GcvB Regulon Revealed by Transcriptomic and Proteomic Analysis in Vibrio alginolyticus
Abstract
:1. Introduction
2. Results
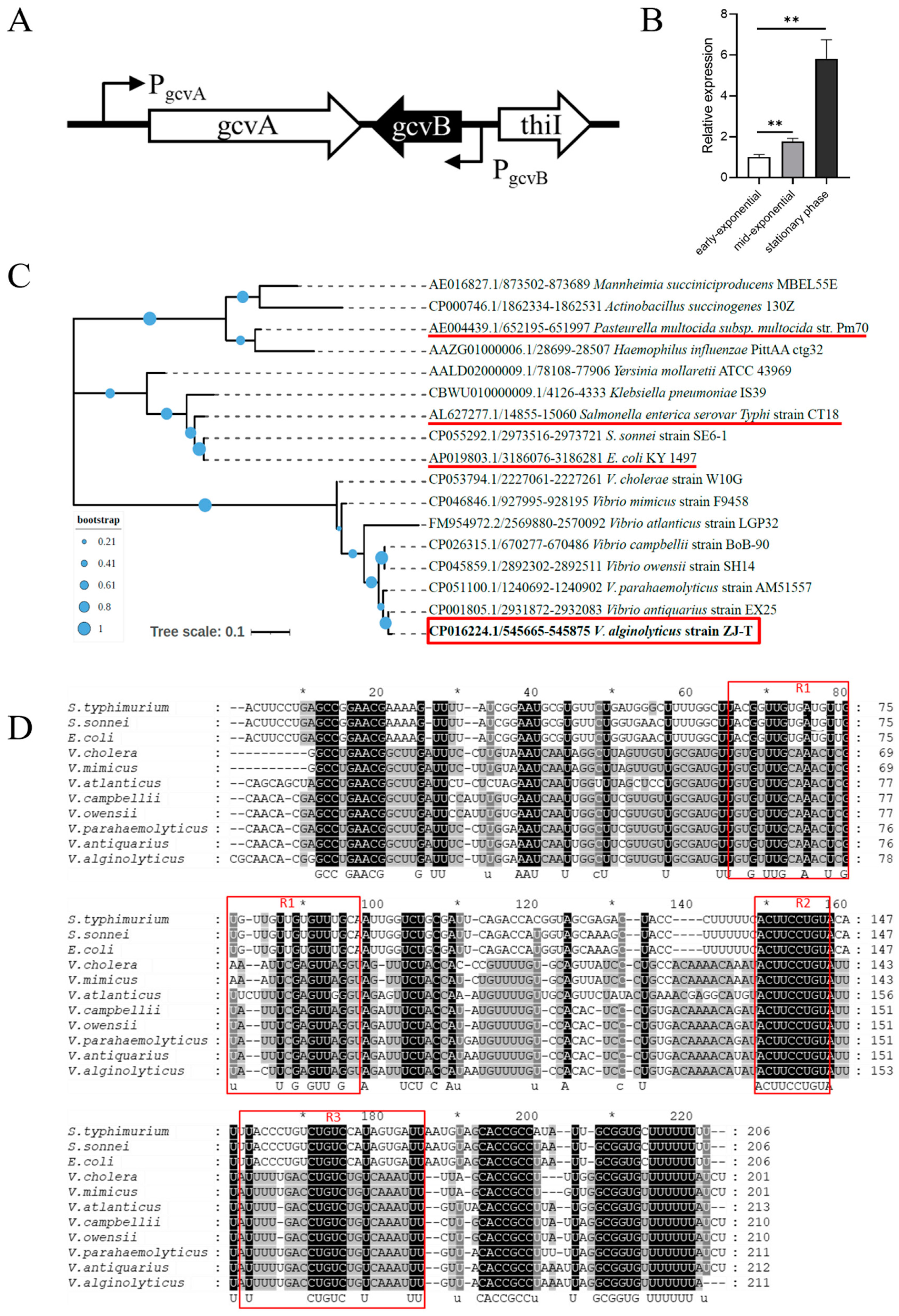
2.1. Bioinformatic Analysis of GcvB Sequence of V. alginolyticus and Its Expression in LBS
2.2. Construction of the Mutant and Complementary Strains and Measurement of Their Growth Ability under Different Conditions
2.3. Integrative Transcriptome and Proteome Analysis of the Wild Type Strain and GcvB Mutant
2.4. Identification of GcvB Regulon and Its Possible Physiological Role in V. alginolyticus ZJ-T
2.4.1. Valine/Leucine/Isoleucine Biosynthetic Pathway
2.4.2. Sulfur and Cysteine Biosynthesis Metabolism
2.4.3. ABC Transporters
2.4.4. Bacterial Secretion Systems
2.5. Effects of Hfq on GcvB
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Media
4.2. Phylogenetic Tree and Sequence Analysis
4.3. Mutant and Complementary Strains Construction
4.4. Growth Measurement
4.5. RNA Extraction and Whole-Genome RNA-Sequencing
4.6. Protein Extraction and Protein Digestion
4.7. High PH Reverse Phase Separation and DIA(Nano-HPLC-MS/MS Analysis)
4.8. Protein Functional Annotation, Enrichment Analysis, and PPI Network Construction and Analysis
4.9. Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
4.10. Translational Fusion
4.11. Hfq Recombinant Protein Construction and Purification
4.12. RNA Electrophoretic Mobility Shift Assays (RNA-EMSA)
4.13. RNA Stability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Urbanowski, M.L.; Stauffer, L.T.; Stauffer, G.V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000, 37, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.M.; Darfeuille, F.; Plantinga, T.H.; Vogel, J. Small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007, 21, 2804–2817. [Google Scholar] [CrossRef] [Green Version]
- Gulliver, E.L.; Wright, A.; Lucas, D.D.; Megroz, M.; Kleifeld, O.; Schittenhelm, R.B.; Powell, D.R.; Seemann, T.; Bulitta, J.B.; Harper, M.; et al. Determination of the small RNA GcvB regulon in the Gram-negative bacterial pathogen Pasteurella multocida and identification of the GcvB seed binding region. Rna 2018, 24, 704–720. [Google Scholar] [CrossRef] [Green Version]
- McArthur, S.D.; Pulvermacher, S.C.; Stauffer, G.V. The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol. 2006, 6, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silveira, A.C.G.; Robertson, K.L.; Lin, B.C.; Wang, Z.; Vora, G.J.; Vasconcelos, A.T.R.; Thompson, F.L. Identification of non-coding RNAs in environmental vibrios. Microbiol. -Sgm 2010, 156, 2452–2458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyakoshi, M.; Okayama, H.; Lejars, M.; Kanda, T.; Tanaka, Y.; Itaya, K.; Okuno, M.; Itoh, T.; Iwai, N.; Wachi, M. Mining RNA-seq data reveals the massive regulon of GcvB small RNA and its physiological significance in maintaining amino acid homeostasis in Escherichia coli. Mol. Microbiol. 2021, 117, 160–178. [Google Scholar] [CrossRef]
- Liu, L.J.; Dong, R.R.; Wang, K.G.; Cheng, Z.T.; Wen, M.; Wen, G.L.; Li, C.; Yang, Q.; Zhou, B.J. mRNA-Seq whole transcriptome analysis of sRNA gcvB deletion background in Salmonella. Acta Veter. Zootech. Sin. 2019, 50, 840–850. [Google Scholar]
- Jorgensen, M.G.; Nielsen, J.S.; Boysen, A.; Franch, T.; Moller-Jensen, J.; Valentin-Hansen, P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012, 84, 36–50. [Google Scholar] [CrossRef] [Green Version]
- Coornaert, A.; Chiaruttini, C.; Springer, M.; Guillier, M. Post-Transcriptional Control of the Escherichia coli PhoQ-PhoP Two-Component System by Multiple sRNAs Involves a Novel Pairing Region of GcvB. PLoS Genet. 2013, 9, e1003156. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Watt, R.M.; Danchin, A.; Huang, J.D. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genom. 2009, 10, 165. [Google Scholar] [CrossRef] [Green Version]
- Ju, X.; Fang, X.X.; Xiao, Y.Z.; Li, B.Y.; Shi, R.P.; Wei, C.L.; You, C.H. Small RNA GcvB Regulates Oxidative Stress Response of Escherichia coli. Antioxidants 2021, 10, 1774. [Google Scholar] [CrossRef] [PubMed]
- Muto, A.; Goto, S.; Kurita, D.; Ushida, C.; Himeno, H. Involvement of GcvB small RNA in intrinsic resistance to multiple aminoglycoside antibiotics in Escherichia coli. J. Biochem. 2021, 169, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Pulvermacher, S.C.; Stauffer, L.T.; Stauffer, G.V. The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. Fems Microbiol. Lett. 2008, 281, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulvermacher, S.C.; Stauffer, L.T.; Stauffer, G.V. The Small RNA GcvB Regulates sstT mRNA Expression in Escherichia coli. J. Bacteriol. 2009, 191, 238–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulvermacher, S.C.; Stauffer, L.T.; Stauffer, G.V. Role of the Escherichia coli Hfq protein in GcvB regulation of oppA and dppA mRNAs. Microbiol. -Sgm 2009, 155, 115–123. [Google Scholar] [CrossRef]
- Sharma, C.M.; Papenfort, K.; Pernitzsch, S.R.; Mollenkopf, H.-J.; Hinton, J.C.D.; Vogel, J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 2011, 81, 1144–1165. [Google Scholar] [CrossRef]
- Yang, Q.; Figueroa-Bossi, N.; Bossi, L. Translation Enhancing ACA Motifs and Their Silencing by a Bacterial Small Regulatory RNA. PLoS Genet. 2014, 10, e1004026. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Gottesman, S. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res. 2016, 44, 6907–6923. [Google Scholar] [CrossRef] [Green Version]
- Lalaouna, D.; Eyraud, A.; Devinck, A.; Prevost, K.; Masse, E. GcvB small RNA uses two distinct seed regions to regulate an extensive targetome. Mol. Microbiol. 2019, 111, 473–486. [Google Scholar] [CrossRef]
- Miyakoshi, M.; Chao, Y.; Vogel, J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. Embo J. 2015, 34, 1478–1492. [Google Scholar] [CrossRef]
- Pulvermacher, S.C.; Stauffer, L.T.; Stauffer, G.V. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiol. -Sgm 2009, 155, 106–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melamed, S.; Peer, A.; Faigenbaum-Romm, R.; Gatt, Y.E.; Reiss, N.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol. Cell 2016, 63, 884–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melamed, S.; Adams, P.P.; Zhang, A.X.; Zhang, H.G.; Storz, G. RNA-RNA Interactomes of ProQ and Hfq Reveal Overlapping and Competing Roles. Mol. Cell 2020, 77, 411. [Google Scholar] [CrossRef] [PubMed]
- Iosub, I.A.; van Nues, R.W.; McKellar, S.W.; Nieken, K.J.; Marchioretto, M.; Sy, B.; Tree, J.J.; Viero, G.; Granneman, S. Hfq CLASH uncovers sRNA-target interaction networks linked to nutrient availability adaptation. eLife 2020, 9, e54655. [Google Scholar] [CrossRef]
- Xie, J.; Bu, L.; Jin, S.; Wang, X.; Zhao, Q.; Zhou, S.; Xu, Y. Outbreak of vibriosis caused by Vibrio harveyi and Vibrio alginolyticus in farmed seahorse Hippocampus kuda in China. Aquaculture 2020, 523, 735168. [Google Scholar] [CrossRef]
- Xie, Z.; Ke, S.; Hu, C.; Zhu, Z.; Wang, S.; Zhou, Y. First Characterization of Bacterial Pathogen, Vibrio alginolyticus, for Porites andrewsi White Syndrome in the South China Sea. PLoS ONE 2013, 8, e75425. [Google Scholar]
- Taylor, R.; Mcdonald, M.; Russ, G.; Carson, M.; Lukaczynski, E. Vibrio alginolyticus peritonitis associated with ambulatory peritoneal dialysis. Br. Med. J. 1981, 283, 275. [Google Scholar] [CrossRef] [Green Version]
- Opal, S.M.; Saxon, J.R.; Opal, S.M.; Saxon, J.R. Intracranial infection by Vibrio alginolyticus following injury in salt water. J. Clin. Microbiol. 1986, 23, 373. [Google Scholar] [CrossRef] [Green Version]
- Barbarossa, V.; Kucisec-Tepes, N.; Aldova, E.; Matek, D.; Stipoljev, F. Ilizarov technique in the treatment of chronic osteomyelitis caused by Vibrio alginolyticus. Croat. Med. J. 2002, 43, 346–349. [Google Scholar]
- Chien, J.; Shih, J.; Hsueh, P.; Yang, P.; Luh, K.J.E.J.C.M.I.D. Vibrio alginolyticus as the cause of pleural empyema and bacteremia in an immunocompromised patient. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 401–403. [Google Scholar]
- Feingold, M.H.; Kumar, M.L. Otitis media associated with Vibrio alginolyticus in a child with pressure-equalizing tubes. Pediatric Infect. Dis. J. 2004, 23, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Xiang, Z.Y.; Xu, X.M.; Yan, W.H.; Ma, J.M. Endophthalmitis Caused by Vibrio alginolyticus. J. Clin. Microbiol. 2009, 47, 3379–3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, G.L.; Dai, X.Q.; Lu, C.J.; Liu, J.M.; Zhao, G.J.; Wu, B.; Li, M.F.; Lu, Z.Q. Temporizing surgical management improves outcome in patients with Vibrio necrotizing fasciitis complicated with septic shock on admission. Burns 2014, 40, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, C.; Zhao, Z.; Zhao, J.; Annick, J.; Huang, X.; Yang, Y.; Mikael, S. The RNA Chaperone Hfq Is Involved in Colony Morphology, Nutrient Utilization and Oxidative and Envelope Stress Response in Vibrio alginolyticus. PLoS ONE 2016, 11, e0163689. [Google Scholar] [CrossRef]
- Liu, B.; Gao, Q.; Zhang, X.; Chen, H.; Zhang, Y.; Sun, Y.; Yang, S.; Chen, C. CsrA Regulates Swarming Motility and Carbohydrate and Amino Acid Metabolism in Vibrio alginolyticus. Microorganisms 2021, 9, 2383. [Google Scholar] [CrossRef]
- Deng, Y.; Chen, C.; Zhao, Z.; Huang, X.; Yang, Y.; Ding, X. Complete Genome Sequence of Vibrio alginolyticus ZJ-T. Genome Announc. 2016, 4, e00912-16. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.Q.; Luo, X.; Xie, M.; Bouloc, P.; Chen, C.; Jacq, A. The ilvGMEDA Operon Is Regulated by Transcription Attenuation in Vibrio alginolyticus ZJ-T. Appl. Environ. Microbiol. 2019, 85, e00880-19. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.H.; Yu, F.; Chen, M.; Zhang, Y.F.; Qu, Q.W.; Wei, Y.R.; Xie, C.M.; Wu, T.; Liu, Y.Y.; Zhang, Z.Y.; et al. Tylosin Inhibits Streptococcus suis Biofilm Formation by Interacting With the O-acetylserine (thiol)-lyase B CysM. Front. Vet. Sci. 2022, 8, 829899. [Google Scholar] [CrossRef]
- Poole, P.S.; Franklin, M.; Glenn, A.R.; Dilworth, M.J. The transport of L-glutamate by Rhizobium-Leguminosarum involves a common amino-acid carrier. J. Gen. Microbiol. 1985, 131, 1441–1448. [Google Scholar] [CrossRef] [Green Version]
- Walshaw, D.L.; Poole, P.S. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol. Microbiol. 1996, 21, 1239–1252. [Google Scholar] [CrossRef]
- Cascales, E. The type VI secretion toolkit. Embo Rep. 2008, 9, 735–741. [Google Scholar] [CrossRef] [Green Version]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Diepold, A.; Kudryashev, M.; Delalez, N.J.; Berry, R.M.; Armitage, J.P. Composition, Formation, and Regulation of the Cytosolic C-ring, a Dynamic Component of the Type III Secretion Injectisome. PLoS Biol. 2015, 13, e1002039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, R.D.; Singh, P.; Hsu, F.S.; Guvener, T.; Carl, M.A.; Trinidad, R.R.S.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.G.; Plemel, R.L.; et al. A Type VI Secretion System of Pseudomonas aeruginosa Targets, a Toxin to Bacteria. Cell Host Microbe 2010, 7, 25–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, T.M.; Unterweger, D.; Bachmann, V.; Kostiuk, B.; Pukatzki, S. Lytic Activity of the Vibrio cholerae Type VI Secretion Toxin VgrG-3 Is Inhibited by the Antitoxin TsaB. J. Biol. Chem. 2013, 288, 7618–7625. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.G.; Ho, B.T.; Yoder-Himes, D.R.; Mekalanos, J.J. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2013, 110, 2623–2628. [Google Scholar] [CrossRef] [Green Version]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Lensmire, J.M.; Hammer, N.D. Nutrient sulfur acquisition strategies employed by bacterial pathogens. Curr. Opin. Microbiol. 2019, 47, 52–58. [Google Scholar] [CrossRef]
- Singh, P.; Brooks, J.F.; Ray, V.A.; Mandel, M.J.; Visick, K.L. CysK Plays a Role in Biofilm Formation and Colonization by Vibrio fischeri. Appl. Environ. Microbiol. 2015, 81, 5223–5234. [Google Scholar] [CrossRef] [Green Version]
- Wasilko, N.P.; Larios-Valencia, J.; Steingard, C.H.; Nunez, B.M.; Verma, S.C.; Miyashiro, T. Sulfur availability for Vibrio fischeri growth during symbiosis establishment depends on biogeography within the squid light organ. Mol. Microbiol. 2019, 111, 621–636. [Google Scholar] [CrossRef]
- Chen, H.; Previero, A.; Deutscher, M.P. A novel mechanism of ribonuclease regulation: GcvB and Hfq stabilize the mRNA that encodes RNase BN/Z during exponential phase. J. Biol. Chem. 2019, 294, 19997–20008. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, C.; Hu, C.-Q.; Ren, C.-H.; Zhao, J.-J.; Zhang, L.-P.; Jiang, X.; Luo, P.; Wang, Q.-B. The type III secretion system of Vibrio alginolyticus induces rapid apoptosis, cell rounding and osmotic lysis of fish cells. Microbiol. -Sgm 2010, 156, 2864–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Liu, J.X.; Deng, Y.Q.; Huang, W.; Ren, C.H.; Call, D.R.; Hu, C.Q. The Vibrio alginolyticus T3SS effectors, Val1686 and Val1680, induce cell rounding, apoptosis and lysis of fish epithelial cells. Virulence 2018, 9, 318–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, K.H.; Corley, J.M.; Djapgne, L.; Cribbs, J.T.; Voelker, D.; Slusher, Z.; Nordell, R.; Regulski, E.E.; Kazmierczak, B.I.; McMackin, E.W.; et al. Hfq and sRNA 179 Inhibit Expression of the Pseudomonas aeruginosa CAMP-Vfr and Type III Secretion Regulons. mBio 2020, 11, e00363-20. [Google Scholar] [CrossRef]
- Chang, C.; Jin, X.; Hu, C. Phenotypic and genetic differences between opaque and translucent colonies of Vibrio alginolyticus. Biofouling 2009, 25, 525–531. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Disconzi, E.; Charriere, G.M.; Destoumieux-Garzon, D.; Bouloc, P.; Le Roux, F.; Jacq, A. csrB Gene Duplication Drives the Evolution of Redundant Regulatory Pathways Controlling Expression of the Major Toxic Secreted Metalloproteases in Vibrio tasmaniensis LGP32. Msphere 2018, 3, e00582-18. [Google Scholar] [CrossRef] [Green Version]
- Val, M.E.; Skovgaard, O.; Ducos-Galand, M.; Bland, M.J.; Mazel, D. Genome engineering in Vibrio cholerae: A feasible approach to address biological issues. PLoS Genet. 2012, 8, e1002472. [Google Scholar] [CrossRef] [Green Version]
- Morales, V.M.; Backman, A.; Bagdasarian, M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 1991, 97, 39–47. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Chen, G.; Lin, H.; Wang, P.; Kuang, B.; Liu, J.; Chen, S. Development of a regulatable expression system for the functional study of Vibrio vulnificus essential genes. Antonie Van Leeuwenhoek 2017, 110, 607–614. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357-U54. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Butz, H.A.; Mey, A.R.; Ciosek, A.L.; Payne, S.M. Vibrio cholerae CsrA Directly Regulates varA To Increase Expression of the Three Nonredundant Csr Small RNAs. mBio 2019, 10, e01042-19. [Google Scholar] [CrossRef] [Green Version]










| Strains or Plasmids | Relevant Characteristics | Source |
|---|---|---|
| Vibrio alginolyticus | ||
| ZJ-T | Apr, translucent/smooth variant of wild strain ZJ51; isolated from diseased Epinephelus coioides off the Southern China coast | [55] |
| ZJ-T-ΔgcvB | Apr; ZJ-T carrying a deletion of gcvB | This study |
| ZJ-T-△gcvB+ | Cmr; ZJ-T carrying a GcvB complementation plasmid pMMB207-gcvB | This study |
| ZJ-T-Δhfq | Apr; ZJ-T carrying a deletion of hfq | [34] |
| ZJ-T/pSCT32-gfp-ilvG-TL | Cmr; ZJ-T carrying a cysK translational fusion plasmid pSCT32-gfp-ilvG-TL | This study |
| ZJ-T-△gcvB/pSCT32-gfp-ilvG-TL | Cmr; ZJ-T-△gcvB carrying a cysK translational fusion plasmid pSCT32-gfp-ilvG-TL | This study |
| ZJ-T/pSCT32-gfp-cysK-TL | Cmr; ZJ-T carrying a cysK translational fusion plasmid pSCT32-gfp-cysK-TL | This study |
| ZJ-T-△gcvB/pSCT32-gfp-cysK-TL | Cmr; ZJ-T-△gcvB carrying a cysK translational fusion plasmid pSCT32-gfp-cysK-TL | This study |
| ZJ-T/pSCT32-gfp-cysN -TL | Cmr; ZJ-T carrying a cysN translational fusion plasmid pSCT32-gfp-cysD-TL | This study |
| ZJ-T-△gcvB/SCT32-gfp-cysN-TL | Cmr; ZJ-T-△gcvB carrying a cysN translational fusion plasmid pSCT32-gfp-cysD-TL | This study |
| E. coli | ||
| GEB883 | WT; E. coli K12 ΔdapA::ermpir RP4-2 ΔrecA gyrA462, zei298::Tn10; donor strain for conjugation | [56] |
| pET28b-Hfq/BL21(DE3) | Kanr; E. coli BL21(DE3) carrying the fusion expression plasmid pET28b-Hfq::His tag | This study |
| Plasmids | ||
| pSW7848 | Cmr; suicide vector with an R6K origin, requiring the Pir protein for its replication, and the ccdB toxin gene | [57] |
| pSW7848-ΔgcvB | Cmr; pSW7848 containing the mutant allele of ΔgcvB | This study |
| pMMB207 | Cmr; RSF1010 derivative, IncQ lacIq Ptac oriT | [58] |
| pMMB207-gcvB | Cmr; pMMB207 containing the wild-type allele of gcvB | This study |
| pSCT32 | Cmr; expression plasmid with a pBR322 and a f1 origin at the same time and a tac promoter | [59] |
| pSCT32-gfp | Cmr; pSCT32 containing reporter gene gfp coding green fluorescent protein | This study |
| pSCT32-gfp-ilvG-TL | Cmr; ilvG sequences (including its promotor and start codon) are translationally fused to pSCT32-gfp | This study |
| pSCT32-gfp-cysK-TL | Cmr; cysK sequences (including its promotor and start codon) are translationally fused to pSCT32-gfp | This study |
| pSCT32-gfp-cysN-TL | Cmr; cysN sequences (including its promotor and start codon) are translationally fused to pSCT32-gfp | This study |
| pET28b | Kanr; expression plasmid with a pBR322 origin, T7 promoter and 6×histag. | Xiaoxue Wang |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Fang, J.; Chen, H.; Sun, Y.; Yang, S.; Gao, Q.; Zhang, Y.; Chen, C. GcvB Regulon Revealed by Transcriptomic and Proteomic Analysis in Vibrio alginolyticus. Int. J. Mol. Sci. 2022, 23, 9399. https://doi.org/10.3390/ijms23169399
Liu B, Fang J, Chen H, Sun Y, Yang S, Gao Q, Zhang Y, Chen C. GcvB Regulon Revealed by Transcriptomic and Proteomic Analysis in Vibrio alginolyticus. International Journal of Molecular Sciences. 2022; 23(16):9399. https://doi.org/10.3390/ijms23169399
Chicago/Turabian StyleLiu, Bing, Jianxiang Fang, Huizhen Chen, Yuehong Sun, Shan Yang, Qian Gao, Ying Zhang, and Chang Chen. 2022. "GcvB Regulon Revealed by Transcriptomic and Proteomic Analysis in Vibrio alginolyticus" International Journal of Molecular Sciences 23, no. 16: 9399. https://doi.org/10.3390/ijms23169399






