Integrative Metabolome and Transcriptome Analysis Reveals the Regulatory Network of Flavonoid Biosynthesis in Response to MeJA in Camelliavietnamensis Huang
Abstract
:1. Introduction
2. Results
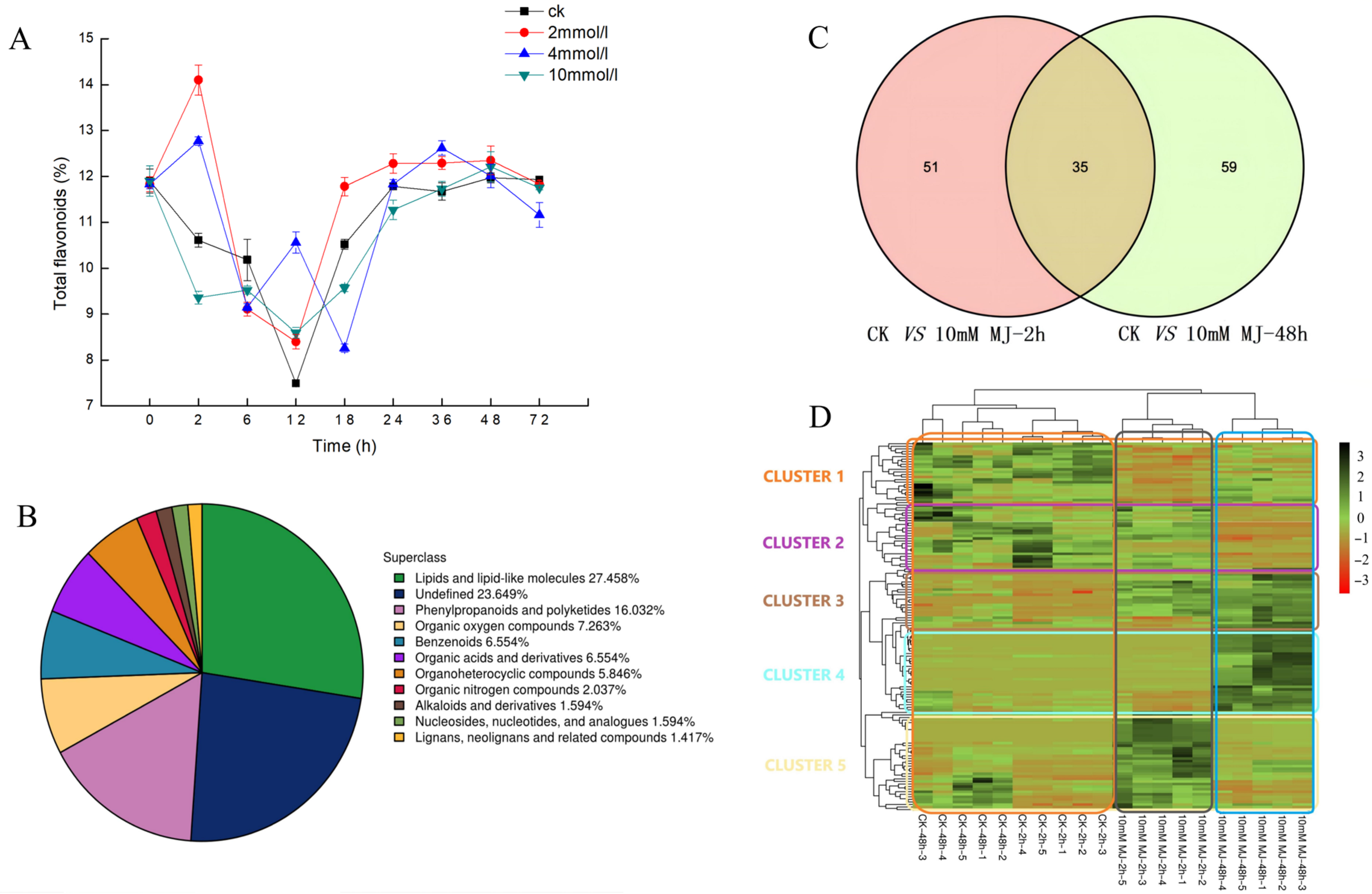
2.1. MeJA Treatment Increased the Content of Flavonoids in C. vietnamensis
2.2. Differentially Accumulated Metabolite Analysis
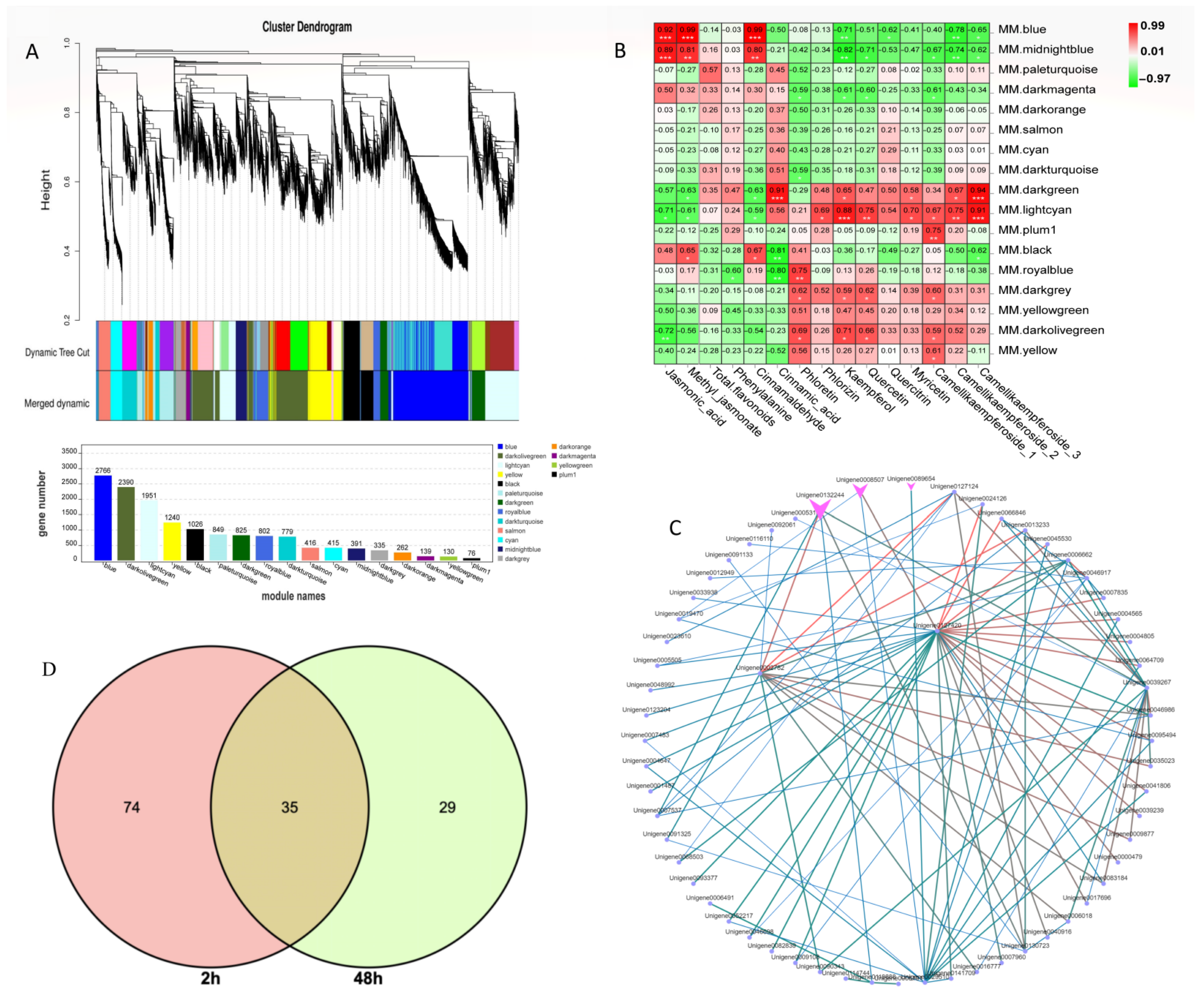
2.3. Hierarchical Cluster Analysis
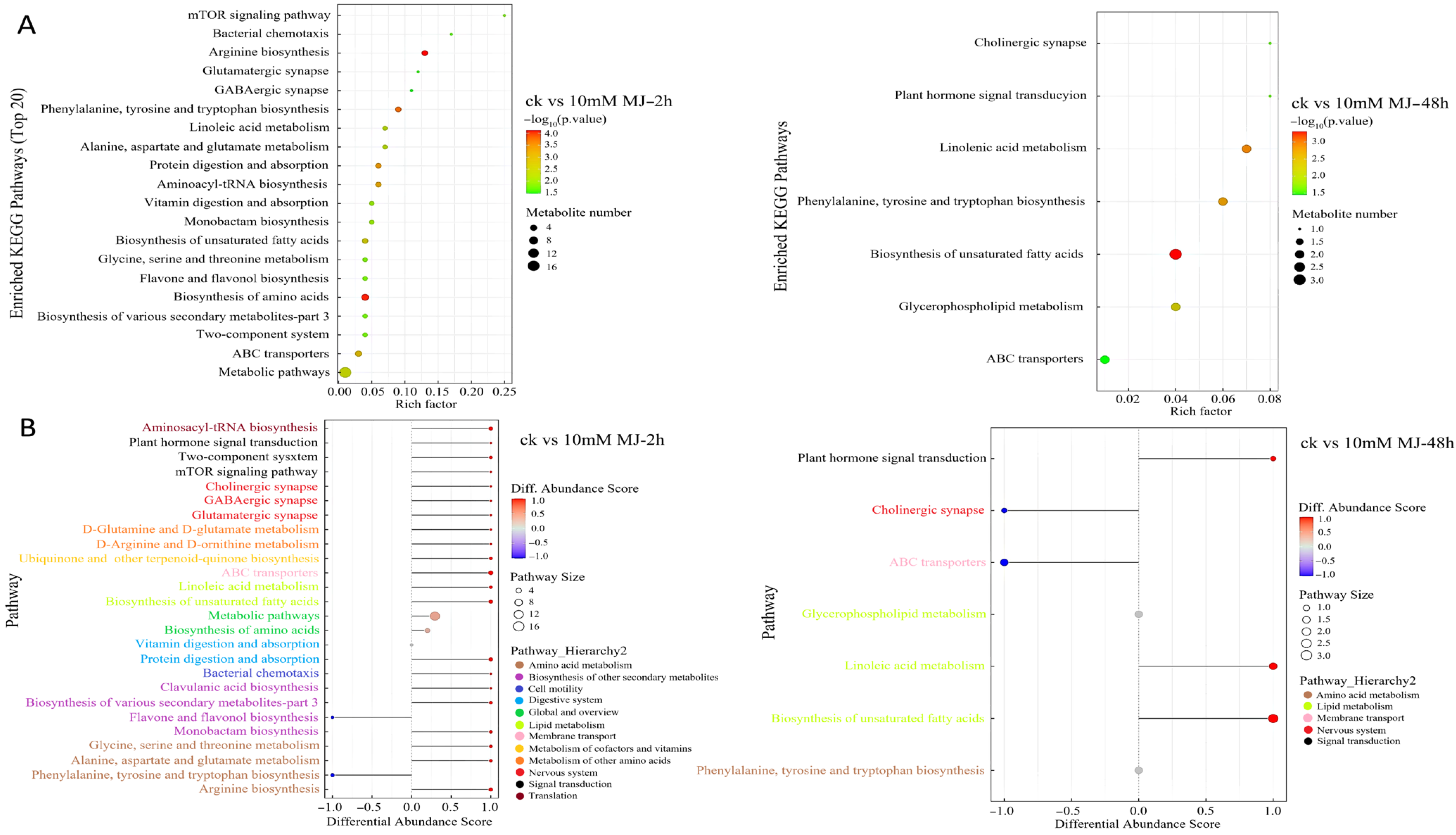
2.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
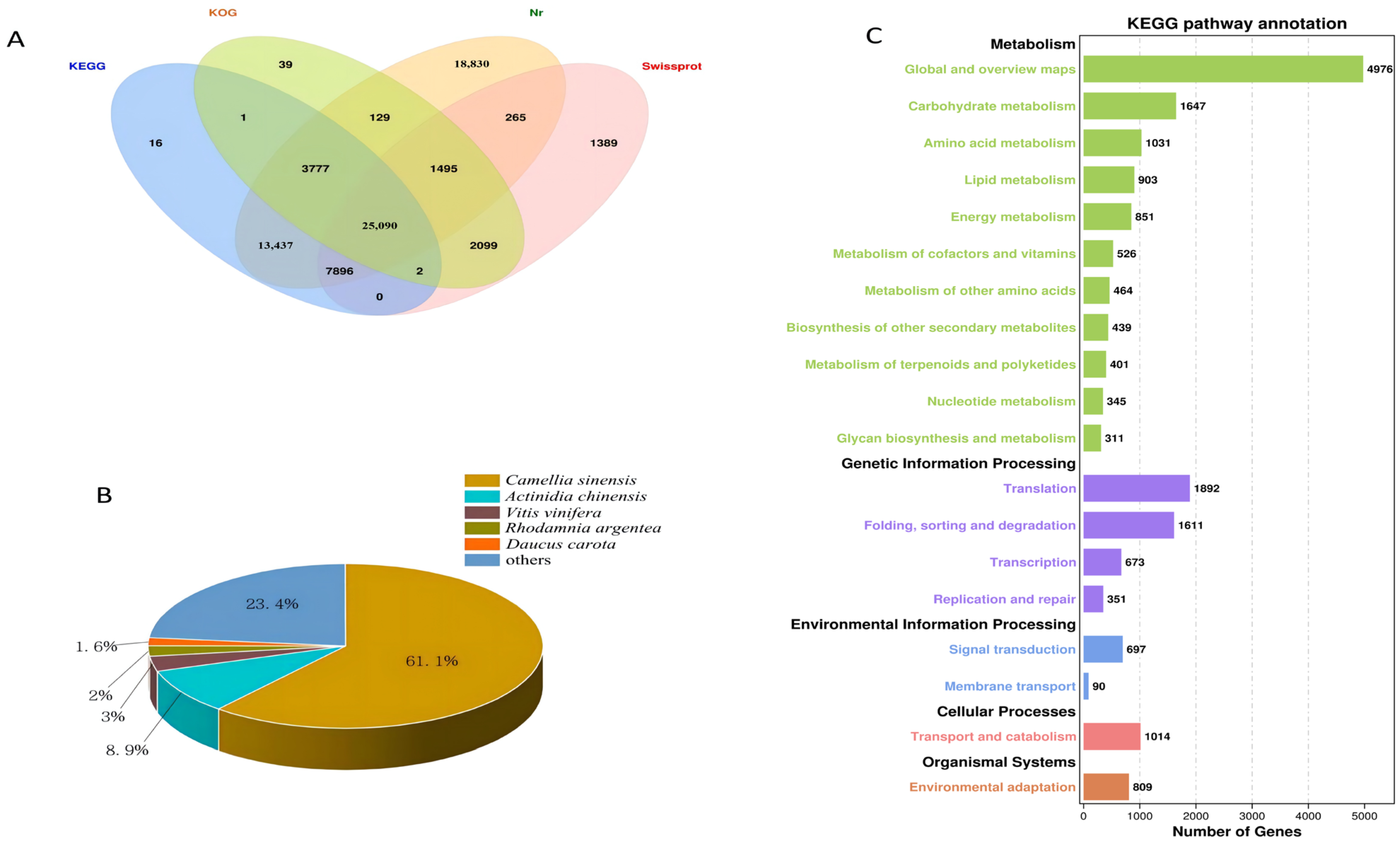
2.5. Transcriptome Sequencing and Gene Functional Annotation
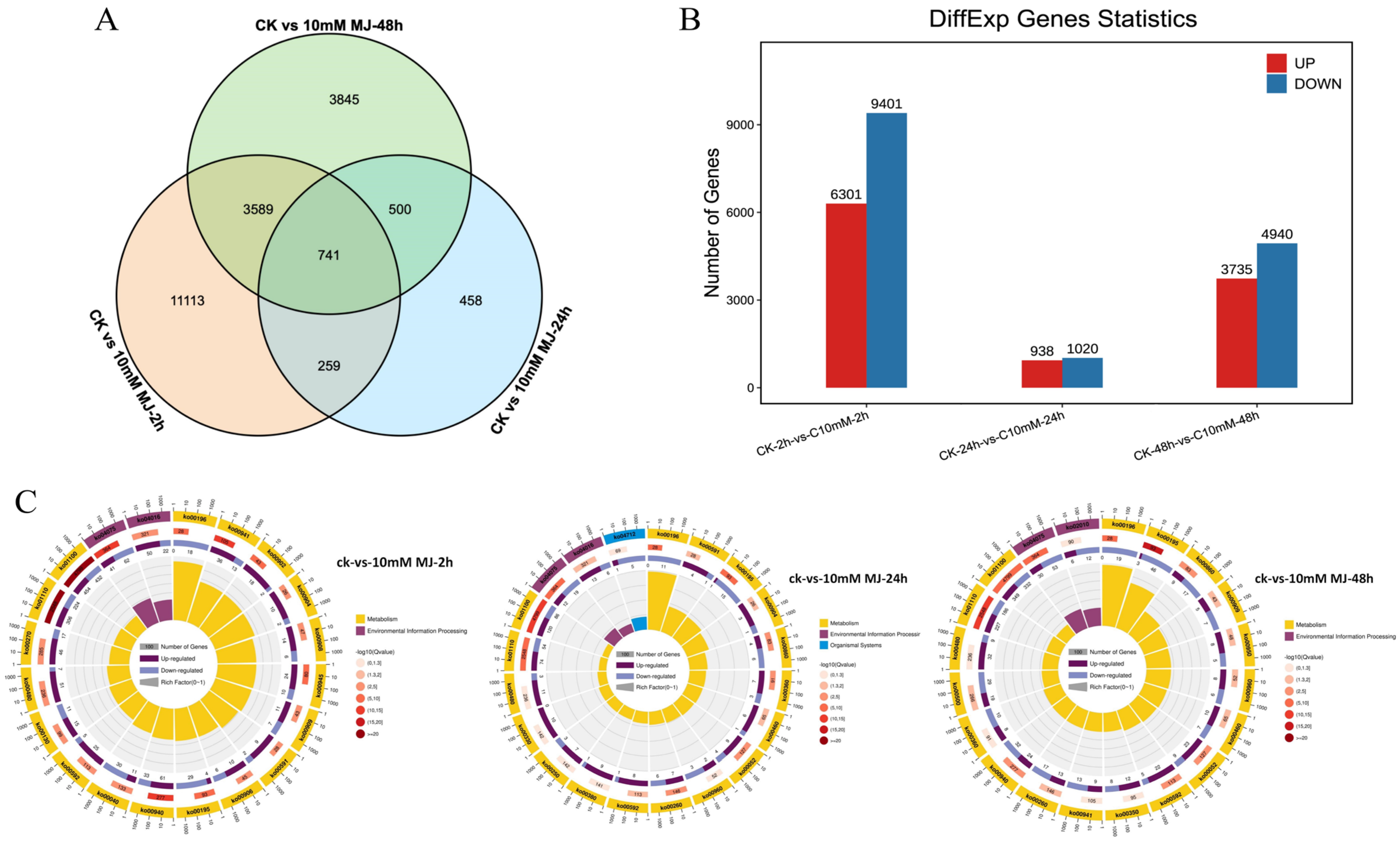
2.6. Identification and Analysis of Differentially Expressed Genes
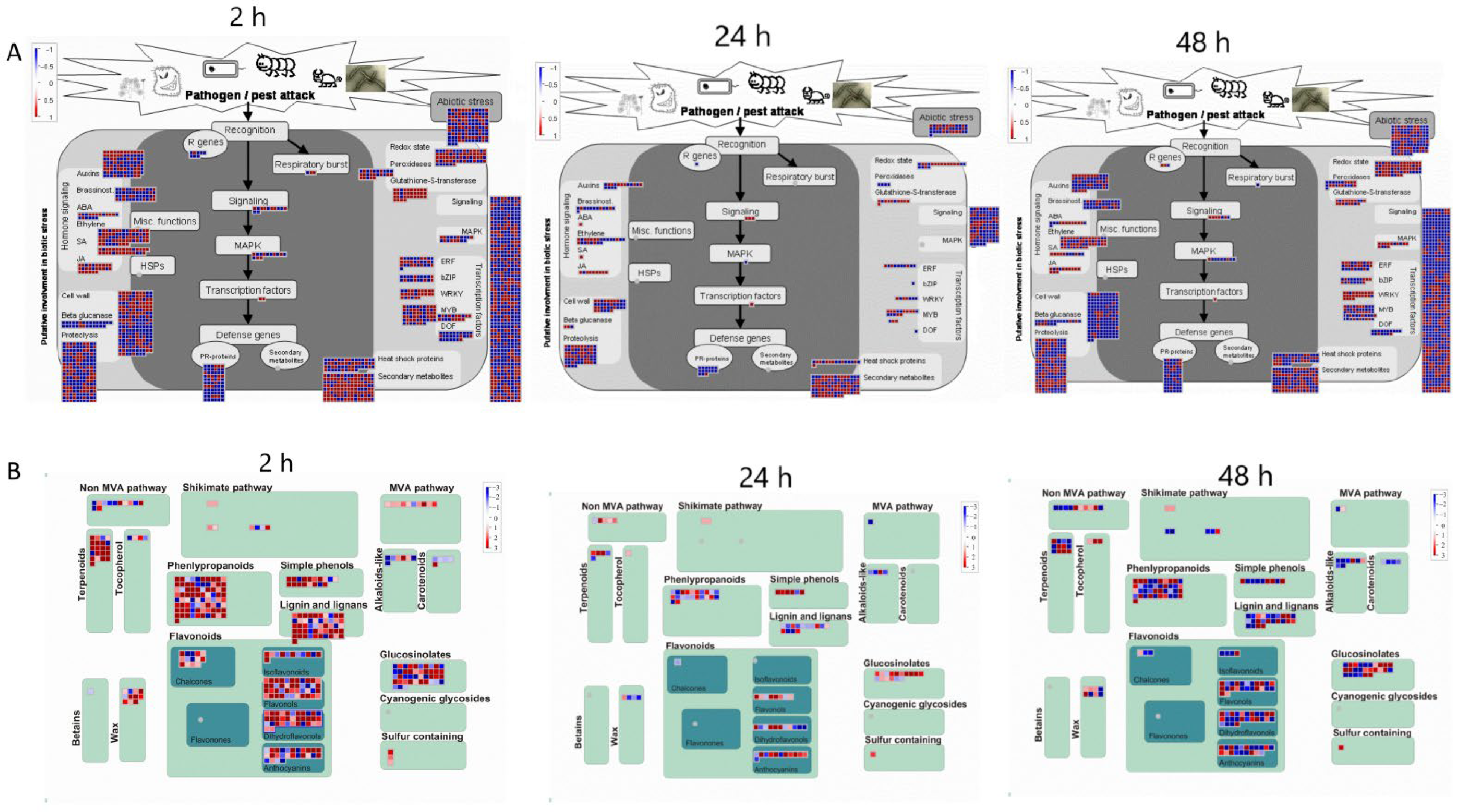
2.7. MapMan Analysis of Differentially Expressed Genes
2.8. Coexpression Network Analysis of Metabolomic and Transcriptomic Data
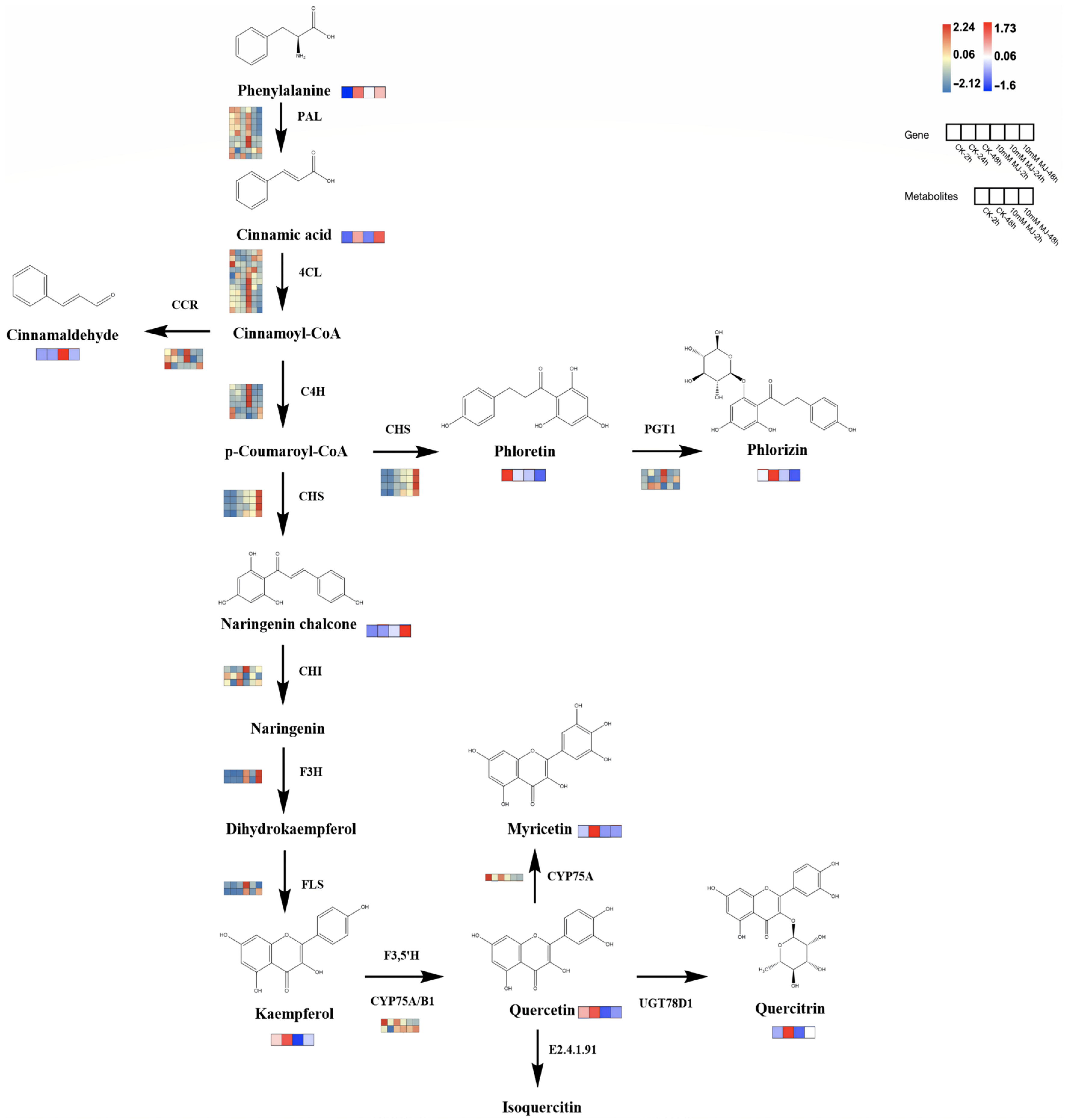
2.9. Combined Analysis of Differentially Accumulated Metabolites and Differentially Expressed Genes Related to Flavonoids
2.10. Expression Analysis of Key Genes through RT–qPCR Validation
3. Discussion
4. Materials and Methods
4.1. Materials, MeJA Treatment and Leaf Sample Preparation
4.2. Determination Conditions, Data Processing and Multivariate Statistical Analysis of UHPLC-QTOF/MS Results
4.3. Transcriptome Sequencing, Gene Expression Analysis, Annotation and Differential Gene Analysis
4.4. MapMan, WGCNA and NCBI Blast + Analysis
4.5. Verification of qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Guo, Y.J.; Hu, X.W.; Zhou, K.B. Comparison of the Chloroplast Genome Sequences of 13 Oil-Tea Camellia Samples and Identification of an Undetermined Oil-Tea Camellia Species from Hainan Province. Front. Plant Sci. 2022, 12, 798581. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.N.; Zheng, W.; Yu, J.; Yan, H.Q.; Wang, Y.; Wu, Y.G.; Hu, X.W.; Lai, H.G. cDNA cloning, prokaryotic expression, and functional analysis of squalene synthase (SQS) in Camellia vietnamensis Huang. Protein Expres. Purif. 2022, 194, 106078. [Google Scholar] [CrossRef] [PubMed]
- Othman, L.; Sleiman, A.; Abdel-Massih, R.M. Antimicrobial activity of polyphenols and alkaloids in middle eastern plants. Front. Microbiol. 2019, 10, 911. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.M.; Yu, X.; Sun, H.Y.; Zhang, W.; Liu, G.; Zhu, L. Flos lonicerae flavonoids attenuate experimental ulcerative colitis in rats via suppression of NF-κB signaling pathway. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 2481–2494. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Li, X.Q.; Jin, S.L.; Zhang, H.Z.; Wang, R.; Pei, L.H.; Zheng, J. The Anti-Proliferative Effect of Flavonoid Nanoparticles on the Human Ovarian Cancer Cell Line SK0V3. J. Nanosci. Nanotechnol. 2020, 20, 6040–6046. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhou, Z.; Mei, Z. Integrative Analysis of Metabolome and Transcriptome Identifies Potential Genes Involved in the Flavonoid Biosynthesis in Entada phaseoloides Stem. Front. Plant Sci. 2022, 13, 792674. [Google Scholar] [CrossRef]
- Khan, J.; Deb, P.K.; Priya, S.; Medina, K.D.; Devi, R.; Walode, S.G.; Rudrapal, M. Dietary flavonoids: Cardioprotective potential with antioxidant effects and their pharmacokinetic, toxicological and therapeutic concerns. Molecules 2021, 26, 4021. [Google Scholar] [CrossRef]
- Qi, Y.; Li, C.; Duan, C.; Gu, C.; Zhang, Q. Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90. Int. J. Mol. Sci. 2021, 22, 8751. [Google Scholar] [CrossRef]
- Mori, K.; Goto-Yamamoto, N.; Kitayama, M.; Hashizume, K. Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 2007, 58, 1935–1945. [Google Scholar] [CrossRef]
- Andersen, J.R.; Zein, I.; Wenzel, G.; Darnhofer, B.; Eder, J.; Ouzunova, M.; Lubberstedt, T. Characterization of phenylpropanoid pathway genes within European maize (Zea mays L.) inbreds. BMC Plant Biol. 2008, 8, 2. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Bioch. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.T.; Jiang, Z.Q.; Lin, Z.M.; Yu, Q.Q.; Song, R.F.; Wang, B. FtUGT79A15 is responsible for rutinosylation in flavonoid diglycoside biosynthesis in Fagopyrum tataricum. Plant Physiol. Bioch. 2022, 181, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Fei, X.; He, B.; Luo, Y.; Qi, Y.; Wei, A. Integrated Analysis of Metabolome and Transcriptome Data for Uncovering Flavonoid Components of Zanthoxylum bungeanum Maxim. Leaves Under Drought Stress. Front. Nutr. 2022, 8, 801244. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Guo, K.Y.; Liu, L.D.; Tian, W.; Xie, X.L.; Wen, S.Q.; Wen, C.X. Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci. Rep. 2020, 10, 16207. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.H.; Xiao, X.; Rao, X.; Dixon, R.A. Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat. Plants 2018, 4, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, W.; Zhang, T.; Jiang, S.; Xu, H.; Wang, Y.; Zhang, Z.; Wang, C.; Chen, X. Transcriptomic analysis of Red-Fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 2018, 66, 7076–7086. [Google Scholar] [CrossRef]
- Ma, S.; Lv, L.; Meng, C.; Zhang, C.; Li, Y. Integrative Analysis of the Metabolome and Transcriptome of Sorghum bicolor Reveals Dynamic Changes in Flavonoids Accumulation under Saline-Alkali Stress. J. Agric. Food Chem. 2020, 68, 14781–14789. [Google Scholar] [CrossRef]
- Park, C.H.; Yeo, H.J.; Park, Y.E.; Kim, Y.J.; Park, S.U. Integrated Analysis of Transcriptome and Metabolome and Evaluation of Antioxidant Activities in Lavandula pubescens. Antioxidants 2021, 10, 1027. [Google Scholar] [CrossRef]
- Song, S.; Tao, Y.; Gao, L.; Liang, H.; Tang, D.; Lin, J.; Wang, Y.; Gmitter, F.G., Jr.; Li, C. An Integrated Metabolome and Transcriptome Analysis Reveal the Regulation Mechanisms of Flavonoid Biosynthesis in a Purple Tea Plant Cultivar. Front. Plant Sci. 2022, 13, 880227. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, X.; Zhang, Y.Y.; Lin, X.R.; Li, B.; Chen, Z.Z. Integrated metabolomic and transcriptomic strategies to understand the effects of dark stress on tea callus flavonoid biosynthesis. Plant Physiol. Biochem. 2020, 155, 549–559. [Google Scholar] [CrossRef]
- Song, Q.L.; Ji, K.; Yu, X.R.; Chen, L.; Wang, L.K.; Gong, W.F.; Yuan, D.Y. Dynamic metabolic and transcriptomic profiling reveal synthetic characters and regulators of flavonoid biosynthesis in Camellia oleifera seeds. Ind. Crop. Prod. 2022, 186, 115295. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Wang, R.; Xian, B.; Pei, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in safflower (Carthamus tinctorius L.) under MeJA treatment. BMC Plant Biol. 2020, 20, 353. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, R.; Ma, G.; Zhang, L.; Hirai, M.; Yahata, M.; Yamawaki, K.; Shimada, T.; Fujii, H.; Endo, T.; Kato, M. Effects of Salicylic Acid and Methyl Jasmonate Treatments on Flavonoid and Carotenoid Accumulation in the Juice Sacs of Satsuma Mandarin In Vitro. Appl. Sci. 2020, 10, 8916. [Google Scholar] [CrossRef]
- Ye, Z.C.; Yu, J.; Yan, W.P.; Zhang, J.F.; Yang, D.M.; Yao, G.L.; Liu, Z.J.; Wu, Y.G.; Hou, X.L. Integrative iTRAQ-based proteomic and transcriptomic analysis reveals the accumulation patterns of key metabolites associated with oil quality during seed ripening of Camellia oleifera. Hortic. Res.-Engl. 2021, 8, 157. [Google Scholar] [CrossRef] [PubMed]
- Repka, V. Evidence for the involvement of cysteine proteases in the regulation of methyl jasmonate-induced cell death in grapevine. Vitis Geilweilerhof 2002, 41, 115–121. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; Coninck, B.; Cammue, B.; Bolle, M. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Bioch. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Wang, C.Y.; Fung, R.W.M.; Ding, C.K. Reducing chilling injury and enhancing transcript levels of heat shock proteins, pr-proteins and alternative oxidase by methyl jasmonate and methyl salicylate in tomatoes and peppers. Acta Hortic. 2005, 682, 481–486. [Google Scholar] [CrossRef]
- Pilaisangsuree, V.; Anuwan, P.; Supdensong, K.; Lumpa, P.; Limmongkon, A. Enhancement of adaptive response in peanut hairy root by exogenous signalling molecules under cadmium stress. J. Plant Physiol. 2020, 254, 153278. [Google Scholar] [CrossRef]
- Jiang, A.L.; Liu, Y.N.; Liu, R.; Ren, A.; Ma, H.Y.; Shu, L.B.; Shi, L.; Zhu, J.; Zhao, M.W. Integrated Proteomics and Metabolomics Analysis Provides Insights into Ganoderic Acid Biosynthesis in Response to Methyl Jasmonate in Ganoderma Lucidum. Int. J. Mol. Sci. 2019, 20, 6116. [Google Scholar] [CrossRef]
- Dixon, D.P.; Skipsey, M.; Edwards, R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 2009, 71, 338–350. [Google Scholar] [CrossRef]
- Estévez, I.H.; Hernández, M.R. Plant Glutathione S-transferases: An overview. Plant Gene 2020, 23, 100233. [Google Scholar] [CrossRef]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined Analysis of the Fruit Metabolome and Transcriptome Reveals Candidate Genes Involved in Flavonoid Biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef] [PubMed]
- Premathilake, A.T.; Ni, J.; Shen, J.; Bai, S.; Teng, Y. Transcriptome analysis provides new insights into the transcriptional regulation of methyl jasmonate-induced flavonoid biosynthesis in pear calli. BMC Plant Biol. 2020, 20, 388. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.Y.; Zhang, Y.X.; Peng, W.; Wang, Z.L.; Xie, D.X. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, Z.; Zhang, L.J.; Wang, J.J.; Wu, C.S. Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features. Comput. Struct. Biotec. 2020, 18, 1383–1390. [Google Scholar] [CrossRef]
- Achnine, L.; Huhman, D.V.; Farag, M.A.; Sumner, L.W.; Blount, J.W.; Dixon, R.A. Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 2005, 41, 875–887. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, K.; Qi, Y.; Lv, G.; Ren, X.; Liu, Z.; Ma, F. Transcriptional regulation of anthocyanin synthesis by MYB-bHLH-WDR complexes in kiwifruit (Actinidia chinensis). J. Agr. Food Chem. 2021, 69, 3677–3691. [Google Scholar] [CrossRef]
- Shi, J.; Wang, L.; Ma, C.Y.; Lv, H.P.; Chen, Z.M.; Lin, Z. Aroma changes of black tea prepared from methyl jasmonate treated tea plants. J. Zhejiang Univ. Sci. B 2014, 15, 313–321. [Google Scholar] [CrossRef]
- Chen, C.; Liu, F.; Zhang, K.X.; Niu, X.L.; Zhao, H.; Liu, Q.X.; Georgiev, M.I.; Xu, X.H.; Zhang, X.Q.; Zhou, M.L. MeJA-responsive bHLH transcription factor LjbHLH7 regulates cyanogenic glucoside biosynthesis in Lotus japonicus. J. Exp. Bot. 2022, 73, 2650–2665. [Google Scholar] [CrossRef]
- Yin, X.J.; Liu, S.; Qin, Y.K.; Xing, R.G.; Li, K.C.; Yu, C.L.; Chen, X.L.; Li, P.C. Metabonomics analysis of drought resistance of wheat seedlings induced by beta-aminobutyric acid-modified chitooligosaccharide derivative. Carbohyd. Polym. 2021, 272, 118437. [Google Scholar] [CrossRef]
- Luo, D.; Deng, T.; Yuan, W.; Deng, H.; Jin, M. Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol. 2017, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Li, L.; Tang, S.; Liu, C.; Fu, X.; Shi, Z.; Mao, H. Metabolomics Reveals that Crossbred Dairy Buffaloes are more Thermotolerant than Holstein cows under Chronic Heat Stress. J. Agric. Food Chem. 2018, 66, 12889–12897. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNAseq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Biogeosciences 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. 2005, 4, 17. [Google Scholar] [CrossRef]

| CK vs. 10 mM MeJA | |
|---|---|
| 2 h | |
| More abundant | 59 |
| Less abundant | 27 |
| 48 h | |
| More abundant | 63 |
| Less abundant | 31 |
| Unigene Number | GC/% | N50 Number | N50 Length/bp | Length/bp | Min Length/bp | Average Length/bp | Total Assembled Bases |
|---|---|---|---|---|---|---|---|
| 148,844 | 38.92 | 24,310 | 1276 | 15,639 | 201 | 776 | 115,572,751 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Zheng, W.; Wang, Y.; Wu, Y.; Yu, J.; Xia, P. Integrative Metabolome and Transcriptome Analysis Reveals the Regulatory Network of Flavonoid Biosynthesis in Response to MeJA in Camelliavietnamensis Huang. Int. J. Mol. Sci. 2022, 23, 9370. https://doi.org/10.3390/ijms23169370
Yan H, Zheng W, Wang Y, Wu Y, Yu J, Xia P. Integrative Metabolome and Transcriptome Analysis Reveals the Regulatory Network of Flavonoid Biosynthesis in Response to MeJA in Camelliavietnamensis Huang. International Journal of Molecular Sciences. 2022; 23(16):9370. https://doi.org/10.3390/ijms23169370
Chicago/Turabian StyleYan, Heqin, Wei Zheng, Yong Wang, Yougen Wu, Jing Yu, and Pengguo Xia. 2022. "Integrative Metabolome and Transcriptome Analysis Reveals the Regulatory Network of Flavonoid Biosynthesis in Response to MeJA in Camelliavietnamensis Huang" International Journal of Molecular Sciences 23, no. 16: 9370. https://doi.org/10.3390/ijms23169370
APA StyleYan, H., Zheng, W., Wang, Y., Wu, Y., Yu, J., & Xia, P. (2022). Integrative Metabolome and Transcriptome Analysis Reveals the Regulatory Network of Flavonoid Biosynthesis in Response to MeJA in Camelliavietnamensis Huang. International Journal of Molecular Sciences, 23(16), 9370. https://doi.org/10.3390/ijms23169370







