The Features of Immune Checkpoint Gene Regulation by microRNA in Cancer
Abstract
:1. Introduction
2. Immune Checkpoints
2.1. PD-1, PD-L1 and CTLA-4
2.2. Gal-9/Tim-3
2.3. VISTA
2.4. BTLA
2.5. B7-H3
2.6. ICOS/ICOSL
2.7. LAG3
3. Simultaneous Inhibition of ICs and Other Targets
3.1. ICs and VEGF
3.2. ICs and c-Met
3.3. ICs and HIF
3.4. ICs and PI3K
3.5. ICs and CXCR4
3.6. ICs and EGFR
3.7. ICs and HER2
4. Regulation of IC Genes by miRNAs
5. miRNAs as Regulators of ICs and Targeted Therapy Genes
6. Prospects for miRNA-Based Therapy
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, S.; Li, D.; Zhu, X. Cancer immunotherapy: Pros, cons and beyond. Biomed. Pharmacother. 2020, 124, 109821. [Google Scholar] [CrossRef]
- Qin, S.; Xu, L.; Yi, M.; Yu, S.; Wu, K.; Luo, S. Novel immune checkpoint targets: Moving beyond PD-1 and CTLA-4. Mol. Cancer 2019, 18, 155. [Google Scholar] [CrossRef]
- Twomey, J.D.; Zhang, B. Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. AAPS J. 2021, 23, 39. [Google Scholar] [CrossRef]
- Vafaei, S.; Zekiy, A.O.; Khanamir, R.A.; Zaman, B.A.; Ghayourvahdat, A.; Azimizonuzi, H.; Zamani, M. Combination therapy with immune checkpoint inhibitors (ICIs); a new frontier. Cancer Cell Int. 2022, 22, 2. [Google Scholar] [CrossRef]
- Esfandiari, A.; Cassidy, S.; Webster, R.M. Bispecific antibodies in oncology. Nat. Rev. Drug Discov. 2022, 21, 411–412. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jin, H.; Guo, K.; Xiang, Y.; Zhang, Y.; Du, W.; Shen, M.; Ruan, S. Results from a Meta-analysis of Combination of PD-1/PD-L1 and CTLA-4 Inhibitors in Malignant Cancer Patients: Does PD-L1 Matter? Front. Pharmacol. 2021, 12, 572845. [Google Scholar] [CrossRef]
- Zhang, Y.; Tanno, T.; Kanellopoulou, C. Cancer therapeutic implications of microRNAs in the regulation of immune checkpoint blockade. ExRNA 2019, 1, 19. [Google Scholar] [CrossRef]
- Shao, L.; He, Q.; Wang, J.; He, F.; Lin, S.; Wu, L.; Gao, Y.; Ma, W.; Dong, J.; Yang, X.; et al. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discov. 2021, 7, 145. [Google Scholar] [CrossRef]
- Xu, W.; Atkins, M.B.; McDermott, D.F. Checkpoint inhibitor immunotherapy in kidney cancer. Nat. Rev. Urol. 2020, 17, 137–150. [Google Scholar] [CrossRef]
- Soleimani, M.; Thi, M.; Saxena, N.; Khalaf, D.J.; Eigl, B.J.; Chi, K.N.; Kollmannsberger, C.K.; Nappi, L. 693P Plasma exosome microRNA-155-3p expression in patients with metastatic renal cell carcinoma treated with immune checkpoint inhibitors: Potential biomarker of response to systemic therapy. Ann. Oncol. 2021, 32, S708. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, L.; Zou, L.; Yang, P.; Wu, R.; Mao, Y.; Zhou, H.; Li, R.; Wang, K.; Wang, W.; et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum. Immunol. 2014, 75, 348–353. [Google Scholar] [CrossRef]
- Cortez, M.A.; Anfossi, S.; Ramapriyan, R.; Menon, H.; Atalar, S.C.; Aliru, M.; Welsh, J.; Calin, G.A. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 2019, 58, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.-H.T.; Luo, Y.-H.; Li, A.-L.; Tsai, J.-C.; Wu, K.-L.; Chung, P.-J.; Ma, N. miRNA as a Modulator of Immunotherapy and Immune Response in Melanoma. Biomolecules 2021, 11, 1648. [Google Scholar] [CrossRef]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Omar, H.A.; El-Serafi, A.T.; Hersi, F.; Arafa, E.A.; Zaher, D.M.; Madkour, M.; Arab, H.H.; Tolba, M.F. Immunomodulatory MicroRNAs in cancer: Targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019, 286, 3540–3557. [Google Scholar] [CrossRef]
- Skafi, N.; Fayyad-Kazan, M.; Badran, B. Immunomodulatory role for MicroRNAs: Regulation of PD-1/PD-L1 and CTLA-4 immune checkpoints expression. Gene 2020, 754, 144888. [Google Scholar] [CrossRef]
- Eichmüller, S.B.; Osen, W.; Mandelboim, O.; Seliger, B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. JNCI J. Natl. Cancer Inst. 2017, 109, djx034. [Google Scholar] [CrossRef]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef]
- Kooshkaki, O.; Derakhshani, A.; Hosseinkhani, N.; Torabi, M.; Safaei, S.; Brunetti, O.; Racanelli, V.; Silvestris, N.; Baradaran, B. Combination of Ipilimumab and Nivolumab in Cancers: From Clinical Practice to Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 4427. [Google Scholar] [CrossRef]
- Acharya, N.; Sabatos-Peyton, C.; Anderson, A.C. Tim-3 finds its place in the cancer immunotherapy landscape. J. Immunother. Cancer 2020, 8, e000911. [Google Scholar] [CrossRef]
- Yuan, J.; Jiang, B.; Zhao, H.; Huang, Q. Prognostic implication of TIM-3 in clear cell renal cell carcinoma. Neoplasma 2014, 61, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Dariane, C.; Combe, P.; Verkarre, V.; Urien, S.; Badoual, C.; Roussel, H.; Mandavit, M.; Ravel, P.; Sibony, M.; et al. Tim-3 Expression on Tumor-Infiltrating PD-1+CD8+ T Cells Correlates with Poor Clinical Outcome in Renal Cell Carcinoma. Cancer Res. 2017, 77, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, V.L.; Heusschen, R.; Caers, J.; Griffioen, A.W. Galectin expression in cancer diagnosis and prognosis: A systematic review. Biochim. Biophys. Acta-Rev. Cancer 2015, 1855, 235–247. [Google Scholar] [CrossRef]
- Yasinska, I.M.; Sakhnevych, S.S.; Pavlova, L.; Teo Hansen Selnø, A.; Teuscher Abeleira, A.M.; Benlaouer, O.; Gonçalves Silva, I.; Mosimann, M.; Varani, L.; Bardelli, M.; et al. The Tim-3-Galectin-9 Pathway and Its Regulatory Mechanisms in Human Breast Cancer. Front. Immunol. 2019, 10, 1594. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Sun, L.; Li, C.-F.; Wang, Y.-H.; Yao, J.; Li, H.; Yan, M.; Chang, W.-C.; Hsu, J.-M.; Cha, J.-H.; et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat. Commun. 2021, 12, 832. [Google Scholar] [CrossRef] [PubMed]
- Jikuya, R.; Kishida, T.; Sakaguchi, M.; Yokose, T.; Yasui, M.; Hashizume, A.; Tatenuma, T.; Mizuno, N.; Muraoka, K.; Umemoto, S.; et al. Galectin-9 expression as a poor prognostic factor in patients with renal cell carcinoma. Cancer Immunol. Immunother. 2020, 69, 2041–2051. [Google Scholar] [CrossRef]
- Fu, H.; Liu, Y.; Xu, L.; Liu, W.; Fu, Q.; Liu, H.; Zhang, W.; Xu, J. Galectin-9 predicts postoperative recurrence and survival of patients with clear-cell renal cell carcinoma. Tumor Biol. 2015, 36, 5791–5799. [Google Scholar] [CrossRef]
- Kawashima, H.; Obayashi, A.; Kawamura, M.; Masaki, S.; Tamada, S.; Iguchi, T.; Uchida, J.; Kuratsukuri, K.; Tanaka, T.; Nakatani, T. Galectin 9 and PINCH, novel immunotherapy targets of renal cell carcinoma: A rationale to find potential tumour antigens and the resulting cytotoxic T lymphocytes induced by the derived peptides. BJU Int. 2014, 113, 320–332. [Google Scholar] [CrossRef]
- Dama, P.; Tang, M.; Fulton, N.; Kline, J.; Liu, H. Gal9/Tim-3 expression level is higher in AML patients who fail chemotherapy. J. Immunother. Cancer 2019, 7, 175. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, L.; Jing, D.; Xu, G.; Zhang, J.; Lin, L.; Zhao, J.; Yao, Z.; Lin, H. Galectin-9 Expression Predicts Favorable Clinical Outcome in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Physiol. 2018, 9, 452. [Google Scholar] [CrossRef]
- Yang, R.; Sun, L.; Li, C.-F.; Wang, Y.-H.; Xia, W.; Liu, B.; Chu, Y.-Y.; Bover, L.; Vien, L.; Hung, M.-C. Development and characterization of anti-galectin-9 antibodies that protect T cells from galectin-9-induced cell death. J. Biol. Chem. 2022, 298, 101821. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Yuan, Q.; Xia, H.; Zhu, G.; Feng, Y.; Wang, Q.; Zhang, Z.; He, W.; Lu, J.; Dong, C.; et al. Analysis of VISTA expression and function in renal cell carcinoma highlights VISTA as a potential target for immunotherapy. Protein Cell 2019, 10, 840–845. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dong, J.; Zheng, Y.; Zhou, J.; Yuan, Y.; Ta, H.M.; Miller, H.E.; Olson, M.; Rajasekaran, K.; Ernstoff, M.S.; et al. Immune-Checkpoint Protein VISTA Regulates Antitumor Immunity by Controlling Myeloid Cell–Mediated Inflammation and Immunosuppression. Cancer Immunol. Res. 2019, 7, 1497–1510. [Google Scholar] [CrossRef]
- Kondo, Y.; Ohno, T.; Nishii, N.; Harada, K.; Yagita, H.; Azuma, M. Differential contribution of three immune checkpoint (VISTA, CTLA-4, PD-1) pathways to antitumor responses against squamous cell carcinoma. Oral Oncol. 2016, 57, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Liu, K.; Xiong, H. Roles of BTLA in Immunity and Immune Disorders. Front. Immunol. 2021, 12, 654960. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Tu, H.; Liang, D.; Chang, D.W.; Ye, Y.; Wu, X. Soluble immune checkpoint-related proteins as predictors of tumor recurrence, survival, and T cell phenotypes in clear cell renal cell carcinoma patients. J. Immunother. Cancer 2019, 7, 334. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Cao, X.; Li, Y.; Li, G.-Q.; He, Q.-H.; Li, S.-J.; Chen, J.; Xu, G.-L.; Zhang, K.-Q. High expression of herpes virus entry mediator is associated with poor prognosis in clear cell renal cell carcinoma. Am. J. Cancer Res. 2019, 9, 975–987. [Google Scholar]
- Hofmeyer, K.A.; Ray, A.; Zang, X. The contrasting role of B7-H3. Proc. Natl. Acad. Sci. USA 2008, 105, 10277–10278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ji, J.; Zhang, G.; Fang, C.; Jiang, F.; Ma, S.; Hou, J. Expression and significance of B7-H3 and Tie-2 in the tumor vasculature of clear cell renal carcinoma. OncoTargets Ther. 2017, 10, 5417–5424. [Google Scholar] [CrossRef]
- Xie, J.; Sun, M.; Zhang, D.; Chen, C.; Lin, S.; Zhang, G. Fibronectin enhances tumor metastasis through B7-H3 in clear cell renal cell carcinoma. FEBS Open Bio 2021, 11, 2977–2987. [Google Scholar] [CrossRef]
- Kontos, F.; Michelakos, T.; Kurokawa, T.; Sadagopan, A.; Schwab, J.H.; Ferrone, C.R.; Ferrone, S. B7-H3: An Attractive Target for Antibody-based Immunotherapy. Clin. Cancer Res. 2021, 27, 1227–1235. [Google Scholar] [CrossRef]
- Xu, Y.; Xiao, Y.; Luo, C.; Liu, Q.; Wei, A.; Yang, Y.; Zhao, L.; Wang, Y. Blocking PD-1/PD-L1 by an ADCC enhanced anti-B7-H3/PD-1 fusion protein engages immune activation and cytotoxicity. Int. Immunopharmacol. 2020, 84, 106584. [Google Scholar] [CrossRef] [PubMed]
- You, G.; Lee, Y.; Kang, Y.-W.; Park, H.W.; Park, K.; Kim, H.; Kim, Y.-M.; Kim, S.; Kim, J.-H.; Moon, D.; et al. B7-H3 × 4-1BB bispecific antibody augments antitumor immunity by enhancing terminally differentiated CD8+ tumor-infiltrating lymphocytes. Sci. Adv. 2021, 7, aax3160. [Google Scholar] [CrossRef]
- Fu, T.; He, Q.; Sharma, P. The ICOS/ICOSL Pathway Is Required for Optimal Antitumor Responses Mediated by Anti–CTLA-4 Therapy. Cancer Res. 2011, 71, 5445–5454. [Google Scholar] [CrossRef]
- Solinas, C.; Gu-Trantien, C.; Willard-Gallo, K. The rationale behind targeting the ICOS-ICOS ligand costimulatory pathway in cancer immunotherapy. ESMO Open 2020, 5, e000544. [Google Scholar] [CrossRef]
- Wang, M.; Du, Q.; Jin, J.; Wei, Y.; Lu, Y.; Li, Q. LAG3 and its emerging role in cancer immunotherapy. Clin. Transl. Med. 2021, 11, e365. [Google Scholar] [CrossRef]
- Zelba, H.; Bedke, J.; Hennenlotter, J.; Mostböck, S.; Zettl, M.; Zichner, T.; Chandran, A.; Stenzl, A.; Rammensee, H.-G.; Gouttefangeas, C. PD-1 and LAG-3 Dominate Checkpoint Receptor–Mediated T-cell Inhibition in Renal Cell Carcinoma. Cancer Immunol. Res. 2019, 7, 1891–1899. [Google Scholar] [CrossRef]
- Wang, J.; Sanmamed, M.F.; Datar, I.; Su, T.T.; Ji, L.; Sun, J.; Chen, L.; Chen, Y.; Zhu, G.; Yin, W.; et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell 2019, 176, 334–347.e12. [Google Scholar] [CrossRef]
- Braun, D.A.; Bakouny, Z.; Hirsch, L.; Flippot, R.; Van Allen, E.M.; Wu, C.J.; Choueiri, T.K. Beyond conventional immune-checkpoint inhibition—Novel immunotherapies for renal cell carcinoma. Nat. Rev. Clin. Oncol. 2021, 18, 199–214. [Google Scholar] [CrossRef]
- Formisano, L.; Jansen, V.M.; Marciano, R.; Bianco, R. From Biology to Therapy: Improvements of Therapeutic Options in Lung Cancer. Anticancer Agents Med. Chem. 2019, 18, 1235–1240. [Google Scholar] [CrossRef]
- Bagegni, N.A.; Park, H.; Kraft, K.; O-Toole, M.; Gao, F.; Waqar, S.N.; Ratner, L.; Morgensztern, D.; Devarakonda, S.; Amin, M.; et al. Phase 1b trial of anti-VEGF/PDGFR vorolanib combined with immune checkpoint inhibitors in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2022, 89, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-H.; Lee, H.J. The Frontline Immunotherapy-Based Treatment of Advanced Clear Cell Renal Cell Carcinoma: Current Evidence and Clinical Perspective. Biomedicines 2022, 10, 251. [Google Scholar] [CrossRef]
- Uchikawa, E.; Chen, Z.; Xiao, G.-Y.; Zhang, X.; Bai, X. Structural basis of the activation of c-MET receptor. Nat. Commun. 2021, 12, 4074. [Google Scholar] [CrossRef]
- Lindner, A.K.; Pichler, M.; Thurnher, M.; Pichler, R. Targeting c-Met to Improve Immune Checkpoint Inhibition in Metastatic Renal Cell Carcinoma. Eur. Urol. 2022, 81, 1–2. [Google Scholar] [CrossRef]
- Lai, Y.; Zeng, T.; Liang, X.; Wu, W.; Zhong, F.; Wu, W. Cell death-related molecules and biomarkers for renal cell carcinoma targeted therapy. Cancer Cell Int. 2019, 19, 221. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Wang, G.; Cui, X. Triple Combination Therapy with PD-1/PD-L1, BRAF, and MEK Inhibitor for Stage III–IV Melanoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 2088. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Long, Q.; Guan, B.; Mu, L. Prognostic Value of High CXCR4 Expression in Renal Cell Carcinoma: A System Review and Meta-Analysis. Dis. Markers 2015, 2015, 568980. [Google Scholar] [CrossRef]
- Vaishampayan, U.N.; McDermott, D.F.; Matrana, M.R.; Rha, S.Y.; Zurita, A.J.; Ho, T.H.; Keam, B.; Lee, J.-L.; Joseph, R.W.; Ali, S.; et al. A phase 1/2 study evaluating the efficacy and safety of the oral CXCR4 inhibitor X4P-001 in combination with axitinib in patients with advanced renal cell carcinoma. J. Clin. Oncol. 2018, 36, 4510. [Google Scholar] [CrossRef]
- Cossu-Rocca, P.; Muroni, M.R.; Sanges, F.; Sotgiu, G.; Asunis, A.; Tanca, L.; Onnis, D.; Pira, G.; Manca, A.; Dore, S.; et al. EGFR kinase-dependent and kinase-independent roles in clear cell renal cell carcinoma. Am. J. Cancer Res. 2016, 6, 71–83. [Google Scholar]
- Li, L.; Deng, L.; Meng, X.; Gu, C.; Meng, L.; Li, K.; Zhang, X.; Meng, Y.; Xu, W.; Zhao, L.; et al. Tumor-targeting anti-EGFR x anti-PD1 bispecific antibody inhibits EGFR-overexpressing tumor growth by combining EGFR blockade and immune activation with direct tumor cell killing. Transl. Oncol. 2021, 14, 100916. [Google Scholar] [CrossRef]
- Matusz-Fisher, A.; Tan, A.R. Combination of HER2-targeted agents with immune checkpoint inhibitors in the treatment of HER2-positive breast cancer. Expert Opin. Biol. Ther. 2022, 22, 385–395. [Google Scholar] [CrossRef]
- Catenacci, D.V.T.; Kang, Y.-K.; Park, H.; Uronis, H.E.; Lee, K.-W.; Ng, M.C.H.; Enzinger, P.C.; Park, S.H.; Gold, P.J.; Lacy, J.; et al. Margetuximab plus pembrolizumab in patients with previously treated, HER2-positive gastro-oesophageal adenocarcinoma (CP-MGAH22–05): A single-arm, phase 1b–2 trial. Lancet Oncol. 2020, 21, 1066–1076. [Google Scholar] [CrossRef]
- Gu, C.; Zhu, H.; Deng, L.; Meng, X.; Li, K.; Xu, W.; Zhao, L.; Liu, Y.; Zhu, Z.; Huang, H. Bispecific antibody simultaneously targeting PD1 and HER2 inhibits tumor growth via direct tumor cell killing in combination with PD1/PDL1 blockade and HER2 inhibition. Acta Pharmacol. Sin. 2022, 43, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Wang, B.; Zeng, J.; Sang, S.; Lei, J.; Lu, Y. MicroRNA-374b inhibits liver cancer progression via down regulating programmed cell death-1 expression on cytokine-induced killer cells. Oncol. Lett. 2018, 15, 4797–4804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, N.; Li, Z.; Zhu, Q.; Li, F.; Yang, C.; Han, Q.; Lv, Y.; Zhou, Z.; Liu, Z. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3′ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget 2015, 6, 18933–18944. [Google Scholar] [CrossRef]
- Zhang, Q.; Di, W.; Dong, Y.; Lu, G.; Yu, J.; Li, J.; Li, P. High serum miR-183 level is associated with poor responsiveness of renal cancer to natural killer cells. Tumor Biol. 2015, 36, 9245–9249. [Google Scholar] [CrossRef]
- Grenda, A.; Krawczyk, P.; Błach, J.; Chmielewska, I.; Kubiatowski, T.; Kieszko, S.; Wojas-Krawczyk, K.; Kucharczyk, T.; Jarosz, B.; Paśnik, I.; et al. Tissue MicroRNA Expression as a Predictor of Response to Immunotherapy in NSCLC Patients. Front. Oncol. 2021, 10, 563613. [Google Scholar] [CrossRef]
- Song, N.; Li, P.; Song, P.; Li, Y.; Zhou, S.; Su, Q.; Li, X.; Yu, Y.; Li, P.; Feng, M.; et al. MicroRNA-138-5p Suppresses Non-small Cell Lung Cancer Cells by Targeting PD-L1/PD-1 to Regulate Tumor Microenvironment. Front. Cell Dev. Biol. 2020, 8, 540. [Google Scholar] [CrossRef]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L.; et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef]
- Aichen, Z.; Kun, W.; Xiaochun, S.; Lingling, T. LncRNA FGD5-AS1 promotes the malignant phenotypes of ovarian cancer cells via targeting miR-142-5p. Apoptosis 2021, 26, 348–360. [Google Scholar] [CrossRef]
- Qu, F.; Ye, J.; Pan, X.; Wang, J.; Gan, S.; Chu, C.; Chu, J.; Zhang, X.; Liu, M.; He, H.; et al. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J. Drug Target. 2019, 27, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yu, H.; Yi, S.; Peng, X.; Su, P.; Xiao, Z.; Liu, R.; Tang, A.; Li, X.; Liu, F.; et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016, 7, 45370–45384. [Google Scholar] [CrossRef] [PubMed]
- Ashizawa, M.; Okayama, H.; Ishigame, T.; Thar Min, A.K.; Saito, K.; Ujiie, D.; Murakami, Y.; Kikuchi, T.; Nakayama, Y.; Noda, M.; et al. miRNA-148a-3p Regulates Immunosuppression in DNA Mismatch Repair–Deficient Colorectal Cancer by Targeting PD-L1. Mol. Cancer Res. 2019, 17, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Zhang, J.; Hou, L.-D.; Zhang, R.; Chen, W.; Fan, H.-N.; Huang, Y.-X.; Liu, H.; Zhu, J.-S. Upregulation of PD-L1 predicts poor prognosis and is associated with miR-191-5p dysregulation in colon adenocarcinoma. Int. J. Immunopathol. Pharmacol. 2018, 32, 205873841879031. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Guo, Q.; Li, X.; Yang, X.; Ni, H.; Wang, T.; Zhao, Q.; Liu, H.; Xing, Y.; Xi, T.; et al. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine 2019, 41, 395–407. [Google Scholar] [CrossRef]
- Yang, L.; Cai, Y.; Zhang, D.; Sun, J.; Xu, C.; Zhao, W.; Jiang, W.; Pan, C. miR-195/miR-497 Regulate CD274 Expression of Immune Regulatory Ligands in Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 371. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, X.; Yang, M.; Kan, Q.; Duan, Z. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp. Cell Res. 2019, 380, 20–28. [Google Scholar] [CrossRef]
- Dastmalchi, N.; Hosseinpourfeizi, M.A.; Khojasteh, S.M.B.; Baradaran, B.; Safaralizadeh, R. Tumor suppressive activity of miR-424-5p in breast cancer cells through targeting PD-L1 and modulating PTEN/PI3K/AKT/mTOR signaling pathway. Life Sci. 2020, 259, 118239. [Google Scholar] [CrossRef]
- Audrito, V.; Serra, S.; Stingi, A.; Orso, F.; Gaudino, F.; Bologna, C.; Neri, F.; Garaffo, G.; Nassini, R.; Baroni, G.; et al. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget 2017, 8, 15894–15911. [Google Scholar] [CrossRef]
- Mastroianni, J.; Stickel, N.; Andrlova, H.; Hanke, K.; Melchinger, W.; Duquesne, S.; Schmidt, D.; Falk, M.; Andrieux, G.; Pfeifer, D.; et al. miR-146a Controls Immune Response in the Melanoma Microenvironment. Cancer Res. 2019, 79, 183–195. [Google Scholar] [CrossRef]
- Kao, S.C.; Cheng, Y.Y.; Williams, M.; Kirschner, M.B.; Madore, J.; Lum, T.; Sarun, K.H.; Linton, A.; McCaughan, B.; Klebe, S.; et al. Tumor Suppressor microRNAs Contribute to the Regulation of PD-L1 Expression in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2017, 12, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Indovina, P.; Mattioli, E.; Forte, I.M.; Iannuzzi, C.A.; Luzzi, L.; Bellan, C.; De Summa, S.; Bucci, E.; Di Marzo, D.; et al. P53-regulated miR-320a targets PDL1 and is downregulated in malignant mesothelioma. Cell Death Dis. 2020, 11, 748. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Yan, F.; Wu, C. Overexpressed miR-195 attenuated immune escape of diffuse large B-cell lymphoma by targeting PD-L1. Biomed. Pharmacother. 2018, 98, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, R.; Zhao, H.-J.; Fu, D.; Zhong, H.-J.; Weng, X.-Q.; Qu, B.; Zhao, Y.; Wang, L.; Zhao, W.-L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer 2019, 18, 54. [Google Scholar] [CrossRef]
- Sun, J.-R.; Zhang, X.; Zhang, Y. MiR-214 prevents the progression of diffuse large B-cell lymphoma by targeting PD-L1. Cell. Mol. Biol. Lett. 2019, 24, 68. [Google Scholar] [CrossRef]
- Tao, Z.; Xu, S.; Ruan, H.; Wang, T.; Song, W.; Qian, L.; Chen, K. MiR-195/-16 Family Enhances Radiotherapy via T Cell Activation in the Tumor Microenvironment by Blocking the PD-L1 Immune Checkpoint. Cell. Physiol. Biochem. 2018, 48, 801–814. [Google Scholar] [CrossRef]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. JNCI J. Natl. Cancer Inst. 2016, 108, djv303. [Google Scholar] [CrossRef]
- Xie, W.-B.; Liang, L.-H.; Wu, K.-G.; Wang, L.-X.; He, X.; Song, C.; Wang, Y.-Q.; Li, Y.-H. MiR-140 Expression Regulates Cell Proliferation and Targets PD-L1 in NSCLC. Cell. Physiol. Biochem. 2018, 46, 654–663. [Google Scholar] [CrossRef]
- Tang, D.; Zhao, D.; Wu, Y.; Yao, R.; Zhou, L.; Lu, L.; Gao, W.; Sun, Y. The miR-3127-5p/p-STAT3 axis up-regulates PD-L1 inducing chemoresistance in non-small-cell lung cancer. J. Cell. Mol. Med. 2018, 22, 3847–3856. [Google Scholar] [CrossRef]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.-H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef]
- Wei, S.; Wang, K.; Huang, X.; Zhao, Z.; Zhao, Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int. J. Immunopathol. Pharmacol. 2019, 33, 205873841985969. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 2015, 27, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y.; et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, hep.30607. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tan, W.; Liu, S.; Huang, X.; Lin, J.; Liang, R.; Su, L.; Su, Q.; Wang, C. MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumor Biol. 2015, 36, 9049–9057. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, Y.; Sun, Y.; Yan, X.; Wang, P. miR-375 inhibits IFN-γ-induced programmed death 1 ligand�1 surface expression in head and neck squamous cell carcinoma cells by blocking JAK2/STAT1 signaling. Oncol. Rep. 2018, 39, 1461–1468. [Google Scholar] [CrossRef]
- Sheng, Q.; Zhang, Y.; Wang, Z.; Ding, J.; Song, Y.; Zhao, W. Cisplatin-mediated down-regulation of miR-145 contributes to up-regulation of PD-L1 via the c-Myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin. Exp. Immunol. 2020, 200, 45–52. [Google Scholar] [CrossRef]
- Zhu, J.; Li, Y.; Luo, Y.; Xu, J.; Liufu, H.; Tian, Z.; Huang, C.; Li, J.; Huang, C. A Feedback Loop Formed by ATG7/Autophagy, FOXO3a/miR-145 and PD-L1 Regulates Stem-Like Properties and Invasion in Human Bladder Cancer. Cancers 2019, 11, 349. [Google Scholar] [CrossRef]
- Wu, L.; Chen, Z.; Zhang, J.; Xing, Y. Effect of miR-513a-5p on etoposide-stimulating B7-H1 expression in retinoblastoma cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2012, 32, 601–606. [Google Scholar] [CrossRef]
- Miliotis, C.; Slack, F.J. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021, 518, 115–126. [Google Scholar] [CrossRef]
- Wang, W.; Li, F.; Mao, Y.; Zhou, H.; Sun, J.; Li, R.; Liu, C.; Chen, W.; Hua, D.; Zhang, X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum. Genet. 2013, 132, 641–648. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Xie, G.; Yin, Y.; Zhao, E.; Tao, K.; Li, R. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget 2017, 8, 28125–28134. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Li, W.; Li, R.; Wu, K.; Zhao, E.; Zhang, Y.; Zhang, P.; Shi, L.; Wang, D.; Yin, Y.; et al. Helicobacter Pylori Promote B7-H1 Expression by Suppressing miR-152 and miR-200b in Gastric Cancer Cells. PLoS ONE 2017, 12, e0168822. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Liu, F.; Chu, Y.; Zhai, Q.; Wei, X.; Shao, J.; Li, R.; Xu, Q.; Yu, L.; Liu, B.; et al. MicroRNA-200c Nanoparticles Sensitized Gastric Cancer Cells to Radiotherapy by Regulating PD-L1 Expression and EMT. Cancer Manag. Res. 2020, 12, 12215–12223. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Yu, J.; Chen, L.; Tao, T.; Yi, S.; Hanley, S.J.B.; Yue, J.; Watari, H.; Sakuragi, N. Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4–miR-18a pathway in cervical cancer. Oncogene 2018, 37, 5257–5268. [Google Scholar] [CrossRef]
- Miao, S.; Mao, X.; Zhao, S.; Song, K.; Xiang, C.; Lv, Y.; Jiang, H.; Wang, L.; Li, B.; Yang, X.; et al. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget 2017, 8, 62143–62153. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Zou, H. microRNA-20b-5p overexpression combing Pembrolizumab potentiates cancer cells to radiation therapy via repressing programmed death-ligand 1. Bioengineered 2022, 13, 917–929. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Zhang, L.; Chen, Y.; Dong, R.; Zhang, J.; Zhao, J.; Guo, X.; Yang, G.; Li, Y.; et al. miR-194-5p down-regulates tumor cell PD-L1 expression and promotes anti-tumor immunity in pancreatic cancer. Int. Immunopharmacol. 2021, 97, 107822. [Google Scholar] [CrossRef]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; OuYang, J.; Chen, J.; et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016, 7, 11406. [Google Scholar] [CrossRef]
- Wei, J.; Nduom, E.K.; Kong, L.-Y.; Hashimoto, Y.; Xu, S.; Gabrusiewicz, K.; Ling, X.; Huang, N.; Qiao, W.; Zhou, S.; et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro. Oncol. 2016, 18, 639–648. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, C.; Wangmo, D.; Subramanian, S. Tumor-Secreted Extracellular Vesicles Regulate T-Cell Costimulation and Can Be Manipulated To Induce Tumor-Specific T-Cell Responses. Gastroenterology 2021, 161, 560–574.e11. [Google Scholar] [CrossRef]
- Boldrini, L.; Giordano, M.; Niccoli, C.; Melfi, F.; Lucchi, M.; Mussi, A.; Fontanini, G. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell Int. 2017, 17, 105. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Johnston, N.; Zheng, X.; Wang, H.; Zhang, X.; Gao, D.; Min, W. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 2016, 7, 53735–53750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-R.; Wang, L.-M.; Shen, J.-J. Overexpression of miR-32 inhibits the proliferation and metastasis of ovarian cancer cells by targeting BTLA. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4671–4678. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, Y.; Andalib, A.; Mohammad-Ganji, M.; Homayouni, V.; Sharifi, M.; Ganjalikhani-Hakemi, M. Evaluation of the effect of TIM-3 suppression by miR-498 and its effect on apoptosis and proliferation rate of HL-60 cell line. Pathol. Res. Pract. 2018, 214, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Lou, Q.; Liu, R.; Yang, X.; Li, W.; Huang, L.; Wei, L.; Tan, H.; Xiang, N.; Chan, K.; Chen, J.; et al. miR-448 targets IDO1 and regulates CD8+ T cell response in human colon cancer. J. Immunother. Cancer 2019, 7, 210. [Google Scholar] [CrossRef]
- Huang, Q.; Xia, J.; Wang, L.; Wang, X.; Ma, X.; Deng, Q.; Lu, Y.; Kumar, M.; Zhou, Z.; Li, L.; et al. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J. Hematol. Oncol. 2018, 11, 58. [Google Scholar] [CrossRef]
- Bieg, D.; Sypniewski, D.; Nowak, E.; Bednarek, I. MiR-424-3p suppresses galectin-3 expression and sensitizes ovarian cancer cells to cisplatin. Arch. Gynecol. Obstet. 2019, 299, 1077–1087. [Google Scholar] [CrossRef]
- Lu, W.; Wang, J.; Yang, G.; Yu, N.; Huang, Z.; Xu, H.; Li, J.; Qiu, J.; Zeng, X.; Chen, S.; et al. Posttranscriptional regulation of Galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget 2017, 8, 15242–15251. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, W.; Zhuang, C.; Geng, Z.; Hou, C.; Huang, D.; Hu, L.; Wang, X. microRNA-22 downregulation of galectin-9 influences lymphocyte apoptosis and tumor cell proliferation in liver cancer. Oncol. Rep. 2015, 34, 1771–1778. [Google Scholar] [CrossRef]
- Yang, Q.; Hou, C.; Huang, D.; Zhuang, C.; Jiang, W.; Geng, Z.; Wang, X.; Hu, L. miR-455-5p functions as a potential oncogene by targeting galectin-9 in colon cancer. Oncol. Lett. 2017, 13, 1958–1964. [Google Scholar] [CrossRef]
- Morishita, A.; Nomura, K.; Tani, J.; Fujita, K.; Iwama, H.; Takuma, K.; Nakahara, M.; Tadokoro, T.; Oura, K.; Chiyo, T.; et al. Galectin-9 suppresses the tumor growth of colon cancer in vitro and in vivo. Oncol. Rep. 2021, 45, 105. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Tang, R.; Zhu, J.; Zou, L.; Wu, R.; Zhou, H.; Mao, Y.; Li, R.; Hua, D.; Wang, W.; et al. A functional variant at miR-24 binding site in B7-H2 alters susceptibility to gastric cancer in a Chinese Han population. Mol. Immunol. 2013, 56, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cheung, I.Y.; Guo, H.-F.; Cheung, N.-K.V. MicroRNA miR-29 Modulates Expression of Immunoinhibitory Molecule B7-H3: Potential Implications for Immune Based Therapy of Human Solid Tumors. Cancer Res. 2009, 69, 6275–6281. [Google Scholar] [CrossRef] [PubMed]
- Huang, L. The expression and clinical significance of B7-H3 and miR-145 in lung cancer patients with malignant pleural effusion. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6759–6766. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, Y.; Xu, M.; Zhou, F.; Yan, J. Serum miR-1301-3p, miR-335-5p, miR-28-5p, and their target B7-H3 may serve as novel biomarkers for colorectal cancer. J. BUON 2019, 24, 1120–1127. [Google Scholar]
- Zhou, X.; Mao, Y.; Zhu, J.; Meng, F.; Chen, Q.; Tao, L.; Li, R.; Fu, F.; Liu, C.; Hu, Y.; et al. TGF-β1 promotes colorectal cancer immune escape by elevating B7-H3 and B7-H4 via the miR-155/miR-143 axis. Oncotarget 2016, 7, 67196–67211. [Google Scholar] [CrossRef]
- Hu, X.; Xu, M.; Hu, Y.; Li, N.; Zhou, L. B7-H3, Negatively Regulated by miR-128, Promotes Colorectal Cancer Cell Proliferation and Migration. Cell Biochem. Biophys. 2021, 79, 397–405. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Xie, C.; Sun, M.; Hu, C.; Zhang, Z.; Luan, L.; Zhou, J.; Zhou, J.; Zhu, X.; et al. MicroRNA miR-29a Inhibits Colon Cancer Progression by Downregulating B7-H3 Expression: Potential Molecular Targets for Colon Cancer Therapy. Mol. Biotechnol. 2021, 63, 849–861. [Google Scholar] [CrossRef]
- Wang, Z.-S.; Zhong, M.; Bian, Y.-H.; Mu, Y.-F.; Qin, S.-L.; Yu, M.-H.; Qin, J. MicroRNA-187 inhibits tumor growth and invasion by directly targeting CD276 in colorectal cancer. Oncotarget 2016, 7, 44266–44276. [Google Scholar] [CrossRef]
- Zhao, J.; Lei, T.; Xu, C.; Li, H.; Ma, W.; Yang, Y.; Fan, S.; Liu, Y. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem. Biophys. Res. Commun. 2013, 438, 439–444. [Google Scholar] [CrossRef]
- Wang, J.; Chong, K.K.; Nakamura, Y.; Nguyen, L.; Huang, S.K.; Kuo, C.; Zhang, W.; Yu, H.; Morton, D.L.; Hoon, D.S.B. B7-H3 Associated with Tumor Progression and Epigenetic Regulatory Activity in Cutaneous Melanoma. J. Investig. Dermatol. 2013, 133, 2050–2058. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mou, J.; Cui, L.; Wang, X.; Zhang, Z. Astragaloside IV inhibits cell proliferation of colorectal cancer cell lines through down-regulation of B7-H3. Biomed. Pharmacother. 2018, 102, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Nygren, M.K.; Tekle, C.; Ingebrigtsen, V.A.; Mäkelä, R.; Krohn, M.; Aure, M.R.; Nunes-Xavier, C.E.; Perälä, M.; Tramm, T.; Alsner, J.; et al. Identifying microRNAs regulating B7-H3 in breast cancer: The clinical impact of microRNA-29c. Br. J. Cancer 2014, 110, 2072–2080. [Google Scholar] [CrossRef] [PubMed]
- Li, R.G.; Gao, Z.; Jiang, Y. B7-H3 repression by miR-539 suppresses cell proliferation in human gliomas. Int. J. Clin. Exp. Pathol. 2017, 10, 4363–4369. [Google Scholar]
- Wang, L.; Kang, F.; Sun, N.; Wang, J.; Chen, W.; Li, D.; Shan, B. The tumor suppressor miR-124 inhibits cell proliferation and invasion by targeting B7-H3 in osteosarcoma. Tumor Biol. 2016, 37, 14939–14947. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, J.; Zhu, M.; Wang, Y.; Yang, S.; Ke, X. MicroRNA-506 inhibits the proliferation and invasion of mantle cell lymphoma cells by targeting B7 H3. Biochem. Biophys. Res. Commun. 2019, 508, 1067–1073. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, P.; Li, W.-J.; Zhang, J.; Wang, G.-P.; Jiang, D.-F.; Chen, F.-P. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol. Immunol. 2020, 117, 20–28. [Google Scholar] [CrossRef]
- Purvis, I.J.; Avilala, J.; Guda, M.R.; Venkataraman, S.; Vibhakar, R.; Tsung, A.J.; Velpula, K.K.; Asuthkar, S. Role of MYC-miR-29-B7-H3 in Medulloblastoma Growth and Angiogenesis. J. Clin. Med. 2019, 8, 1158. [Google Scholar] [CrossRef]
- Kanchan, R.K.; Perumal, N.; Atri, P.; Chirravuri Venkata, R.; Thapa, I.; Klinkebiel, D.L.; Donson, A.M.; Perry, D.; Punsoni, M.; Talmon, G.A.; et al. MiR-1253 exerts tumor-suppressive effects in medulloblastoma via inhibition of CDK6 and CD276 (B7-H3). Brain Pathol. 2020, 30, 732–745. [Google Scholar] [CrossRef]
- Yang, X.; Feng, K.-X.; Li, H.; Wang, L.; Xia, H. MicroRNA-199a Inhibits Cell Proliferation, Migration, and Invasion and Activates AKT/mTOR Signaling Pathway by Targeting B7-H3 in Cervical Cancer. Technol. Cancer Res. Treat. 2020, 19, 153303382094224. [Google Scholar] [CrossRef]
- Oliveira, P.; Carvalho, J.; Rocha, S.; Azevedo, M.; Reis, I.; Camilo, V.; Sousa, B.; Valente, S.; Paredes, J.; Almeida, R.; et al. Dies1/VISTA expression loss is a recurrent event in gastric cancer due to epigenetic regulation. Sci. Rep. 2016, 6, 34860. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Tang, R.; Qi, Q.; Zhou, X.; Zhou, H.; Mao, Y.; Li, R.; Liu, C.; Wang, W.; Hua, D.; et al. Five functional polymorphisms of B7/CD28 co-signaling molecules alter susceptibility to colorectal cancer. Cell. Immunol. 2015, 293, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Feng, L.; Wu, W.; Weng, T.; Hu, C.; Hong, B.; Wang, F.X.C.; Shen, L.; Wang, Q.; Jin, X.; et al. MicroRNA Expression Profiling of Pancreatic Cancer Cell Line L3.6p1 Following B7-H4 Knockdown. Cell. Physiol. Biochem. 2017, 44, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Ran, Z.; Liu, M.; Ou, Y. Prognostic Significance of Potential Immune Checkpoint Member HHLA2 in Human Tumors: A Comprehensive Analysis. Front. Immunol. 2019, 10, 1573. [Google Scholar] [CrossRef]
- Korotaeva, A.; Mansorunov, D.; Apanovich, N.; Kuzevanova, A.; Karpukhin, A. MiRNA Expression in Neuroendocrine Neoplasms of Frequent Localizations. Non-Coding RNA 2021, 7, 38. [Google Scholar] [CrossRef]
- Kipkeeva, F.; Muzaffarova, T.; Korotaeva, A.; Nikulin, M.; Grishina, K.; Mansorunov, D.; Apanovich, P.; Karpukhin, A. MicroRNA in Gastric Cancer Development: Mechanisms and Biomarkers. Diagnostics 2020, 10, 891. [Google Scholar] [CrossRef]
- Reid, G.; Pel, M.E.; Kirschner, M.B.; Cheng, Y.Y.; Mugridge, N.; Weiss, J.; Williams, M.; Wright, C.; Edelman, J.J.B.; Vallely, M.P.; et al. Restoring expression of miR-16: A novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. 2013, 24, 3128–3135. [Google Scholar] [CrossRef]
- Yeh, M.; Wang, Y.-Y.; Yoo, J.Y.; Oh, C.; Otani, Y.; Kang, J.M.; Park, E.S.; Kim, E.; Chung, S.; Jeon, Y.-J.; et al. MicroRNA-138 suppresses glioblastoma proliferation through downregulation of CD44. Sci. Rep. 2021, 11, 9219. [Google Scholar] [CrossRef]
- He, Z.; Ruan, X.; Liu, X.; Zheng, J.; Liu, Y.; Liu, L.; Ma, J.; Shao, L.; Wang, D.; Shen, S.; et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J. Exp. Clin. Cancer Res. 2019, 38, 65. [Google Scholar] [CrossRef]
- Kang, X.; Kong, B.; Chen, Q.; Zhao, S. Low expression of miR-138 inhibit the proliferation, migration and invasion of colorectal cancer and affect patient survival by targeting SIRT1. Transl. Cancer Res. 2021, 10, 3548–3559. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Slack, F.J. miRNA-34 Prevents Cancer Initiation and Progression in a Therapeutically Resistant K-ras and p53-Induced Mouse Model of Lung Adenocarcinoma. Cancer Res. 2012, 72, 5576–5587. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Liu, X.-M.; Zhang, C.-Y.; Zhou, J.-B.; Shao, Y.; Liang, C.; Wang, H.-M.; Hua, Z.-Y.; Lu, S.-D.; Ma, Z.-L. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis 2017, 6, e372. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Huang, C.; Li, S.; Yang, C.; Xi, Y.; Wang, L.; Zhang, F.; Fu, Y.; Li, D. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget 2016, 7, 8341–8359. [Google Scholar] [CrossRef]
- Cai, M.; Wang, Z.; Zhang, J.; Zhou, H.; Jin, L.; Bai, R.; Weng, Y. Adam17, a Target of Mir-326, Promotes Emt-Induced Cells Invasion in Lung Adenocarcinoma. Cell. Physiol. Biochem. 2015, 36, 1175–1185. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, Q.; Wang, C.; Wang, X.; Huang, J.; Zhao, J.; Mao, S.; Zhang, G.; Xu, X.; Zhang, N. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer 2011, 117, 2842–2852. [Google Scholar] [CrossRef]
- Bruning, U.; Cerone, L.; Neufeld, Z.; Fitzpatrick, S.F.; Cheong, A.; Scholz, C.C.; Simpson, D.A.; Leonard, M.O.; Tambuwala, M.M.; Cummins, E.P.; et al. MicroRNA-155 Promotes Resolution of Hypoxia-Inducible Factor 1α Activity during Prolonged Hypoxia. Mol. Cell. Biol. 2011, 31, 4087–4096. [Google Scholar] [CrossRef]
- Su, J.; Liang, H.; Yao, W.; Wang, N.; Zhang, S.; Yan, X.; Feng, H.; Pang, W.; Wang, Y.; Wang, X.; et al. MiR-143 and MiR-145 Regulate IGF1R to Suppress Cell Proliferation in Colorectal Cancer. PLoS ONE 2014, 9, e114420. [Google Scholar] [CrossRef]
- Qian, X.; Yu, J.; Yin, Y.; He, J.; Wang, L.; Li, Q.; Zhang, L.-Q.; Li, C.-Y.; Shi, Z.-M.; Xu, Q.; et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle 2013, 12, 1385–1394. [Google Scholar] [CrossRef]
- Yin, Y.; Yan, Z.-P.; Lu, N.-N.; Xu, Q.; He, J.; Qian, X.; Yu, J.; Guan, X.; Jiang, B.-H.; Liu, L.-Z. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim. Biophys. Acta-Gene Regul. Mech. 2013, 1829, 239–247. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, Z.; Liu, Z.; Qiu, B.; Liu, K.; Dong, G. MicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2. Med. Oncol. 2014, 31, 982. [Google Scholar] [CrossRef]
- Lian, B.; Yang, D.; Liu, Y.; Shi, G.; Li, J.; Yan, X.; Jin, K.; Liu, X.; Zhao, J.; Shang, W.; et al. miR-128 Targets the SIRT1/ROS/DR5 Pathway to Sensitize Colorectal Cancer to TRAIL-Induced Apoptosis. Cell. Physiol. Biochem. 2018, 49, 2151–2162. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.I.; Nicoloso, M.S.; Zeng, L.; Ivan, C.; Spizzo, R.; Gafà, R.; Xiao, L.; Zhang, X.; Vannini, I.; Fanini, F.; et al. Strand-Specific miR-28-5p and miR-28-3p Have Distinct Effects in Colorectal Cancer Cells. Gastroenterology 2012, 142, 886–896.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zeng, Y.; Zhou, J.M.; Nie, S.L.; Peng, Q.; Gong, J.; Huo, J.R. MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction. Mol. Med. Rep. 2016, 13, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.; Miao, Z.; Chen, Y.; Huang, C.; Yeh, Y.; Yang, I.; Wang, J. miR-148a inhibits early relapsed colorectal cancers and the secretion of VEGF by indirectly targeting HIF-1α under non-hypoxia/hypoxia conditions. J. Cell. Mol. Med. 2019, 23, 3572–3582. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Huang, Q.; Ren, X.; Hu, H.; Sheng, H.; Lai, M. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011, 18, 1702–1710. [Google Scholar] [CrossRef]
- Zhao, W.; Zheng, J.; Wei, G.; Yang, K.; Wang, G.; Sun, X. miR-148a inhibits cell proliferation and migration through targeting ErbB3 in colorectal cancer. Oncol. Lett. 2019, 18, 2530–2536. [Google Scholar] [CrossRef]
- Shi, L.; Xi, J.; Xu, X.; Peng, B.; Zhang, B. MiR-148a suppressed cell invasion and migration via targeting WNT10b and modulating β-catenin signaling in cisplatin-resistant colorectal cancer cells. Biomed. Pharmacother. 2019, 109, 902–909. [Google Scholar] [CrossRef]
- Zheng, J.; Yang, T.; Gao, S.; Cheng, M.; Shao, Y.; Xi, Y.; Guo, L.; Zhang, D.; Gao, W.; Zhang, G.; et al. miR-148a-3p silences the CANX/MHC-I pathway and impairs CD8+ T cell-mediated immune attack in colorectal cancer. FASEB J. 2021, 35, e2177. [Google Scholar] [CrossRef]
- Rodriguez-Barrueco, R.; Nekritz, E.A.; Bertucci, F.; Yu, J.; Sanchez-Garcia, F.; Zeleke, T.Z.; Gorbatenko, A.; Birnbaum, D.; Ezhkova, E.; Cordon-Cardo, C.; et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017, 31, 553–566. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Chen, Z.; Zhu, M.; Gu, Y. MicroRNA-214 acts as a potential oncogene in breast cancer by targeting the PTEN-PI3K/Akt signaling pathway. Int. J. Mol. Med. 2016, 37, 1421–1428. [Google Scholar] [CrossRef]
- Lu, Y.-Y.; Zheng, J.-Y.; Liu, J.; Huang, C.-L.; Zhang, W.; Zeng, Y. miR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed. Pharmacother. 2015, 70, 151–157. [Google Scholar] [CrossRef]
- Li, X.; Chen, W.; Jin, Y.; Xue, R.; Su, J.; Mu, Z.; Li, J.; Jiang, S. miR-142-5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Biochem. Pharmacol. 2019, 161, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, G.; Luo, X.; Wang, D.; Zhou, W.; Zhang, Y.; Zhang, W.; Chen, J.; Meng, Q.; Chen, E.; et al. Co-delivery of 5-fluorouracil and miRNA-34a mimics by host-guest self-assembly nanocarriers for efficacious targeted therapy in colorectal cancer patient-derived tumor xenografts. Theranostics 2021, 11, 2475–2489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, X.; Lv, Y.; Xin, X.; Qin, C.; Han, X.; Yang, L.; He, W.; Yin, L. Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Sci. Rep. 2017, 7, 46186. [Google Scholar] [CrossRef] [PubMed]
- Amri, J.; Molaee, N.; Baazm, M.; Karami, H. Targeting Epidermal Growth Factor Receptor by MiRNA-145 Inhibits Cell Growth and Sensitizes NSCLC Cells to Erlotinib. Asian Pac. J. Cancer Prev. 2019, 20, 2781–2787. [Google Scholar] [CrossRef] [PubMed]
- Karaayvaz, M.; Zhai, H.; Ju, J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013, 4, e659. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Phung, C.D.; Tran, T.H.; Pham, T.T.; Pham, L.M.; Nguyen, T.T.; Jeong, J.-H.; Choi, H.-G.; Ku, S.K.; Yong, C.S.; et al. Manipulating immune system using nanoparticles for an effective cancer treatment: Combination of targeted therapy and checkpoint blockage miRNA. J. Control. Release 2021, 329, 524–537. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Kelnar, K.; Stahlhut, C.; Orellana, E.; Zhao, J.; Shimer, E.; Dysart, S.; Chen, X.; Bader, A.G.; Slack, F.J. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene 2015, 34, 3547–3555. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Akiyama, Y.; Yuasa, Y. Multiple-to-Multiple Relationships between MicroRNAs and Target Genes in Gastric Cancer. PLoS ONE 2013, 8, e62589. [Google Scholar] [CrossRef]
- Ahir, M.; Upadhyay, P.; Ghosh, A.; Sarker, S.; Bhattacharya, S.; Gupta, P.; Ghosh, S.; Chattopadhyay, S.; Adhikary, A. Delivery of dual miRNA through CD44-targeted mesoporous silica nanoparticles for enhanced and effective triple-negative breast cancer therapy. Biomater. Sci. 2020, 8, 2939–2954. [Google Scholar] [CrossRef]
- Byun, Y.; Choi, Y.-C.; Jeong, Y.; Lee, G.; Yoon, S.; Jeong, Y.; Yoon, J.; Baek, K. MiR-200c downregulates HIF-1α and inhibits migration of lung cancer cells. Cell. Mol. Biol. Lett. 2019, 24, 28. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, S.; Wu, H.; Zhang, L.; Dai, X.; Hu, J.; Xue, J.; Liu, T.; Liang, Y.; Wu, G. MiR-200c Increases the Radiosensitivity of Non-Small-Cell Lung Cancer Cell Line A549 by Targeting VEGF-VEGFR2 Pathway. PLoS ONE 2013, 8, e78344. [Google Scholar] [CrossRef] [PubMed]
- McArthur, K.; Feng, B.; Wu, Y.; Chen, S.; Chakrabarti, S. MicroRNA-200b Regulates Vascular Endothelial Growth Factor–Mediated Alterations in Diabetic Retinopathy. Diabetes 2011, 60, 1314–1323. [Google Scholar] [CrossRef]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Viteri, S.; Rosell, R. An innovative mesothelioma treatment based on miR-16 mimic loaded EGFR targeted minicells (TargomiRs). Transl. Lung Cancer Res. 2018, 7, S1–S4. [Google Scholar] [CrossRef]
- Jebbawi, F.; Fayyad-Kazan, H.; Merimi, M.; Lewalle, P.; Verougstraete, J.-C.; Leo, O.; Romero, P.; Burny, A.; Badran, B.; Martiat, P.; et al. A microRNA profile of human CD8+ regulatory T cells and characterization of the effects of microRNAs on Treg cell-associated genes. J. Transl. Med. 2014, 12, 218. [Google Scholar] [CrossRef]
- Liu, Y.; Nie, W.; Jin, Y.; Zhuo, A.; Zang, Y.; Xiu, Q. B and T Lymphocyte Attenuator is a Target of miR-155 during Naïve CD4+ T Cell Activation. Iran. J. Immunol. 2016, 13, 89–99. [Google Scholar]
- Huffaker, T.B.; Lee, S.-H.; Tang, W.W.; Wallace, J.A.; Alexander, M.; Runtsch, M.C.; Larsen, D.K.; Thompson, J.; Ramstead, A.G.; Voth, W.P.; et al. Antitumor immunity is defective in T cell–specific microRNA-155–deficient mice and is rescued by immune checkpoint blockade. J. Biol. Chem. 2017, 292, 18530–18541. [Google Scholar] [CrossRef]
- Singh, A.; Srivastava, N.; Yadav, A.; Ateeq, B. Targeting AGTR1/NF-κB/CXCR4 axis by miR-155 attenuates oncogenesis in glioblastoma. Neoplasia 2020, 22, 497–510. [Google Scholar] [CrossRef]
- Zhang, T.; Davidson-Moncada, J.K.; Mukherjee, P.; Furman, R.R.; Bhavsar, E.; Chen, Z.; Hakimpour, P.; Papavasiliou, N.; Tam, W. MicroRNA-155 regulates casein kinase 1 gamma 2: A potential pathogenetic role in chronic lymphocytic leukemia. Blood Cancer J. 2017, 7, e606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Bill, K.; Liu, J.; Young, E.; Peng, T.; Bolshakov, S.; Hoffman, A.; Song, Y.; Demicco, E.G.; Terrada, D.L.; et al. MiR-155 Is a Liposarcoma Oncogene That Targets Casein Kinase-1α and Enhances β-Catenin Signaling. Cancer Res. 2012, 72, 1751–1762. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Witten, L.; Slack, F.J. miR-155 as a novel clinical target for hematological malignancies. Carcinogenesis 2020, 41, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.-H.; Wang, D.-D.; Chen, D.; Liu, S.-W.; Wang, Z.; Yan, D.-L.; Dong, S.-C.; Feng, J.-F. MiR-138: A promising therapeutic target for cancer. Tumor Biol. 2017, 39, 101042831769757. [Google Scholar] [CrossRef] [PubMed]
- Shadbad, M.A.; Safaei, S.; Brunetti, O.; Derakhshani, A.; Lotfinejad, P.; Mokhtarzadeh, A.; Hemmat, N.; Racanelli, V.; Solimando, A.G.; Argentiero, A.; et al. A Systematic Review on the Therapeutic Potentiality of PD-L1-Inhibiting MicroRNAs for Triple-Negative Breast Cancer: Toward Single-Cell Sequencing-Guided Biomimetic Delivery. Genes 2021, 12, 1206. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Orellana, E.A.; Li, C.; Lisevick, A.; Kasinski, A.L. Identification and validation of microRNAs that synergize with miR-34a—A basis for combinatorial microRNA therapeutics. Cell Cycle 2019, 18, 1798–1811. [Google Scholar] [CrossRef]
- Meng, F.; Chen, Y.; Yang, M.; Zhang, H.; Wang, W. Concomitant inhibition of B7-H3 and PD-L1 expression by a novel and synthetic microRNA delivers potent antitumor activities in colorectal tumor models. Investig. New Drugs 2021, 39, 1267–1274. [Google Scholar] [CrossRef]
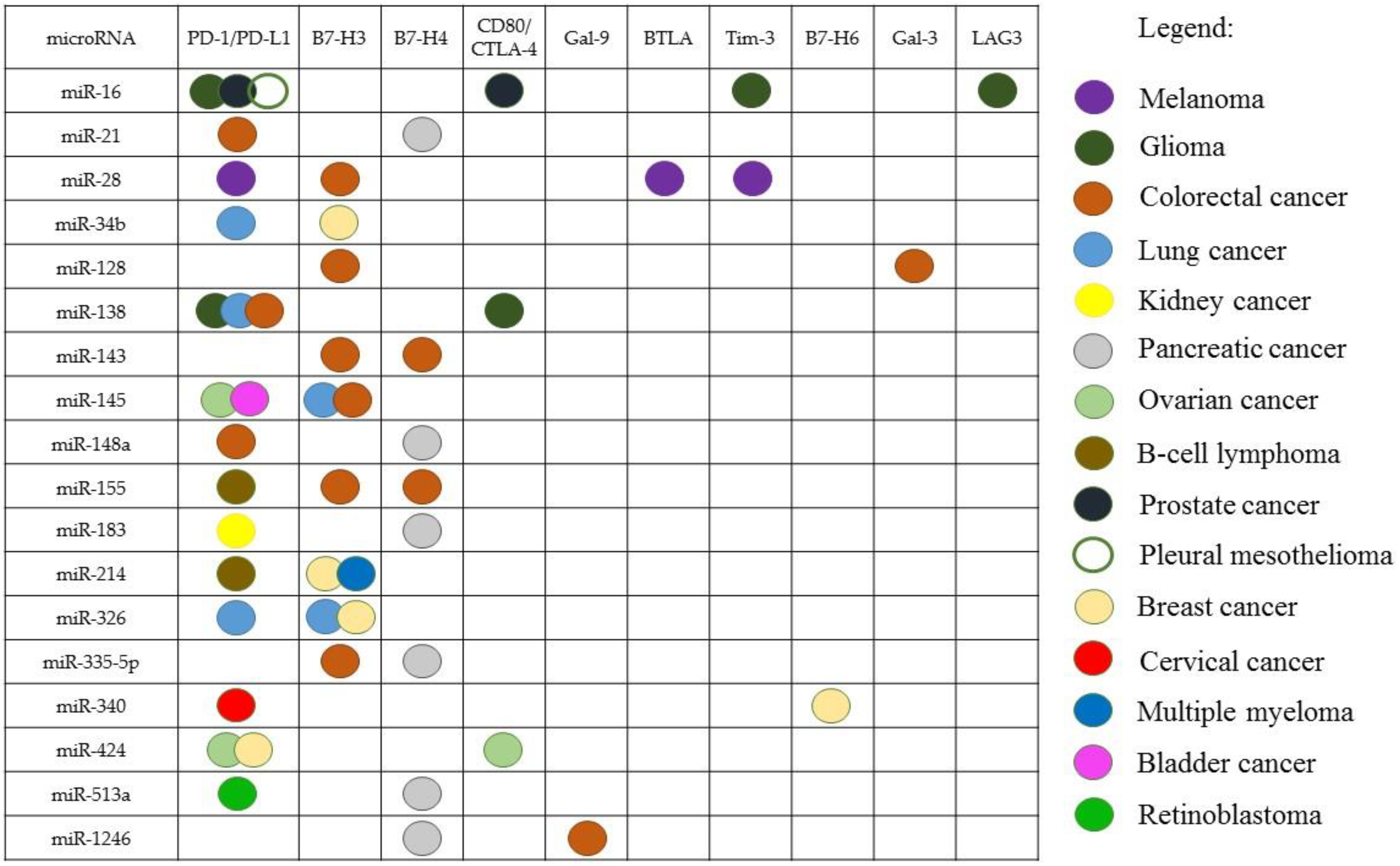
| Immune Checkpoint | microRNA | Cancer | Reference |
|---|---|---|---|
| PD-1 | miR-374b, miR-4717 | Liver cancer | [64,65] |
| PD-1/PD-L1 | miR-183 | RCC | [66] |
| miR-138-5p, miR-200b, miR-429, miR-508 | Lung cancer | [67,68] | |
| PD-L1 | miR-142-5p | PC, OC | [69,70] |
| miR-497-5p | ccRCC | [71] | |
| miR-20-b, miR-21, miR-130b, miR-138-5p, miR-148a-3p, miR-191-5p | CRC | [11,72,73,74] | |
| miR-195, miR-424-5p, miR-497, miR-873, miR-3609 | BC | [75,76,77,78] | |
| miR-17-5p, miR-146a | Melanoma | [79,80] | |
| miR-15a, miR-15b, miR-16, miR-193a-3p, miR-320a | Pleural Mesothelioma | [81,82] | |
| miR-155, miR-195, miR-214 | B-cell lymphoma | [83,84,85] | |
| miR-16, miR-195 | Prostate cancer | [86] | |
| miR-34a, miR-34b, miR-34c, miR-140, miR-200, miR-200a-3p, miR-3127-5p | Lung cancer | [87,88,89,90,91] | |
| miR-34a | AML | [92] | |
| miR-23a-3p, miR-570 | Liver cancer | [93,94] | |
| miR-375 | HNSCC | [95] | |
| miR-145 | OC, bladder cancer | [96,97] | |
| miR-513a-5p | Retinoblastoma | [98] | |
| miR-105-5p, miR-152, miR-200b, miR-200c, miR-570 | GC | [99,100,101,102,103] | |
| miR-18a, miR-140, miR-142, miR-340, miR-383 | Cervical cancer | [104] | |
| miR-217 | Laryngeal cancer | [105] | |
| miR-20b-5p | Models of lung and BC | [106] | |
| miR-194-5p | PC | [107] | |
| PD-L1+B7-H3 | miR-326 | Lung cancer | [8] |
| PD-1, CTLA-4 | miR-424 | OC | [108] |
| miR-138-5p | Glioma | [109] | |
| CD80/CTLA-4 | miR-424 | CRC | [110] |
| PD-1, PD-L1, CTLA-4 | miR-33a | Lung cancer | [111] |
| PD-1, BTLA, Tim-3 | miR-28 | Melanoma mouse model | [112] |
| BTLA | miR-32 | OC | [113] |
| Tim-3 | miR-498 | AML | [114] |
| IDO1 | miR-153, miR-448 | CRC | [115,116] |
| Gal-3 | miR-424-3p | OC | [117] |
| miR-128 | CRC | [118] | |
| Gal-9 | miR-22 | Liver cancer | [119] |
| miR-15b-5p, miR-455-5p, miR-1237, miR-1246 | CRC | [120,121] | |
| ICOS (B7-H2)/ICOSL | miR-24 | GC | [122] |
| B7-H3 | miR-29 (a, b and c) | Neuroblastoma, sarcoma, brain tumors | [123] |
| miR-145 | Lung cancer | [124] | |
| miR-28-5p, miR-29a, miR-128, miR-145, miR-155/miR-143, miR-187, miR-192, miR-335-5p, miR-378, miR-1301-3p | CRC | [125,126,127,128,129] | |
| miR-187 | ccRCC | [130] | |
| miR-29c | Melanoma, CRC | [131,132] | |
| miR-29c, miR-34b, miR-124a, miR-125b-2, miR-214, miR-297, miR-326, miR-363, miR-380-5p, miR-506, miR-555, miR-567, miR-593, miR-601, miR-665, miR-708, miR-885-3p, miR-940 | BC | [133] | |
| miR-539 | Glioma | [134] | |
| miR-124 | Osteosarcoma | [135] | |
| miR-506 | Mantle cell lymphoma | [136] | |
| miR-214 | Multiple myeloma | [137] | |
| miR-29, miR-1253 | Medulloblastoma | [138,139] | |
| miR-199a | Cervical cancer | [140] | |
| B7-H5 (VISTA, BTNL2) | miR-125a-5p | GC | [141] |
| B7-H4 (VTCN1) | miR-155/miR-143, miR-1207 | CRC | [126,142] |
| miR-7–5p, hsa-let-7c, hsa-let-7f-5p, miR-17–3p, miR-21–3p, miR-21–5p, miR-24–1-5p, miR-27b-3p, miR-31–3p, miR-31–5p, miR-33a-5p, miR-33b-5p, miR-122–3p, miR-130b-3p, miR-138–1-3p, miR-148a-3p, miR-149–3p, miR-183–3p, miR-186–5p, miR-196a-5p, hsa-miR-204–3p, miR-299–5p, miR-302a-3p, miR-302e, miR-335–3p, miR-335–5p, miR-361–5p, miR-374c-5p, miR-483–3p, miR-513a-5p, miR-519e-3p, miR-520d-5p, miR-525–5p, miR-615–3p, miR-642a-5p, miR-744–5p, miR-937, miR-1246, miRPlus-G1246–3p, miR-1260a, miR-1265, miR-1284, miR-1290, miR-1973, miR-2115–3p, miR-2116–5p, miR-3178, miR-3202, miR-3646, miR-3651, miR-3676–3p, miR-3685, miR-3686, miR-4258, miR-4279, miR-4284, miR-4288, miR-4290, miR-4306, miR-4324 | PC | [143] | |
| B7-H6 (NCR3LG1) | miR-93, miR-195, miR-340 | BC | [76] |
| B7-H7 (HHLA2) | miR-3116, miR-6870-5p | ccRCC | [144] |
| microRNA | Immune Checkpoints | Other Targets | Cancer | Reference |
|---|---|---|---|---|
| miR-16 | PD-1/PD-L1 | BCL2, CCND1 | Pleural mesothelioma | [81,147] |
| miR-138 | PD-1/PD-L1, CD80/CTLA-4 | CD44, SOX13 | Glioma | [109,148,149] |
| PD-1/PD-L1 | SIRT1 | CRC | [72,150] | |
| miR-34a | PD-1/PD-L1 | EGFR, BCL-2, Met | Lung cancer | [87,151,152] |
| miR-326 | PD-1/PD-L1, B7-H3 | CCND1, ADAM17 | [8,153,154] | |
| miR-340 | B7-H6 | MET | BC | [76,155] |
| miR-155 | B7-H3, B7-H4 | HIF-1 | CRC | [126,156] |
| miR-143 and miR-145 | B7-H3, B7-H4 | VEGF/VEGFR, HIF-1, IRS-1/IGF-IR | [126,157,158,159] | |
| miR-335 | B7-H3 | ZEB2 | [125,160] | |
| miR-128 | B7-H3, Gal-3 | SIRT1 | [118,127,161] | |
| miR-28 | B7-H3 | CCND1 | [125,162] | |
| miR-1246 | Gal-9 | CCNG2 | [121,163] | |
| miR-21 | PD-1/PD-L1 | PTEN | [11] | |
| miR-148a | PD-1/PD-L1 | HER3, WNT10b, VEGF/VEGFR, HIF-1, BCL-2, CANX | [73,164,165,166,167,168] | |
| miR-424 | PD-1/PD-L1 | PTEN, IRS-1/IGF-IR, BCL-2 | BC | [78,169] |
| miR-214 | B7-H3 | PTEN | [133,170] | |
| miR-183 | B7-H4 | PDCD4 | PC | [143,171] |
| miR-142 | PD-1/PD-L1 | BIRC3, BCL2, BCL2L2, MCL1, XIAP | OC | [70,172] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kipkeeva, F.; Muzaffarova, T.; Korotaeva, A.; Mansorunov, D.; Apanovich, P.; Nikulin, M.; Malikhova, O.; Stilidi, I.; Karpukhin, A. The Features of Immune Checkpoint Gene Regulation by microRNA in Cancer. Int. J. Mol. Sci. 2022, 23, 9324. https://doi.org/10.3390/ijms23169324
Kipkeeva F, Muzaffarova T, Korotaeva A, Mansorunov D, Apanovich P, Nikulin M, Malikhova O, Stilidi I, Karpukhin A. The Features of Immune Checkpoint Gene Regulation by microRNA in Cancer. International Journal of Molecular Sciences. 2022; 23(16):9324. https://doi.org/10.3390/ijms23169324
Chicago/Turabian StyleKipkeeva, Fatimat, Tatyana Muzaffarova, Alexandra Korotaeva, Danzan Mansorunov, Pavel Apanovich, Maxim Nikulin, Olga Malikhova, Ivan Stilidi, and Alexander Karpukhin. 2022. "The Features of Immune Checkpoint Gene Regulation by microRNA in Cancer" International Journal of Molecular Sciences 23, no. 16: 9324. https://doi.org/10.3390/ijms23169324






