor, Vg = vitellogenin, Kr-h1 = krüppel-homolog 1, Met = methoprene-tolerant (JH receptor).

Table A2.
Oligonucleotide sequences of primers used for dsRNA construct design. Underlined sequences are the T7 promoter sequences.
Table A2.
Oligonucleotide sequences of primers used for dsRNA construct design. Underlined sequences are the T7 promoter sequences.
| Target Genes | Forward Primer | Reverse Primer |
|---|---|---|
| SchgrSpo | 5′-TAATACGACTCACTATAGGGAGA GTGGACTTCATGCCGTGGCT-3′ | 5′-TAATACGACTCACTATAGGGAGA AGGATGGTGGCCTCGGTGAA-3′ |
| SchgrPhm | 5′-TAATACGACTCACTATAGGGAGA GCGCAACCTGGGCATGGTGAAGGC-3′ | 5′-TAATACGACTCACTATAGGGAGA CGGCGACGCCGATGAGCCTGGTGC-3′ |
| SchgrDib | 5′-TAATACGACTCACTATAGGGAGA TAGCTGGAATGGACACAACATC-3′ | 5′-TAATACGACTCACTATAGGGAGA CTGGGTCTGGGAAATACTCTGG-3′ |
| SchgrSad | 5′-TAATACGACTCACTATAGGGAGA CCTTCCTGTCGCGATACCT-3′ | 5′- TAATACGACTCACTATAGGGAGA GTCTCGGCGAGCTTCTGG-3′ |
| SchgrShd | 5′-TAATACGACTCACTATAGGGAGA CTAGTGCCTCATGGCGCTC-3′ | 5′-TAATACGACTCACTATAGGGAGA TGAGGAGTTCAGGACTGTGGTTT-3′ |
| GFP | 5′-TAATACGACTCACTATAGGGAGA AAGGTGATGCTACATACGGAA-3′ | 5′-TAATACGACTCACTATAGGGAGA ATCCCAGCAGCAGTTACAAAC-3′ |
References
- Delbecque, J.-P.; Weidner, K.; Hoffmann, K.H. Alternative Sites for Ecdysteroid Production in Insects. Invertebr. Reprod. Dev. 1990, 18, 29–42. [Google Scholar] [CrossRef]
- Gilbert, L.I.; Rybczynski, R.; Warren, J.T. Control and Biochemical Nature of the Ecdysteroidogenic Pathway. Annu. Rev. Entomol. 2002, 47, 883–916. [Google Scholar] [CrossRef] [PubMed]
- Lafont, R.; Koolman, J. Diversity of Ecdysteroids in Animal Species. In Ecdysone: Structures and Functions; Springer: Dordrecht, The Netherlands, 2009; pp. 47–71. [Google Scholar]
- Yao, T.P.; Forman, B.M.; Jiang, Z.; Cherbas, L.; Chen, J.D.; McKeown, M.; Cherbas, P.; Evans, R.M. Functional Ecdysone Receptor Is the Product of EcR and Ultraspiracle Genes. Nature 1993, 366, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Richards, G. Sequential Gene Activation by Ecdysone in Polytene Chromosomes of Drosophila melanogaster, III. Consequences of Ecdysone Withdrawal. Dev. Biol. 1976, 54, 241–255. [Google Scholar] [CrossRef]
- Urness, L.D.; Thummel, C.S. Molecular Interactions within the Ecdysone Regulatory Hierarchy: DNA Binding Properties of the Drosophila Ecdysone-Inducible E74A Protein. Cell 1990, 63, 47–61. [Google Scholar] [CrossRef]
- Hill, R.J.; Segraves, W.A.; Choi, D.; Underwood, P.A.; Macavoy, E. The Reaction with Polytene Chromosomes of Antibodies Raised against Drosophila E75A Protein. Insect Biochem. Mol. Biol. 1993, 23, 99–104. [Google Scholar] [CrossRef]
- Guay, P.S.; Guild, G.M. The Ecdysone-Induced Puffing Cascade in Drosophila Salivary Glands: A Broad-Comples Early Gene Regulates Intermolt and Late Gene Transcription. Genetics 1991, 129, 169–175. [Google Scholar] [CrossRef]
- Stone, B.L.; Thummel, C.S. The Drosophila 78C Early Late Puff Contains E78, an Ecdysone-Inducible Gene That Encodes a Novel Member of the Nuclear Hormone Receptor Superfamily. Cell 1993, 75, 307–320. [Google Scholar] [CrossRef]
- Koelle, M.R.; Segraves, W.A.; Hogness, D.S. DHR3: A Drosophila Steroid Receptor Homolog. Proc. Natl. Acad. Sci. USA 1992, 89, 6167–6171. [Google Scholar] [CrossRef]
- Huet, F.; Ruiz, C.; Richards, G. Sequential Gene Activation by Ecdysone in Drosophila melanogaster: The Hierarchical Equivalence of Early and Early Late Genes. Development 1995, 121, 1195–1204. [Google Scholar] [CrossRef]
- White, K.P.; Hurban, P.; Watanabe, T.; Hogness, D.S. Coordination of Drosophila Metamorphosis by Two Ecdysone-Induced Nuclear Receptors. Science 1997, 276, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, A.; Martín, F.A.; Martín, D.; Ferrús, A. Ligand-Independent Requirements of Steroid Receptors EcR and USP for Cell Survival. Cell Death Differ. 2016, 23, 405–416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ono, H.; Rewitz, K.F.; Shinoda, T.; Itoyama, K.; Petryk, A.; Rybczynski, R.; Jarcho, M.; Warren, J.T.; Marqués, G.; Shimell, M.J.; et al. Spook and Spookier Code for Stage-Specific Components of the Ecdysone Biosynthetic Pathway in Diptera. Dev. Biol. 2006, 298, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.T.; O’Connor, M.B.; Gilbert, L.I. Studies on the Black Box: Incorporation of 3-Oxo-7-Dehydrocholesterol into Ecdysteroids by Drosophila melanogaster and Manduca sexta. Insect Biochem. Mol. Biol. 2009, 39, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Matsuda, T.; Yoshiyama, T.; Namiki, T.; Mita, K.; Fujimoto, Y.; Kataoka, H. CYP306A1, a Cytochrome P450 Enzyme, Is Essential for Ecdysteroid Biosynthesis in the Prothoracic Glands of Bombyx and Drosophila. J. Biol. Chem. 2004, 279, 35942–35949. [Google Scholar] [CrossRef]
- Warren, J.T.; Petryk, A.; Marqués, G.; Parvy, J.P.; Shinoda, T.; Itoyama, K.; Kobayashi, J.; Jarcho, M.; Li, Y.; O’Connor, M.B.; et al. Phantom Encodes the 25-Hydroxylase of Drosophila melanogaster and Bombyx mori: A P450 Enzyme Critical in Ecdysone Biosynthesis. Insect Biochem. Mol. Biol. 2004, 34, 991–1010. [Google Scholar] [CrossRef]
- Chávez, V.M.; Marqués, G.; Delbecque, J.P.; Kobayashi, K.; Hollingsworth, M.; Burr, J.; Natzle, J.E.; O’Connor, M.B. The Drosophila Disembodied Gene Controls Late Embryonic Morphogenesis and Codes for a Cytochrome P450 Enzyme That Regulates Embryonic Ecdysone Levels. Development 2000, 127, 4115–4126. [Google Scholar] [CrossRef]
- Warren, J.T.; Petryk, A.; Marqués, G.; Jarcho, M.; Parvy, J.; Dauphin-villemant, C.; O’Connor, M.B.; Gilbert, L.I. Molecular and Biochemical Characterization of Two P450 Enzymes in the Ecdysteroidogenic Pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 11043–11048. [Google Scholar] [CrossRef]
- Petryk, A.; Warren, J.T.; Marqués, G.; Jarcho, M.P.; Gilbert, L.I.; Kahler, J.; Parvy, J.P.; Li, Y.; Dauphin-Villemant, C.; O’Connor, M.B. Shade Is the Drosophila P450 Enzyme That Mediates the Hydroxylation of Ecdysone to the Steroid Insect Molting Hormone 20-Hydroxyecdysone. Proc. Natl. Acad. Sci. USA 2003, 100, 13773–13778. [Google Scholar] [CrossRef]
- Gilbert, L.I.; Warren, J.T. A Molecular Genetic Approach to the Biosynthesis of the Insect Steroid Molting Hormone. Vitam. Horm. 2005, 73, 31–57. [Google Scholar] [CrossRef]
- Lafont, R.; Dauphin–Villemant, C.; Warren, J.T.; Rees, H. Ecdysteroid Chemistry and Biochemistry. In Comprehensive Molecular Insect Science; Gilbert, L.I., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 125–195. ISBN 978-0-444-51924-5. [Google Scholar]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. The Halloween Genes Code for Cytochrome P450 Enzymes Mediating Synthesis of the Insect Moulting Hormone. Biochem. Soc. Trans. 2006, 34, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Girardie, J.; Girardie, A. Endocrine Regulation of Oogenesis in Insects. Ann. N. Y. Acad. Sci. 1998, 839, 118–122. [Google Scholar] [CrossRef]
- Gancz, D.; Lengil, T.; Gilboa, L. Coordinated Regulation of Niche and Stem Cell Precursors by Hormonal Signaling. PLoS Biol. 2011, 9, e1001202. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.X.; Spradling, A.C. Steroid Signaling within Drosophila Ovarian Epithelial Cells Sex-Specifically Modulates Early Germ Cell Development and Meiotic Entry. PLoS ONE 2012, 7, e46109. [Google Scholar] [CrossRef]
- Terashima, J.; Bownes, M. Translating Available Food into the Number of Eggs Laid by Drosophila melanogaster. Genet. Soc. Am. 2004, 167, 1711–1719. [Google Scholar] [CrossRef]
- Buszczak, M.; Freeman, M.R.; Carlson, J.R.; Bender, M.; Cooley, L.; Segraves, W.A. Ecdysone Response Genes Govern Egg Chamber Development during Mid-Oogenesis in Drosophila. Development 1999, 126, 4581–4589. [Google Scholar] [CrossRef]
- Carney, G.E.; Bender, M. The Drosophila Ecdysone Receptor (EcR) Gene Is Required Maternally for Normal Oogenesis. Genetics 2000, 154, 1203–1211. [Google Scholar] [CrossRef]
- Oro, A.E.; McKeown, M.; Evans, R.M. The Drosophila Retinoid X Receptor Homolog Ultraspiracle Functions in Both Female Reproduction and Eye Morphogenesis. Development 1992, 115, 449–462. [Google Scholar] [CrossRef]
- Belles, X.; Piulachs, M.-D. Ecdysone Signalling and Ovarian Development in Insects: From Stem Cells to Ovarian Follicle Formation. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2015, 1849, 181–186. [Google Scholar] [CrossRef]
- Swevers, L. An Update on Ecdysone Signaling during Insect Oogenesis. Curr. Opin. Insect Sci. 2018, 31, 8–13. [Google Scholar] [CrossRef]
- Lagueux, M.; Hirn, M.; Hoffmann, J.A. Ecdysone during Ovarian Development in Locusta migratoria. J. Insect Physiol. 1977, 23, 109–119. [Google Scholar] [CrossRef]
- Lanot, R.; Thiebold, J.; Lagueux, M.; Goltzene, F.; Hoffmann, J.A. Involvement of Ecdysone in the Control of Meiotic Reinitiation in Oocytes of Locusta migratoria (Insecta, Orthoptera). Dev. Biol. 1987, 121, 174–181. [Google Scholar] [CrossRef]
- Cruz, J.; Mané-Padrós, D.; Bellés, X.; Martín, D. Functions of the Ecdysone Receptor Isoform-A in the Hemimetabolous Insect Blattella Germanica Revealed by Systemic RNAi in Vivo. Dev. Biol. 2006, 297, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Bellés, X.; Cassier, P.; Cerdá, X.; Pascual, N.; André, M.; Rósso, Y.; Piulachs, M.D. Induction of Choriogenesis by 20-Hydroxyecdysone in the German Cockroach. Tissue Cell 1993, 25, 195–204. [Google Scholar] [CrossRef]
- Lenaerts, C.; Marchal, E.; Peeters, P.; Vanden Broeck, J. The Ecdysone Receptor Complex Is Essential for the Reproductive Success in the Female Desert Locust, Schistocerca gregaria. Sci. Rep. 2019, 9, 15. [Google Scholar] [CrossRef]
- Marchal, E.; Verlinden, H.; Badisco, L.; Van Wielendaele, P.; Vanden Broeck, J. RNAi-Mediated Knockdown of Shade Negatively Affects Ecdysone-20-Hydroxylation in the Desert Locust, Schistocerca gregaria. J. Insect Physiol. 2012, 58, 890–896. [Google Scholar] [CrossRef]
- Marchal, E.; Badisco, L.; Verlinden, H.; Vandersmissen, T.; Van Soest, S.; Van Wielendaele, P.; Vanden Broeck, J. Role of the Halloween Genes, Spook and Phantom in Ecdysteroidogenesis in the Desert Locust, Schistocerca gregaria. J. Insect Physiol. 2011, 57, 1240–1248. [Google Scholar] [CrossRef]
- Verlinden, H.; Sterck, L.; Li, J.; Li, Z.; Yssel, A.; Gansemans, Y.; Verdonck, R.; Holtof, M.; Song, H.; Behmer, S.T.; et al. First Draft Genome Assembly of the Desert Locust, Schistocerca gregaria. F1000Res 2020, 9, 775. [Google Scholar] [CrossRef]
- Zhang, X.; Kang, X.; Wu, H.; Silver, K.; Zhang, J.; Ma, E.; Yan, K. Transcriptome-Wide Survey, Gene Expression Profiling and Exogenous Chemical-Induced Transcriptional Responses of Cytochrome P450 Superfamily Genes in Migratory Locust (Locusta migratoria). Insect Biochem. Mol. Biol. 2018, 100, 66–77. [Google Scholar] [CrossRef]
- Niwa, R.; Sakudoh, T.; Namiki, T.; Saida, K.; Fujimoto, Y.; Kataoka, H. The Ecdysteroidogenic P450 Cyp302a1/Disembodied from the Silkworm, Bombyx mori, Is Transcriptionally Regulated by Prothoracicotropic Hormone. Insect Mol. Biol. 2005, 14, 563–571. [Google Scholar] [CrossRef]
- Pondeville, E.; Maria, A.; Jacques, J.C.; Bourgouin, C.; Dauphin-Villemant, C. Anopheles Gambiae Males Produce and Transfer the Vitellogenic Steroid Hormone 20-Hydroxyecdysone to Females during Mating. Proc. Natl. Acad. Sci. USA 2008, 105, 19631–19636. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, Y.; Zhao, Q.; Tang, S.; Huang, J.; Shen, X. Expression Profile of Several Genes on Ecdysteroidogenic Pathway Related to Diapause in Pupal Stage of Bombyx mori Bivoltine Strain. Gene 2019, 707, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Rewitz, K.F.; O’Connor, M.B.; Gilbert, L.I. Molecular Evolution of the Insect Halloween Family of Cytochrome P450s: Phylogeny, Gene Organization and Functional Conservation. Insect Biochem. Mol. Biol. 2007, 37, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Piepho, H. Letters in Mean Comparisons: What They Do and Don’t Mean. Agron. J. 2018, 110, 431–434. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect Cytochrome P450. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatrou, K., Gill, S., Eds.; Elsevier: Oxford, UK, 2005; Volume 4, pp. 1–77. ISBN 0-44-451646-8. [Google Scholar]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A Success Story. Genome Biol 2000, 1, 3003.1–3003.9. [Google Scholar] [CrossRef]
- Tawfik, A.I.; Vedrová, A.; Li, W.; Sehnal, F.; Obeng-Ofori, D. Haemolymph Ecdysteroids and the Prothoracic Glands in the Solitary and Gregarious Adults of Schistocerca gregaria. J. Insect Physiol. 1997, 43, 485–493. [Google Scholar] [CrossRef]
- Huang, X.; Warren, J.T.; Gilbert, L.I. New Players in the Regulation of Ecdysone Biosynthesis. J. Genet. Genom. 2008, 35, 1–10. [Google Scholar] [CrossRef]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. Identification, Characterization and Developmental Expression of Halloween Genes Encoding P450 Enzymes Mediating Ecdysone Biosynthesis in the Tobacco Hornworm, Manduca Sexta. Insect Biochem. Mol. Biol. 2006, 36, 188–199. [Google Scholar] [CrossRef]
- Sieglaff, D.H.; Duncan, K.A.; Brown, M.R. Expression of Genes Encoding Proteins Involved in Ecdysteroidogenesis in the Female Mosquito, Aedes Aegypti. Insect Biochem. Mol. Biol. 2005, 35, 471–490. [Google Scholar] [CrossRef]
- Zhou, X.; Ye, Y.-Z.; Ogihara, M.H.; Takeshima, M.; Fujinaga, D.; Liu, C.-W.; Zhu, Z.; Kataoka, H.; Bao, Y.-Y. Functional Analysis of Ecdysteroid Biosynthetic Enzymes of the Rice Planthopper, Nilaparvata Lugens. Insect Biochem. Mol. Biol. 2020, 123, 103428. [Google Scholar] [CrossRef]
- Ramos, S.; Chelemen, F.; Pagone, V.; Elshaer, N.; Irles, P.; Piulachs, M.D. Eyes Absent in the Cockroach Panoistic Ovaries Regulates Proliferation and Differentiation through Ecdysone Signalling. Insect Biochem. Mol. Biol. 2020, 123, 103407. [Google Scholar] [CrossRef] [PubMed]
- Irles, P.; Elshaer, N.; Piulachs, M.-D. The Notch Pathway Regulates Both the Proliferation and Differentiation of Follicular Cells in the Panoistic Ovary of Blattella Germanica. Open Biol. 2016, 6, 150197. [Google Scholar] [CrossRef] [PubMed]
- Bäumer, D.; Ströhlein, N.M.; Schoppmeier, M. Opposing Effects of Notch-Signaling in Maintaining the Proliferative State of Follicle Cells in the Telotrophic Ovary of the Beetle Tribolium. Front. Zool. 2012, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C. Notch Signaling: Control of Cell Communication and Cell Fate. Development 2004, 131, 965 LP–973 LP. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.K.; Deng, W.-M.; Holder, K.; Tworoger, M.; Clegg, N.; Ruohola-Baker, H. Role of Notch Pathway in Terminal Follicle Cell Differentiation during Drosophila Oogenesis. Dev. Genes Evol. 1999, 209, 301–311. [Google Scholar] [CrossRef]
- Ruohola, H.; Bremer, K.A.; Baker, D.; Swedlow, J.R.; Jan, L.Y.; Jan, Y.N. Role of Neurogenic Genes in Establishment of Follicle Cell Fate and Oocyte Polarity during Oogenesis in Drosophila. Cell 1991, 66, 433–449. [Google Scholar] [CrossRef]
- Wilson, M.J.; Helen, A.; Dearden, P.K. The Evolution of Oocyte Patterning in Insects: Multiple Cell-signaling Pathways Are Active during Honeybee Oogenesis and Are Likely to Play a Role in Axis Patterning. Evol. Dev. 2011, 13, 127–137. [Google Scholar] [CrossRef]
- Yatsenko, A.S.; Shcherbata, H.R. Stereotypical Architecture of the Stem Cell Niche Is Spatiotemporally Established by MiR-125-Dependent Coordination of Notch and Steroid Signaling. Development 2018, 145, dev159178. [Google Scholar] [CrossRef]
- Kumar, R.; Mota, L.C.; Litoff, E.J.; Rooney, J.P.; Boswell, W.T.; Courter, E.; Henderson, C.M.; Hernandez, J.P.; Corton, J.C.; Moore, D.D.; et al. Compensatory Changes in CYP Expression in Three Different Toxicology Mouse Models: CAR-Null, Cyp3a-Null, and Cyp2b9/10/13-Null Mice. PLoS ONE 2017, 12, e0174355. [Google Scholar] [CrossRef]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Hölper, S.; Krüger, M.; Stainier, D.Y.R. Genetic Compensation Induced by Deleterious Mutations but Not Gene Knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, W.; Li, Z.; Ma, L.; Yu, J.; Wang, H.; Liu, Z.; Xu, B. Identification and Characterization of Three New Cytochrome P450 Genes and the Use of RNA Interference to Evaluate Their Roles in Antioxidant Defense in Apis Cerana Cerana Fabricius. Front. Physiol. 2018, 9, 1608. [Google Scholar] [CrossRef] [PubMed]
- Berrigan, D. The Allometry of Egg Size and Number in Insects. Oikos 1991, 60, 313. [Google Scholar] [CrossRef]
- Maeno, K.O.; Piou, C.; Ghaout, S. The Desert Locust, Schistocerca gregaria, Plastically Manipulates Egg Size by Regulating Both Egg Numbers and Production Rate According to Population Density. J. Insect Physiol. 2020, 122, 104020. [Google Scholar] [CrossRef]
- Tawfik, A.I.; Vedrová, A.; Sehnal, F. Ecdysteroids during Ovarian Development and Embryogenesis in Solitary and Gregarious Schistocerca gregaria. Arch. Insect Biochem. Physiol. 1999, 41, 134–143. [Google Scholar] [CrossRef]
- Parthasarathy, R.; Sheng, Z.; Sun, Z.; Palli, S.R. Ecdysteroid Regulation of Ovarian Growth and Oocyte Maturation in the Red Flour Beetle, Tribolium Castaneum. Insect Biochem. Mol. Biol. 2010, 40, 429–439. [Google Scholar] [CrossRef]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.T.; Orteho, F.; et al. The Genome of Tetranychus Urticae Reveals Herbivorous Pest Adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef]
- Tawfik, A.I.; Tanaka, Y.; Tanaka, S. Possible Involvement of Ecdysteroids in Photoperiodically Induced Suppresion of Ovarian Development in a Japanese Strain of the Migratory Locust, Locusta migratoria. J. Insect Physiol. 2002, 48, 411–418. [Google Scholar] [CrossRef]
- Bownes, M.; Shirras, A.; Blair, M.; Collins, J.; Coulson, A. Evidence That Insect Embryogenesis Is Regulated by Ecdysteroids Released from Yolk Proteins. Proc. Natl. Acad. Sci. USA 1988, 85, 1554–1557. [Google Scholar] [CrossRef]
- Isaac, R.E.; Rees, H.H. Isolation and Identification of Ecdysteroid Phosphates and Acetylecdysteroid Phosphates from Developing Eggs of the Locust, Schistocerca gregaria. Biochem. J. 1984, 221, 459–464. [Google Scholar] [CrossRef]
- Lenaerts, C.; Van Wielendaele, P.; Peeters, P.; Vanden Broeck, J.; Marchal, E. Ecdysteroid Signalling Components in Metamorphosis and Development of the Desert Locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2016, 75, 10–23. [Google Scholar] [CrossRef]
- Hult, E.F.; Huang, J.; Marchal, E.; Lam, J.; Tobe, S.S. RXR/USP and EcR Are Critical for the Regulation of Reproduction and the Control of JH Biosynthesis in Diploptera Punctata. J. Insect Physiol. 2015, 80, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, M.H.; Hikiba, J.; Iga, M.; Kataoka, H. Negative Regulation of Juvenile Hormone Analog for Ecdysteroidogenic Enzymes. J. Insect Physiol. 2015, 80, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Konopova, B.; Smykal, V.; Jindra, M. Common and Distinct Roles of Juvenile Hormone Signaling Genes in Metamorphosis of Holometabolous and Hemimetabolous Insects. PLoS ONE 2011, 6, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Lozano, J.; Belles, X. Conserved Repressive Function of Krüppel Homolog 1 on Insect Metamorphosis in Hemimetabolous and Holometabolous Species. Sci. Rep. 2011, 1, 1–7. [Google Scholar] [CrossRef]
- Minakuchi, C.; Namiki, T.; Shinoda, T. Krüppel Homolog 1, an Early Juvenile Hormone-Response Gene Downstream of Methoprene-Tolerant, Mediates Its Anti-Metamorphic Action in the Red Flour BeetleTribolium Castaneum. Dev. Biol. 2009, 325, 341–350. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, C.; Liu, C.; Song, Q.; Zhou, S. Rhythmic Change of Adipokinetic Hormones Diurnally Regulates Locust Vitellogenesis and Egg Development. Insect Mol. Biol. 2020, 29, 283–292. [Google Scholar] [CrossRef]
- Wallace, I.M.; O’Sullivan, O.; Higgins, D.G.; Notredame, C. M-Coffee: Combining Multiple Sequence Alignment Methods with T-Coffee. Nucleic Acids Res. 2006, 34, 1692–1699. [Google Scholar] [CrossRef]
- Lenaerts, C.; Palmans, J.; Marchal, E.; Verdonck, R.; Vanden Broeck, J. Role of the Venus Kinase Receptor in the Female Reproductive Physiology of the Desert Locust, Schistocerca gregaria. Sci. Rep. 2017, 7, 11730. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internat Control Genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Van Hiel, M.B.; Van Wielendaele, P.; Temmerman, L.; Van Soest, S.; Vuerinckx, K.; Huybrechts, R.; Vanden Broeck, J.; Simonet, G. Identification and Validation of Housekeeping Genes in Brains of the Desert Locust Schistocerca gregaria under Different Developmental Conditions. BMC Mol. Biol. 2009, 10, 56. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. QBase Relative Quantification Framework and Software for Management and Automated Analysis of Real-Time Quantitative PCR Data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).