Blocking Rice Shoot Gravitropism by Altering One Amino Acid in LAZY1
Abstract
:1. Introduction
2. Results
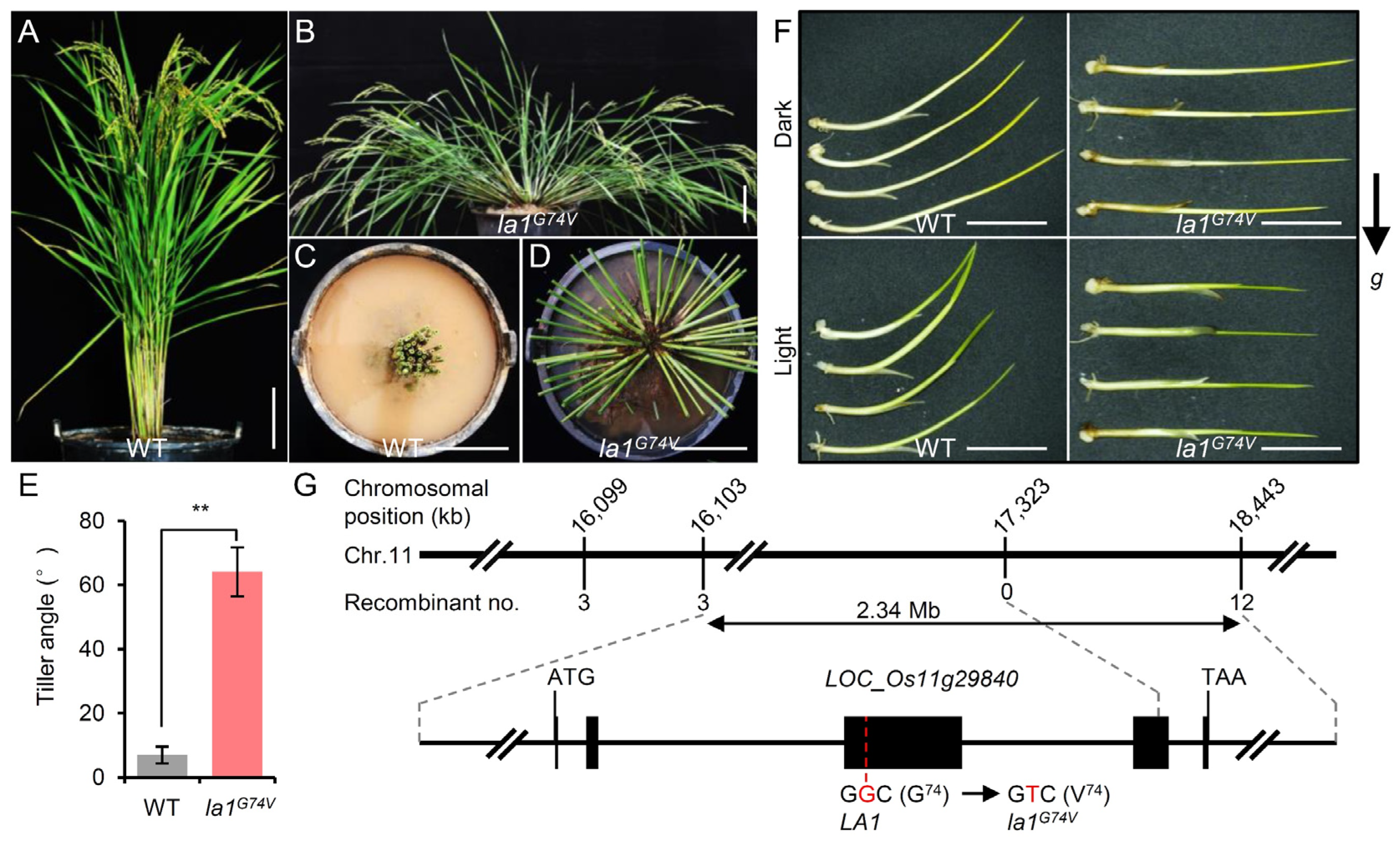
2.1. Characterization of the Rice Spreading-Tiller Mutant la1G74V
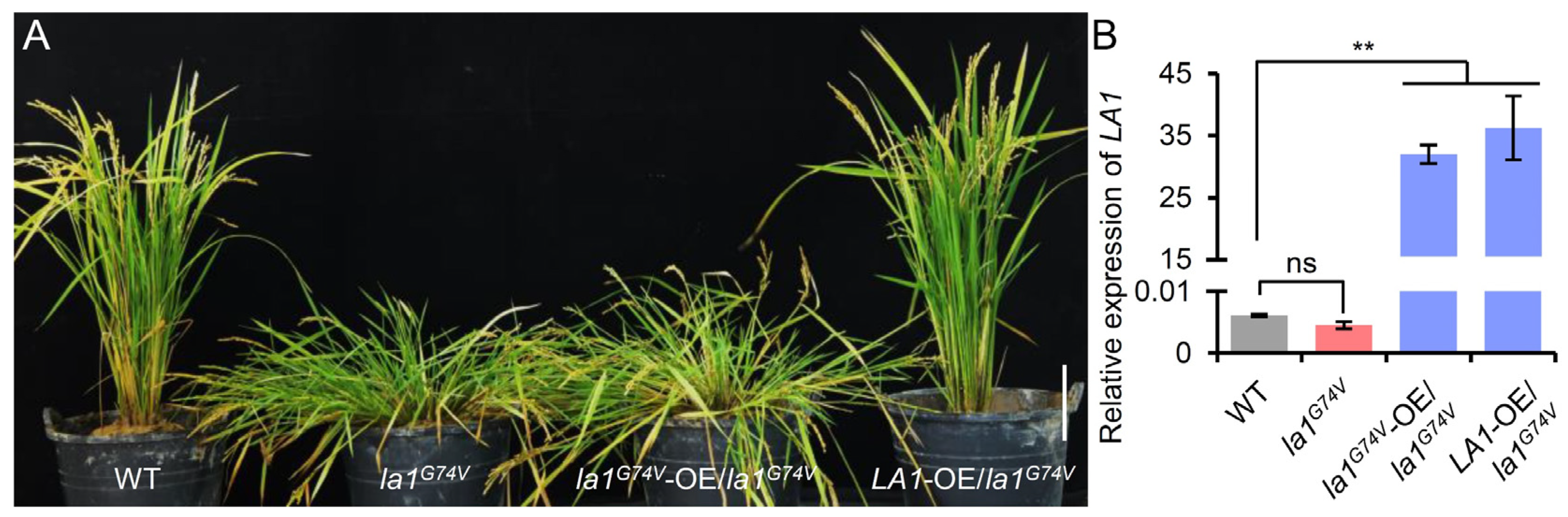
2.2. Map-Based Cloning of la1G74V
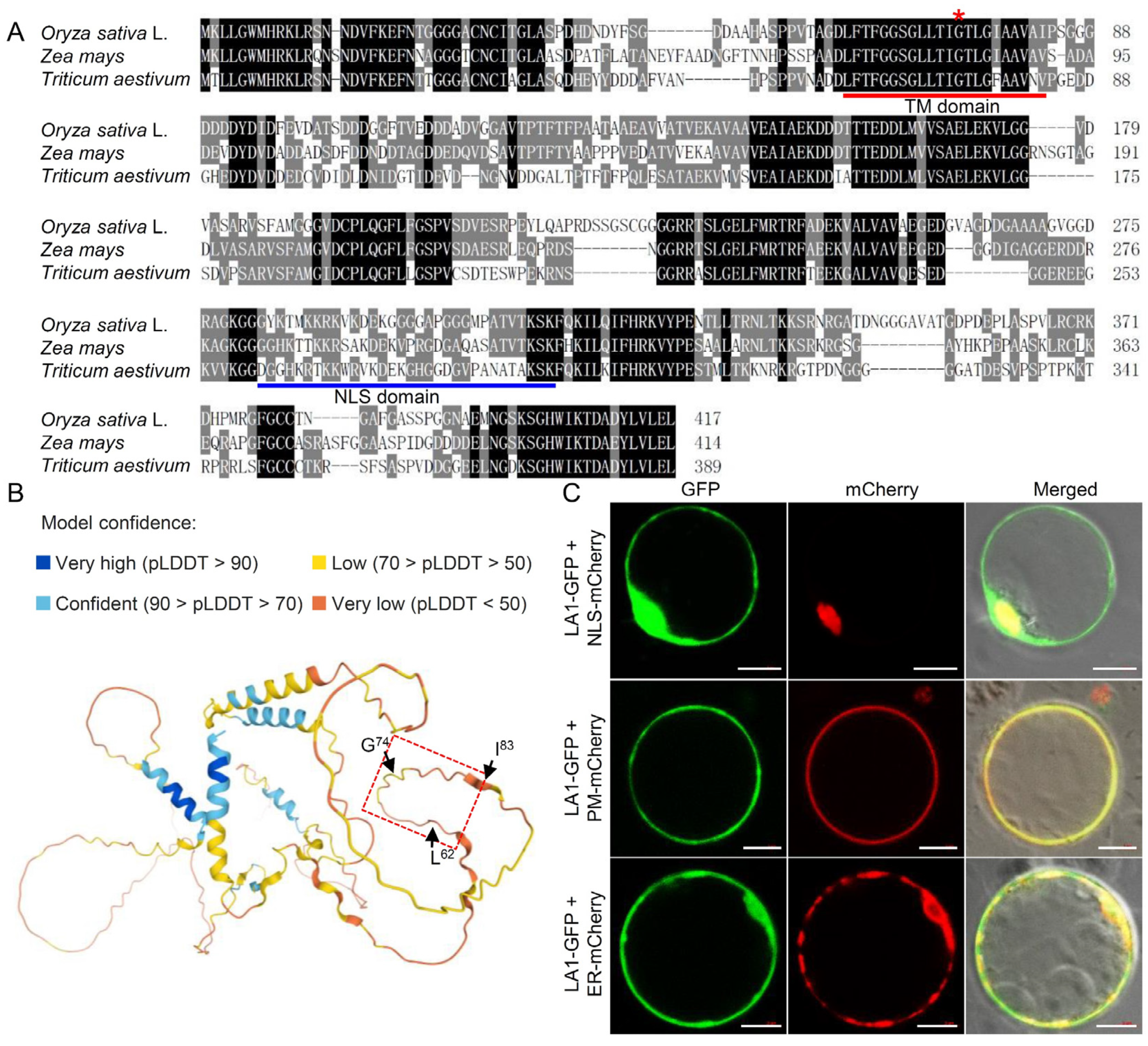
2.3. The Effect of G74V Mutation on the Conformation and Subcellular Localization of LA1 Protein
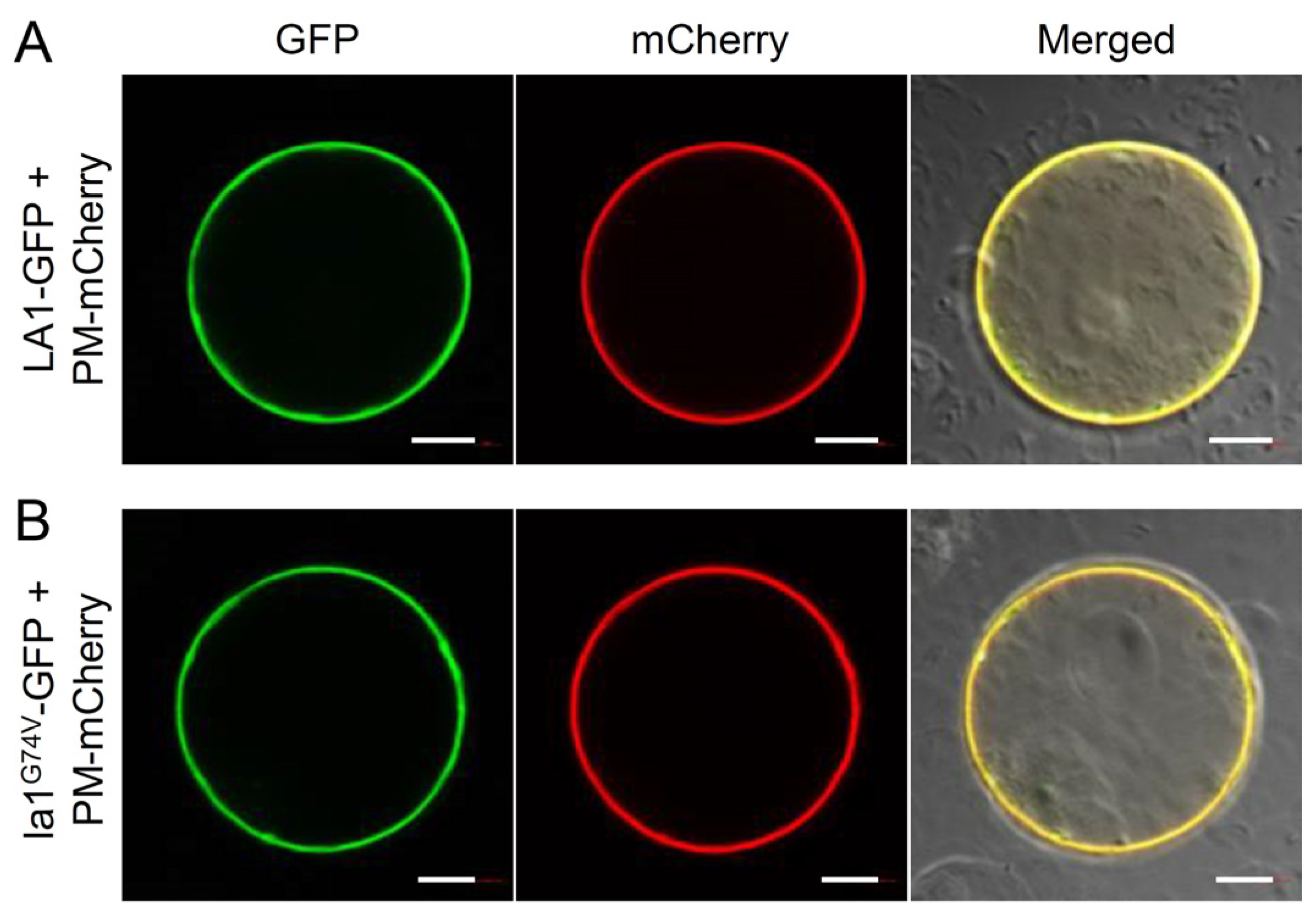
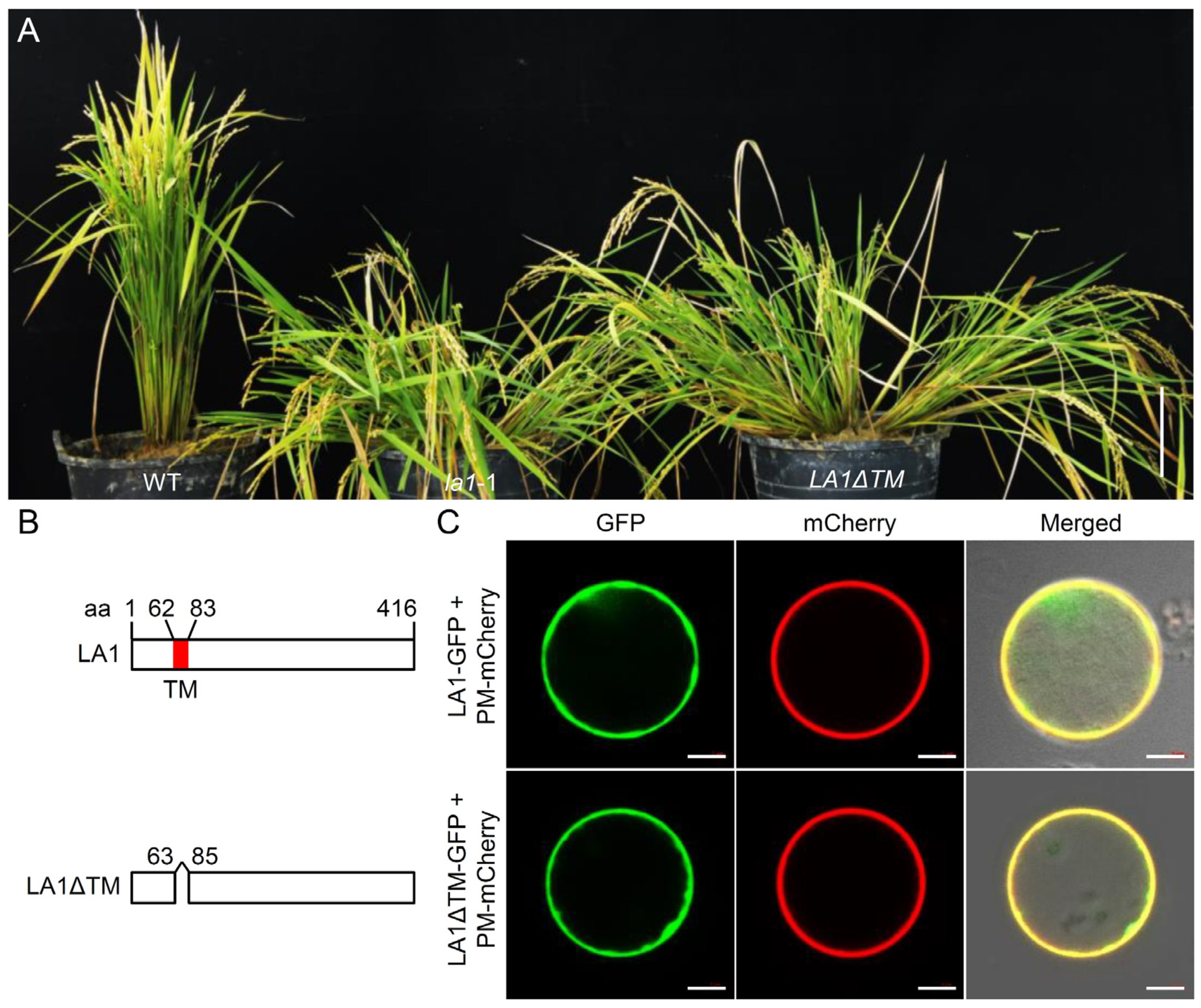
2.4. The TM Domain of LA1 Is Essential for Its Biological Function Rather Than Its Plasma Membrane Localization
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Analysis of Plant Gravity Response
4.3. Sequence Alignment and 3D Structural Modeling of Proteins
4.4. RNA Extraction and qRT-PCR
4.5. Vector Construction and Rice Transformation
4.6. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, Y.; Li, J. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Du, H.; Chen, S.; Huang, W.; Ge, L. LAZY gene family in plant gravitropism. Front. Plant Sci. 2021, 11, 606241. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Chen, L.; Herrera-Estrella, L.; Cao, D.; Tran, L.P. Altering plant architecture to improve performance and resistance. Trends Plant Sci. 2020, 25, 1154–1170. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Wing, R.A.; Purugganan, M.D.; Zhang, Q. The rice genome revolution: From an ancient grain to green super rice. Nat. Rev. Genet. 2018, 19, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Parida, S.K. Restructuring plant types for developing tailor-made crops. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Qian, Q.; Fu, Z.; Wang, M.; Zeng, D.; Li, B.; Wang, X.; Li, J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007, 17, 402–410. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, T.; Iino, M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 2007, 48, 678–688. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef]
- Dong, H.; Zhao, H.; Xie, W.; Han, Z.; Li, G.; Yao, W.; Bai, X.; Hu, Y.; Guo, Z.; Lu, K.; et al. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 2016, 12, e1006412. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Sun, H.; Jiang, J.; Sun, X.; Tan, L.; Sun, C. TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnol. J. 2021, 19, 64–73. [Google Scholar] [CrossRef]
- Jin, J.; Huang, W.; Gao, J.; Yang, J.; Shi, M.; Zhu, M.; Luo, D.; Lin, H. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef]
- Hu, M.; Lv, S.; Wu, W.; Fu, Y.; Liu, F.; Wang, B.; Li, W.; Gu, P.; Cai, H.; Sun, C.; et al. The domestication of plant architecture in African rice. Plant J. 2018, 94, 661–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Tang, D.; Wang, K.; Cheng, Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Piao, W.; Lim, J.; Han, S.; Kim, Y.; An, G.; Paek, N. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tan, L.; Zhu, Z.; Xiao, L.; Xie, D.; Sun, C. PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 2015, 83, 528–536. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Tan, L.; Sun, H.; Zhao, X.; Liu, F.; Cai, H.; Fu, Y.; Sun, X.; Gu, P.; Zhu, Z.; et al. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol. Plant 2019, 12, 1075–1089. [Google Scholar] [CrossRef]
- Lopez, D.; Tocquard, K.; Venisse, J.; Legué, V.; Drevet-Roeckel, P. Gravity sensing, a largely misunderstood trigger of plant orientated growth. Front. Plant Sci. 2014, 5, 610. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Nishimura, T.; Morita, M.T. Gravity sensing and signal conversion in plant gravitropism. J. Exp. Bot. 2019, 70, 3495–3506. [Google Scholar] [CrossRef]
- Dong, Z.; Jiang, C.; Chen, X.; Zhang, T.; Ding, L.; Song, W.; Luo, H.; Lai, J.; Chen, H.; Liu, R.; et al. Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol. 2013, 163, 1306–1322. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Spalding, E.P. LAZY genes mediate the effects of gravity on auxin gradients and plant architecture. Plant Physiol. 2017, 175, 959–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef]
- Kitomi, Y.; Hanzawa, E.; Kuya, N.; Inoue, H.; Hara, N.; Kawai, S.; Kanno, N.; Endo, M.; Sugimoto, K.; Yamazaki, T.; et al. Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proc. Natl. Acad. Sic. USA 2020, 114, 21242–21250. [Google Scholar] [CrossRef]
- Yoshihara, T.; Spalding, E.P.; Iino, M. AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J. 2013, 74, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Furutani, M.; Nishimura, T.; Nakamura, M.; Fushita, T.; Iijima, K.; Baba, K.; Tanaka, H.; Toyota, M.; Tasaka, M.; et al. The Arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell 2017, 29, 1984–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strohm, A.K.; Baldwin, K.L.; Masson, P.H. Multiple roles for membrane-associated protein trafficking and signaling in gravitropism. Front. Plant Sci. 2012, 3, 274. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, K.L.; Strohm, A.K.; Masson, P.H. Gravity sensing and signal transduction in vascular plant primary roots. Am. J. Bot. 2013, 100, 126–142. [Google Scholar] [CrossRef]
- Sang, D.; Chen, D.; Liu, G.; Liang, Y.; Huang, L.; Meng, X.; Chu, J.; Sun, X.; Dong, G.; Yuan, Y.; et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Pro. Natl. Acad. Sci. USA 2014, 111, 11199–11204. [Google Scholar] [CrossRef] [Green Version]
- Waite, M.J.; Dardick, C. The roles of the IGT gene family in plant architecture: Past, present, and future. Cur. Opin. Plant Biol. 2021, 59, 101983. [Google Scholar] [CrossRef]
- Wang, W.; Gao, H.; Liang, Y.; Li, J.; Wang, Y. Molecular basis underlying rice tiller angle: Current progress and future perspectives. Mol. Plant 2022, 15, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Li, S.; Fan, X.; Song, S.; Zhou, X.; Weng, X.; Xiao, J.; Li, X.; Xiong, L.; You, A.; et al. OsHOX1 and OsHOX28 redundantly shape rice tiller angle by reducing HSFA2D expression and auxin content. Plant Physiol. 2020, 184, 1424–1437. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, S.; Tian, L.; Liu, L.; Huang, M.; Xu, X.; Song, G.; Wu, P.; Sato, S.; Jiang, H.; et al. LAZY3 plays a pivotal role in positive root gravitropism in Lotus japonicus. J. Exp. Bot. 2020, 71, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liang, Y.; Yuan, Y.; Wang, L.; Meng, X.; Xiong, G.; Zhou, J.; Cai, Y.; Han, N.; Hua, L.; et al. OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization. Mol. Plant 2019, 12, 1143–1156. [Google Scholar] [CrossRef]
- Chen, Y.; Liang, H.; Ma, X.; Lou, S.; Xie, Y.; Liu, Z.; Chen, L.; Liu, Y. An efficient rice mutagenesis system based on suspension-cultured cells. J. Integr. Plant Biol. 2013, 55, 122–130. [Google Scholar] [CrossRef]
- Hill, J.L., Jr.; Hollender, C.A. Branching out: New insights into the genetic regulation of shoot architecture in trees. Curr. Opin. Plant Biol. 2019, 47, 73–80. [Google Scholar] [CrossRef]
- Zheng, H.Q.; Staehelin, L.A. Nodal endoplasmic reticulum, a specialized form of endoplasmic reticulum found in gravity-sensing root tip columella cells. Plant Physiol. 2001, 125, 252–265. [Google Scholar] [CrossRef] [Green Version]
- Leitz, G.; Kang, B.H.; Schoenwaelder, M.E.; Staehelin, L.A. Statolith sedimentation kinetics and force transduction to the cortical endoplasmic reticulum in gravity-sensing Arabidopsis columella cells. Plant Cell 2009, 21, 843–860. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Nishimura, T.; Morita, M.T. Bridging the gap between amyloplasts and directional auxin transport in plant gravitropism. Curr. Opin. Plant Biol. 2019, 52, 54–60. [Google Scholar] [CrossRef]
- Ge, L.; Chen, R. Negative gravitropic response of roots directs auxin flow to control root gravitropism. Plant Cell Environ. 2019, 42, 2372–2383. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Spalding, E.P. Switching the direction of stem gravitropism by altering two amino acids in AtLAZY1. Plant Physiol. 2020, 182, 1039–1051. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Hu, Y.; Tong, A.; Yan, B.; Lv, Y.; Wang, S.; Ma, W.; Cui, Z.; Wang, X. LAZY1 controls tiller angle and shoot gravitropism by regulating the expression of auxin transporters and signaling factors in rice. Plant Cell Physiol. 2021, 61, 2111–2125. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Gupta, A.; Laxmi, A. Striking the right chord: Signaling enigma during root gravitropism. Front. Plant Sci. 2017, 8, 1304. [Google Scholar] [CrossRef] [Green Version]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [Green Version]
- García Bossi, J.; Kumar, K.; Barberini, M.L.; Domínguez, G.D.; Rondón Guerrero, Y.D.C.; Marino-Buslje, C.; Obertello, M.; Muschietti, J.P.; Estevez, J.M. The role of P-type IIA and P-type IIB Ca2+-ATPases in plant development and growth. J. Exp. Bot. 2020, 71, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Rössner, N.; Hoang, M.T.T.; Alejandro, S.; Peiter, E. Transport, functions, and interaction of calcium and manganese in plant organellar compartments. Plant Physiol. 2021, 187, 1940–1972. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, H.; Yan, J.; Tian, F. Natural variation in crops: Realized understanding, continuing promise. Annu. Rev. Plant Biol. 2021, 72, 357–385. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, S.; Li, X.; Zhang, B.; Jiang, L.; Tang, Y.; Zhao, J.; Ma, X.; Cai, H.; Sun, C.; et al. Deletions linked to PROG1 gene participate in plant architecture domestication in Asian and African rice. Nat. Commun. 2018, 9, 4157. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xie, Y.; Zhu, Q.; Liu, Y. Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol. Plant 2017, 10, 1368–1370. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Liu, Y.; Chen, Y. Genome-editing technologies: The gap between application and policy. Sci. China Life Sci. 2019, 62, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, Z.; Ding, Z.; Meng, H.; Shen, R.; Tang, H.; Liu, Y.; Chen, L. Public-transcriptome-database-assisted selection and validation of reliable reference genes for qRT-PCR in rice. Sci. China Life Sci. 2020, 63, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Schimittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient; high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Ma, K.; Li, H.; Su, J.; Zhou, L.; Tang, J.; Zhang, S.; Hou, Y.; Chen, L.; Liu, Y.; et al. All-in-one: A robust fluorescent fusion protein vector toolbox for protein localization and BiFC analyses in plants. Plant Biotechnol. J. 2022, 20, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Huang, Y.; Han, J.; Zhang, S.; Yang, Q.; Li, Z.; Zhang, Y.; Mao, R.; Fan, L.; Liu, Y.; et al. Blocking Rice Shoot Gravitropism by Altering One Amino Acid in LAZY1. Int. J. Mol. Sci. 2022, 23, 9452. https://doi.org/10.3390/ijms23169452
Chen S, Huang Y, Han J, Zhang S, Yang Q, Li Z, Zhang Y, Mao R, Fan L, Liu Y, et al. Blocking Rice Shoot Gravitropism by Altering One Amino Acid in LAZY1. International Journal of Molecular Sciences. 2022; 23(16):9452. https://doi.org/10.3390/ijms23169452
Chicago/Turabian StyleChen, Shuifu, Yuqun Huang, Jingluan Han, Shijuan Zhang, Qiaoyu Yang, Zhijie Li, Ya Zhang, Runyuan Mao, Ling Fan, Yaoguang Liu, and et al. 2022. "Blocking Rice Shoot Gravitropism by Altering One Amino Acid in LAZY1" International Journal of Molecular Sciences 23, no. 16: 9452. https://doi.org/10.3390/ijms23169452
APA StyleChen, S., Huang, Y., Han, J., Zhang, S., Yang, Q., Li, Z., Zhang, Y., Mao, R., Fan, L., Liu, Y., Chen, Y., & Xie, X. (2022). Blocking Rice Shoot Gravitropism by Altering One Amino Acid in LAZY1. International Journal of Molecular Sciences, 23(16), 9452. https://doi.org/10.3390/ijms23169452




