Effects of Exogenous Melatonin on Root Physiology, Transcriptome and Metabolome of Cotton Seedlings under Salt Stress
Abstract
:1. Introduction
2. Results
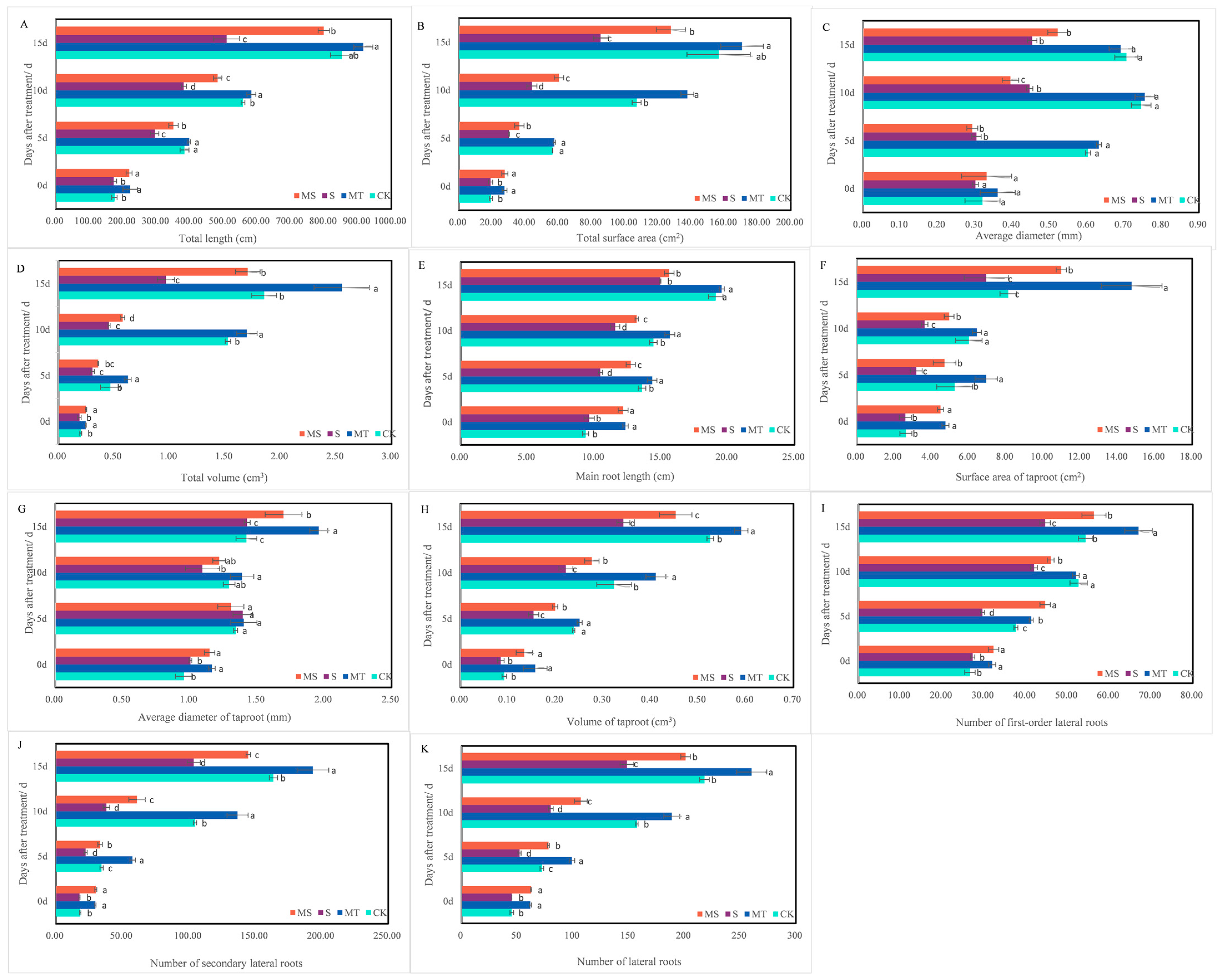
2.1. Effects of Exogenous Melatonin on Cotton Seedlings and Their Root Morphology under Salt Stress
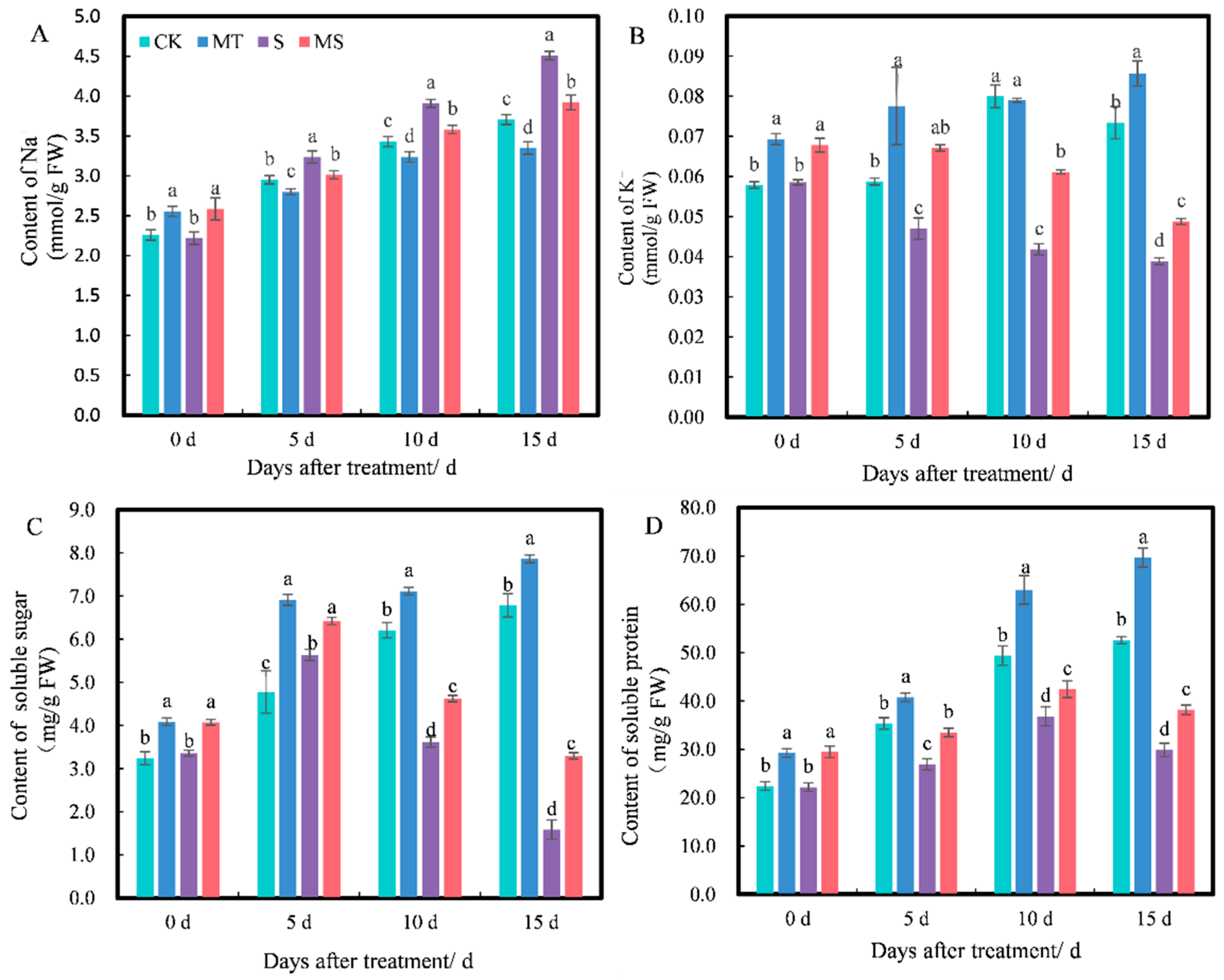
2.2. Effects of Exogenous Melatonin on the Root Antioxidant System of Cotton Seedlings under Salt Stress
2.3. Effects of Exogenous Melatonin on Osmotic Substance Contents of Cotton Seedling Roots under Salt Stress
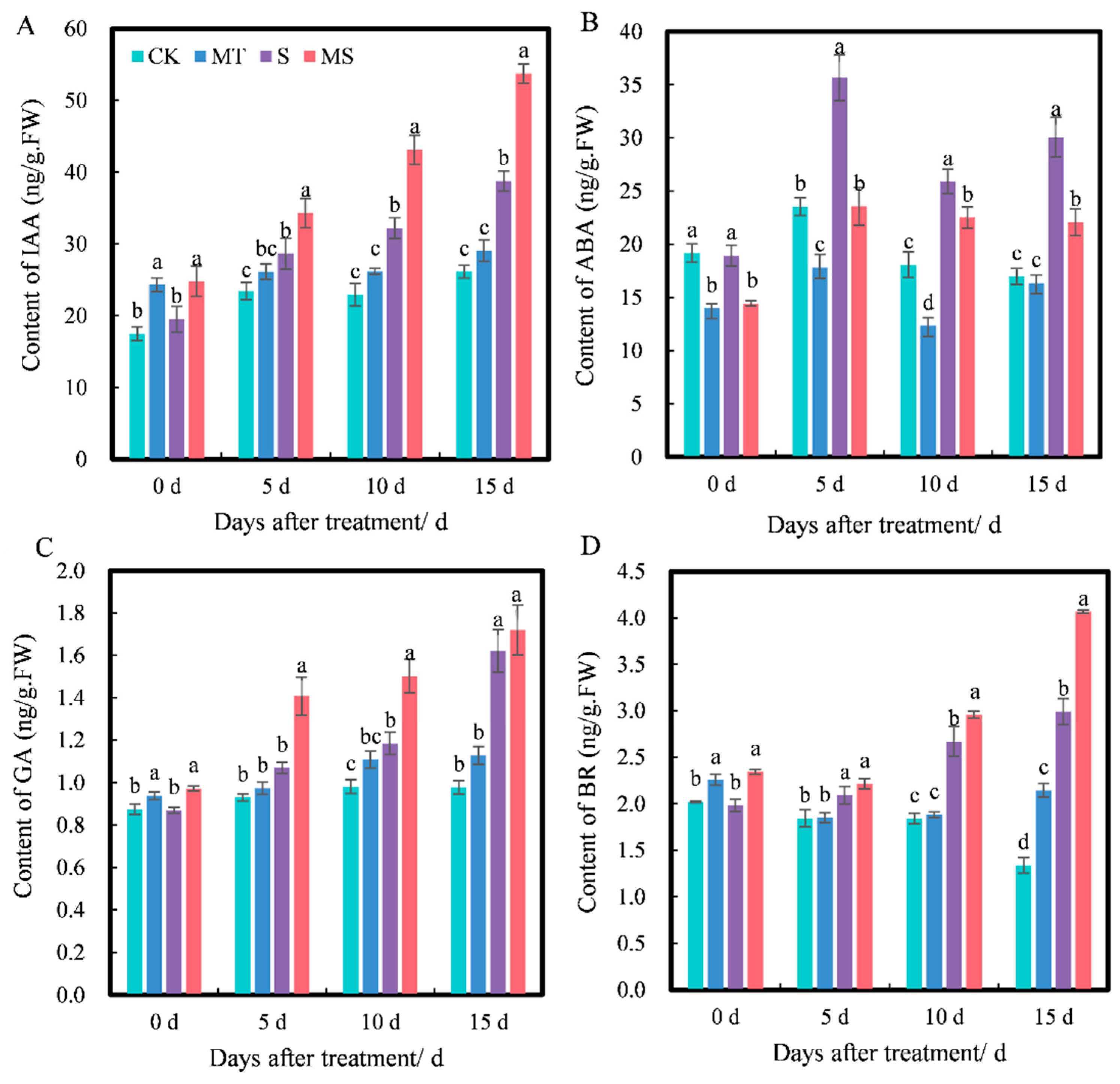
2.4. Effects of Exogenous Melatonin on Root Hormones of Cotton Seedlings under Salt Stress
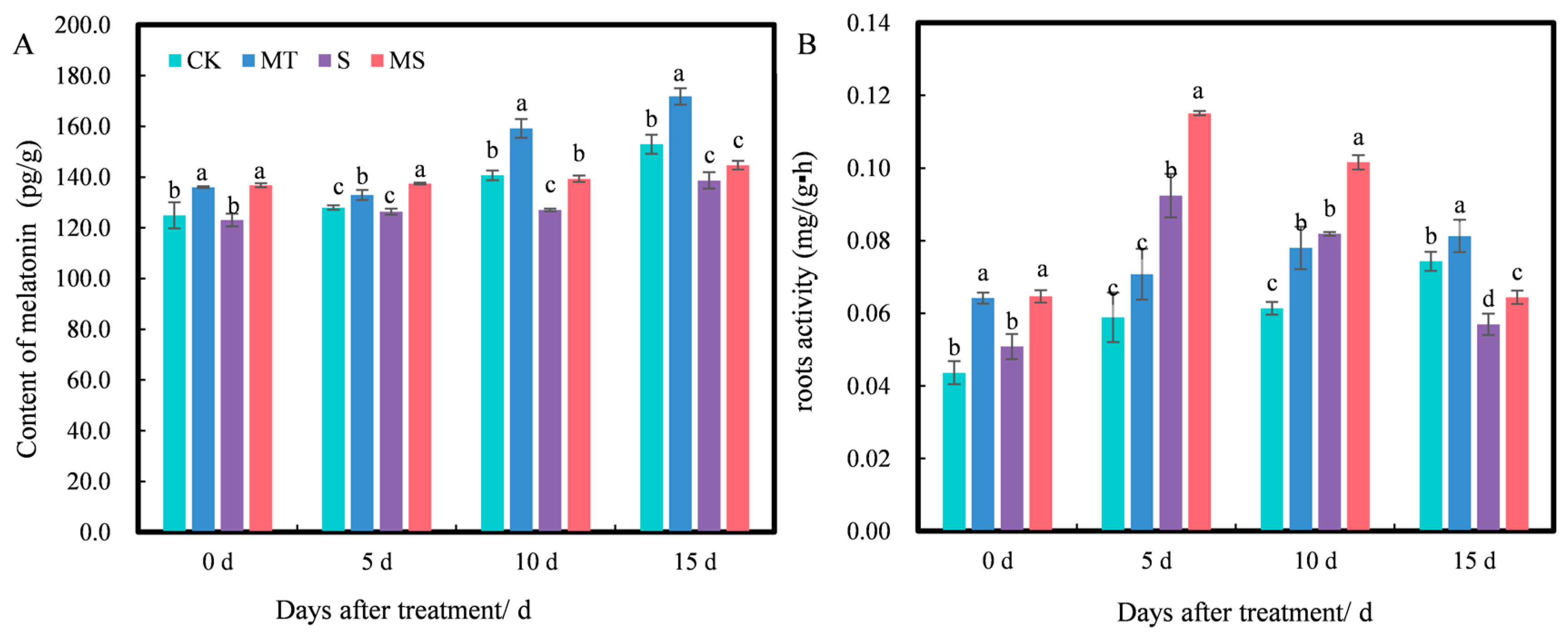
2.5. Effects of Exogenous Melatonin on Endogenous Melatonin Content and Root Activity of Cotton Seedlings under Salt Stress
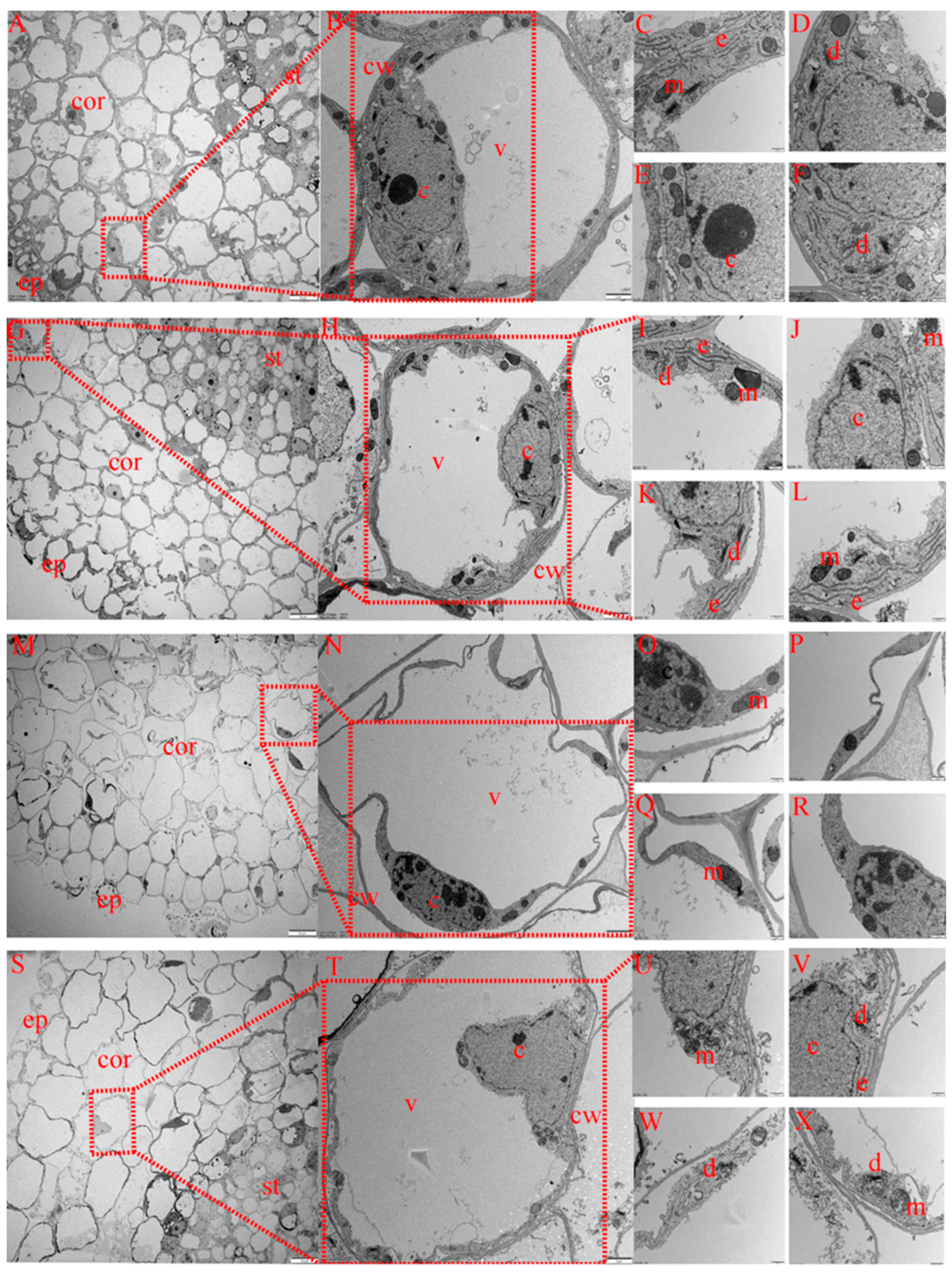
2.6. Effects of Exogenous Melatonin on Taproot Tip Ultrastructure of Cotton Seedlings under Salt Stress
2.6.1. Differences in Root Epidermal Cell Characteristics of Cotton Seedlings among Treatments
2.6.2. Differences in Root Cortex Cell Characteristics of Cotton Seedlings among Treatments
2.6.3. Differences in Root Phloem Cell Characteristics of Cotton Seedlings among Treatments
2.7. Transcriptomic Analysis of Cotton Seedling Root Systems under Melatonin and Salt Stress
2.7.1. Functional Analysis of Gene Expression Changes in Roots of Melatonin-Treated Cotton Seedlings under Salt Stress
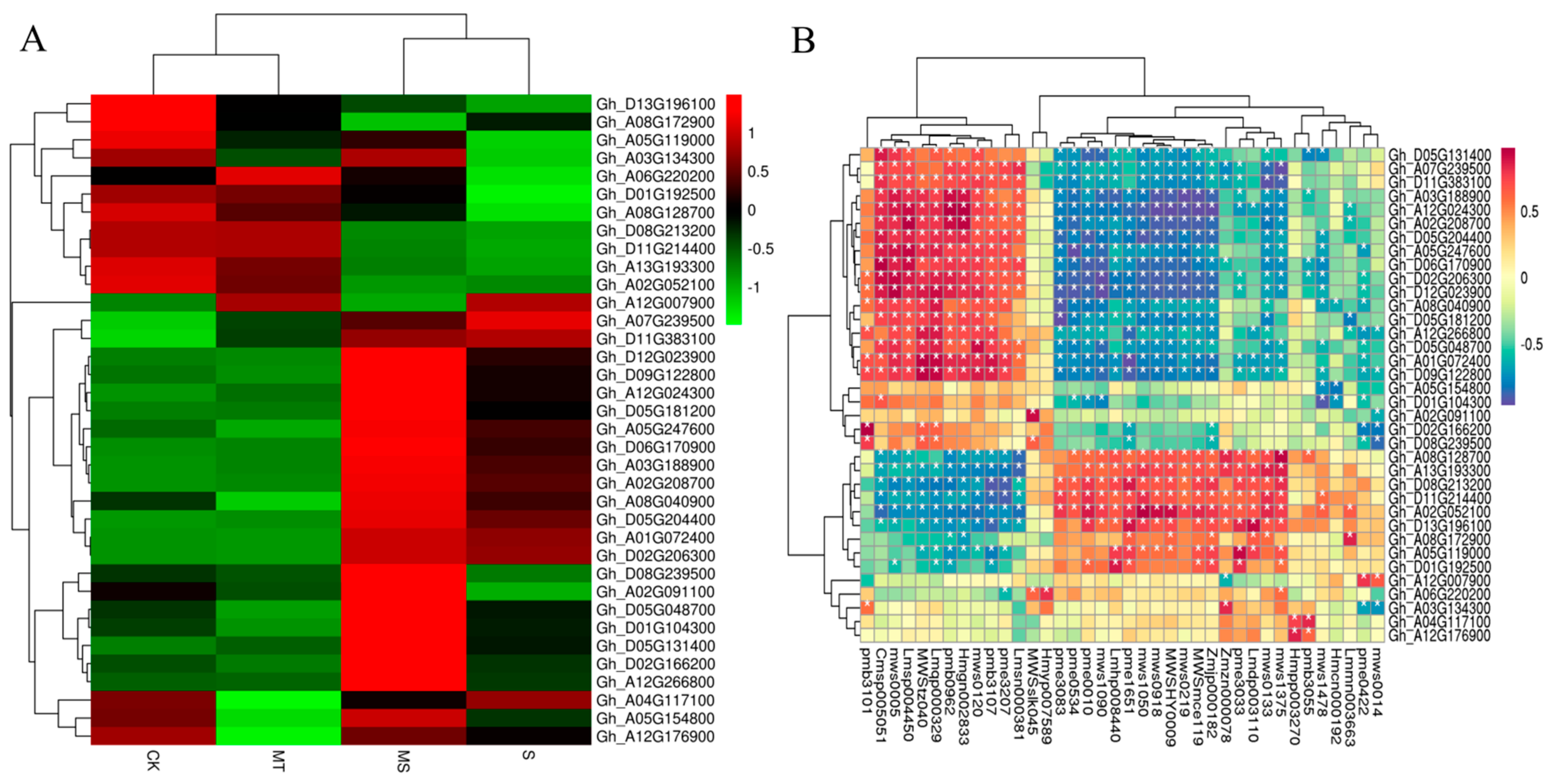
2.7.2. Quantitative Analysis of the Effect of Exogenous Melatonin on Differentially Expressed Genes in Cotton Seedling Roots under Salt Stress
2.8. Metabolomic Effects of Exogenous Melatonin on the Root Systems of Cotton Seedlings under Salt Stress
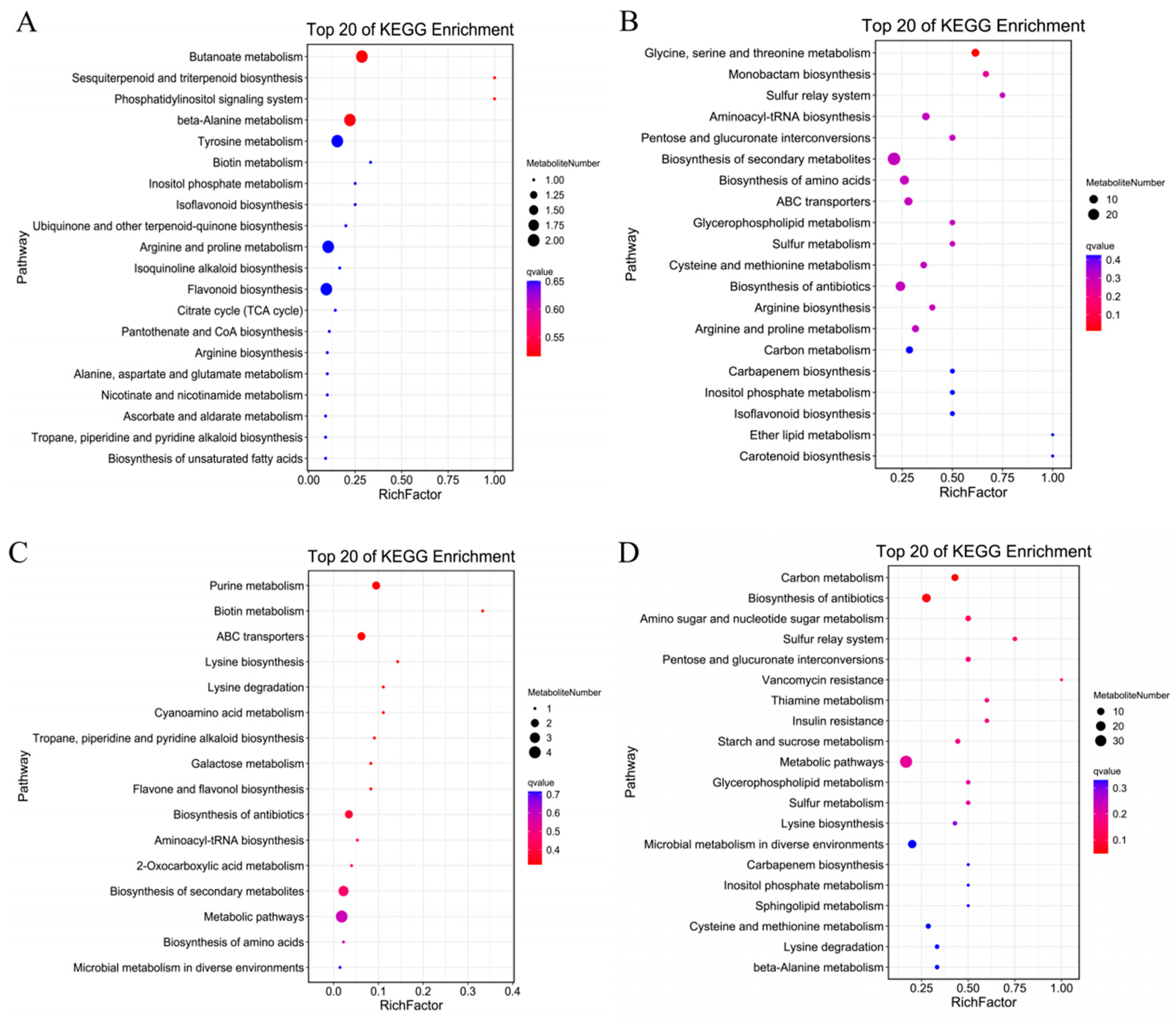
2.8.1. Analysis of Differentially Accumulated Metabolites in Roots of Cotton Seedlings under Salt Stress and Exogenous Melatonin Treatment
2.8.2. Combined Transcriptomic and Metabolomic Analysis
3. Discussion
3.1. Effect of Exogenous Melatonin on the Antioxidant System of Cotton Seedling Roots under Salt Stress
3.2. Effect of Exogenous Melatonin on Osmotic Substances in Cotton Seedling Roots under Salt Stress
3.3. Effect of Exogenous Melatonin on Root Hormones of Cotton Seedlings under Salt Stress
3.4. Effect of Exogenous Melatonin on Root Microstructure of Cotton Seedlings under Salt Stress
3.5. Effects of Exogenous Melatonin on Root Metabolic Pathways of Cotton Seedlings under Salt Stress
3.6. Regulation Mode of Exogenous Melatonin on Root Growth of Cotton under Salt Stress
4. Materials and Methods
4.1. Plant Materials
4.2. Methods
4.2.1. Experimental Treatment
4.2.2. Determination Method
Determination of Antioxidant Enzyme Activity, Contents of Osmotic Regulatory Substances and Root Activity
Determination of Hormone Content
Ultrastructure Observations
Transcriptome Analysis
RNA Extraction, Library Construction and Sequencing
qRT-PCR
Metabolic Analysis
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Morton, M.J.L.; Awlia, M.; Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel-Dissecting the genetics of salt tolerance. Plant J. 2018, 97, 148–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenrick, P.; Crane, P.R. The origin and early evolution of plants on land. Nature 1997, 389, 33–39. [Google Scholar] [CrossRef]
- Ghosh, D.; Xu, J. Abiotic stress responses in plant roots: A proteomics perspective. Front. Plant Sci. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasim, I.; Mosaddek, A.I.; Chen, X.H.; Wu, F.B. Genotype-dependent alleviation effects of exogenous GSH on salinity stress in cotton is related to improvement in chlorophyll content, photosynthetic performance, and leaf/root ultrastructure. Environ. Sci. Pollut. Res. 2017, 24, 9417–9427. [Google Scholar]
- Jbir, N.; Chaïbi, W.; Ammar, S.; Jemmali, A.; Ayadi, A. Root growth and lignification of two wheat species differing in their sensitivity to NaCl in response to salt stress. CR Acad. Sci. III 2001, 324, 863–868. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Z.; Ge, S.; Peng, B.; Zhang, K.; Hu, M.; Mai, W.; Tian, C. Root Morphology and Rhizosphere Characteristics Are Related to Salt Tolerance of Suaeda salsa and Beta vulgaris L. Front. Plant. Sci. 2021, 12, 677767. [Google Scholar] [CrossRef]
- Ulrich, D.; Aaron, B.S.; Tomoaki, H.; Wei, L.; Guohua, X.; Julian, I.S. Plant salt-tolerance mechanisms. Trends in Plant Sci. 2014, 19, 371–379. [Google Scholar]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2009, 33, 453–467. [Google Scholar] [CrossRef]
- West, G.; Inzé, D.; Beemster, G.T. Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 2004, 135, 1050–1058. [Google Scholar] [CrossRef]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Li, Y.; Zhu, Y.M.; Bai, X.; Lv, D.K.; Guo, D.J.; Ji, W.; Cai, H. Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 2010, 10, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaura, K.; Mochida, K.; Ogihara, Y. Genome-wide analysis for identification of salt responsive genes in common wheat. Funct. Integr. Genom. 2008, 8, 277–286. [Google Scholar] [CrossRef]
- Gao, C.Q.; Wang, Y.C.; Liu, G.F.; Wang, C.; Jiang, J.; Yang, C.P. Cloning of ten peroxidase (POD) genes from Tamarix hispida and characterization of their responses to abiotic stress. Plant Mol. Biol. Rep. 2010, 28, 77–89. [Google Scholar] [CrossRef]
- Wang, Y.C.; Yang, C.P.; Liu, G.F.; Jiang, J. Development of a cDNA microarray to identify gene expression of Puccinellia tenuiflora under salinealkali stress. Plant Physiol. Biochem. 2007, 45, 567–576. [Google Scholar] [CrossRef]
- ALerner, B.; Case, J.D.; Takahashi, Y. Isolation of melatonin, apinealg land factor that lightens melanocytes. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Manchester, L.C.; Coto-Montes, A.; Boga, J.A.; Peter, H.L.A.; Zhou, Z.; Galano, A.; Vriend, J.; Tan, D.X.; Reiter, R.J. Melatonin: An ancient molecule that makes oxygen metabolically tolerable. J. Pineal Res. 2015, 59, 403–419. [Google Scholar] [CrossRef]
- Murch, S.J.; Campbell, S.S.B.; Saxena, P.K. The role of serotonin and melatonin in plant morphogenesis: Regulation of auxin induced root organogenesis in in vitro-cultured explants of St. John’s wort (Hypericum perforatum L.). In Vitro Cell Dev. Biol. Plant 2001, 37, 786–793. [Google Scholar] [CrossRef]
- Ramón, P.F.; Francisco, R.H.L.; José, L.B. Serotonin modulates Arabidopsis root growth via changes in reactive oxygen species and jasmonic acid-ethylene signaling. Physiol. Plant. 2016, 158, 92–105. [Google Scholar]
- Back, K.W.; Tan, D.X.; Reiter, R.J. Melatonin biosynthesis in plants: Multiple pathway scatalyze tryptophan to melatonin in the cytoplasmor chloroplasts. J. Pineal Res. 2016, 61, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.Z.; Li, A.F.; Yu, H.; Li, W.Z.; Liang, C.Z.; Guo, S.D.; Zhang, R.; Chu, C.C. Melatonin Regulates Root Architecture by Modulating Auxin Response in Rice. Front. Plants Sci. 2017, 8, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.Q.; Wang, Y.; Zhang, J.K.; Gong, X.J.; Zhang, Z.; Zhang, Z.; Sun, J.J.; Chen, X.S.; Wang, Y.L. Exogenous Melatonin Improves Physiological Characteristics and Promotes Growth of Strawberry Seedlings Under Cadmium Stress. Hortic. Plant J. 2020, 7, 13–22. [Google Scholar] [CrossRef]
- ElSayed, A.; Rafudeen, M.; Gomaa, A.; Hasanuzzaman, M. Exogenous melatonin enhances the reactive oxygen species metabolism, antioxidant defense-related gene expression, and photosynthetic capacity of Phaseolus vulgaris L. to confer salt stress tolerance. Physiol. Plant 2021, 173, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Manchester, L.C.; Liu, X.Y.; Sergio, A.; Corral, R.; Acuna-Castroviejo, D.; Reiter, R.J. Mitochondria and chloroplasts as the original sites of melatonin synthesis: Ahypo thesis related to melatonin’s primary function and evolution in eukaryotes. J. Pineal Res. 2013, 54, 127–138. [Google Scholar] [CrossRef]
- Chen, L.; Lu, B.; Liu, L.T.; Duan, W.J.; Jiang, D.; Li, J.; Zhang, K.; Sun, H.C.; Zhang, Y.J.; Li, C.D.; et al. Melatonin promotes seed germination under salt stress by regulating ABA and GA3 in cotton (Gossypium hirsutum L.). Plant Physiol. Biochem. 2021, 162, 506–516. [Google Scholar] [CrossRef]
- Wang, D.K.; Hao, Z.Q.; Zhao, J.S.; Yao, J.; Huang, J.; Zhou, R.Q.; Wu, C.D. Comparative physiological and transcriptomic analyses reveal salt tolerance mechanisms of Zygosaccharomyces rouxii. Process Biochem. 2019, 82, 59–67. [Google Scholar] [CrossRef]
- Turk, K.; Erdal, S.; Genisel, M.; Atici, O.; Demir, Y.; Yanmis, D. The regulatory effect of melatonin on physiological, biochemical and molecular parameters in cold-stressed wheat seedlings. Plant Growth Regul. 2014, 74, 139–152. [Google Scholar] [CrossRef]
- Chen, L.; Liu, L.T.; Lu, B.; Ma, T.T.; Jiang, D.; Li, J.; Zhang, K.; Sun, H.C.; Zhang, Y.J.; Bai, Z.Y.; et al. Exogenous melatonin promotes seed germination and osmotic regulation under salt stress in cotton (Gossypium hirsutum L.). PLoS ONE 2020, 15, e0228241. [Google Scholar]
- Ma, Y.; Wei, Z.; Liu, J.; Liu, X.; Liu, F. Growth and physiological responses of cotton plants to salt stress. J. Agron. Crop Sci. 2021, 207, 565–576. [Google Scholar] [CrossRef]
- Jiang, D.; Lu, B.; Liu, L.T.; Duan, W.J.; Meng, Y.J.; Li, J.; Zhang, K.; Sun, H.C.; Zhang, Y.J.; Dong, H.Z.; et al. Exogenous melatonin improves the salt tolerance of cotton by removing active oxygen and protecting photosynthetic organs. BMC Plant Biol. 2021, 21, 331. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.J.; Meng, Y.J.; Jiang, D.; Liu, L.T.; Zhang, K.; Zhang, Y.J.; Sun, H.C.; Bai, Z.Y.; Li, C.D. Effects of exogenous melatonin on morphology and antioxidant enzyme activities of cotton seedlings under salt stress. Chin. J. Eco-Agric. 2022, 30, 92–104. (In Chinese) [Google Scholar]
- StuartJ, R.; Sónia, N.; Mark, T. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar]
- Ashrafm, N. Assessment of variation in antioxidative defense system in salt-treated pea (Pisum sativum) cultivars and its putativeuseas salinity tolerance markers. J. Plant Physiol. 2009, 166, 1764–1774. [Google Scholar]
- Witze, L.K.; Weidner, A.; Surabhi, G.K.; Börner, A.; Mock, H.P. Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response to wards salinity. J. Exp. Bot. 2009, 60, 3545–3557. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Ren, J.; Shi, H.; Chen, X.D.; Zhang, M.M.; Pan, Y.; Fan, J.B.; Nevo, E.; Sun, D.F.; Fu, J.M.; et al. Physiological and Molecular Responses to Salt Stress in Wild Emmer and Cultivated Wheat. Plant Mol. Biol. Rep. 2013, 31, 1212–1219. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Alamri, S.; Al-Khaishany, M.Y.; Khan, M.N.; Al-Amri, A.; Ali, H.M.; Alaraidh, I.A.; Alsahli, A.A. Exogenous Melatonin Counteracts NaCl-Induced Damage by Regulating the Antioxidant System, Proline and Carbohydrates Metabolism in Tomato Seedlings. Nat. Rev. Neurosci. 2019, 20, 353. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Cai, J.S.; Li, J.J.; Lu, G.Y.; Li, C.S.; Fu, G.P.; Zhang, X.K.; Ma, H.Q.; Liu, Q.Y.; Zou, X.L.; et al. Exogenous application of a low concentration of melatonin enhances salt tolerance in rapeseed (Brassicanapus L.) seedlings. J. Integr. Agric. 2018, 17, 328–335. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.P.; Xie, Y.J.; Gu, Q.; Zhao, G.; Zhang, Y.H.; Cui, W.T.; Xu, S.; Wang, R.; Shen, W.B. The AtrbohF-dependent regulation of ROS signaling is required for melatonin-induced salinity tolerance in Arabidopsis. Free Radic. Biol. Med. 2017, 108, 465–477. [Google Scholar] [CrossRef]
- Alscher, R.G.; Donahue, J.L.; Cramer, C.L. Reactive oxygen species and antioxidants: Relationships in green cells. Physiol. Plant. 1997, 100, 224–233. [Google Scholar] [CrossRef]
- Rao, M.V.; Hale, B.A.; Ormrod, D.P. Amelio ration of ozone-induced oxidative damage in Wheat plants grown under high carbon dioxide (Role of antioxidant enzymes). Plant Physiol. 1995, 109, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Liu, J.L.; Wang, W.X.; Sun, Y. Exogenous melatonin improves growth and photosynthetic capacity of cucumber under salinity-induced stress. Photo Synth. 2016, 54, 19–27. [Google Scholar] [CrossRef]
- Kostopoulou, Z.; Therios, I.; Roumeliotis, E.; Kanellis, A.K.; Molassiotis, A. Melatonin combined with ascorbic acid provides salt adaptation in Citrusaurantium L. Seedlings. Plant Physiol. Biochem. 2015, 86, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Citterio, S.; Sparvoli, E. Ascorbic acid effect on the onset of cell proliferation in pea root. Physiol. Plantarum. 1994, 92, 601–607. [Google Scholar] [CrossRef]
- Cristina, O.V.; Rubén, R.A.; Campo, F.F.D.; Carpena-Ruiz, R.O.; Hernández, L.E. Cellular damage induced by cadmium and mercury in Medicagosativa. J. Exp. Bot. 2005, 56, 2239–2251. [Google Scholar]
- Afzal, M.Z.; Jia, Q.; Ibrahim, A.K.; Niyitanga, S.; Zhang, L.W. Mechanisms and Signaling Pathways of Salt Tolerance in Crops: Understanding from the Transgenic Plants. Trop. Plant Biol. 2020, 13, 297–320. [Google Scholar] [CrossRef]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.X. Mechanisms of Plant Salt Response Insights from Proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef]
- Galano, A.; Reiter, R.J. Melatonin and its Metabolites versus Oxidative Stress: From Individual Actions to Collective Protection. J. Pineal Res. 2018, 65, e12514. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188. [Google Scholar] [CrossRef]
- Ahmad, M.S.A.; Ali, Q.; Ashraf, M.; Haider, M.Z.; Abbas, Q. Involvement of polyamines, abscisic acid and antioxidative enzymes inadaptation of Blue Panic grass (Panicum antidotale Retz.) to saline environments. Environ. Exp. Bot. 2009, 66, 409–417. [Google Scholar] [CrossRef]
- Li, Z.G.; Xu, Y.; Bai, L.K.; Zhang, S.Y.; Wang, Y. Melatonin enhances thermotolerance of maize seedlings (Zea mays L.) by modulating antioxidant defense, methylglyoxal detoxification, and osmoregulation systems. Protoplasma 2019, 256, 471–490. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.Y.; Feng, Z.; Bai, Q.Q.; He, J.J.; Wang, Y.J. Melatonin-mediated regulation of growth and antioxidant capacity in salt-tolerant Naked Oat under salt Stress. Int. J. Mol. Sci. 2019, 20, 1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, Action, andInteraction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.P.; Duan, X.B.; Luo, L.; Dai, S.J.; Ding, Z.J.; Xia, G.M. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Shinozaki, K. Type2C protein phosphatases directly regulate abscisicacid-activated proteinkinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [Green Version]
- Su, Q.F.; Zheng, X.D.; Tian, Y.K.; Wang, C.H. Exogenous brassinolide alleviates salt stress in Malus hupehensis Rehd. By regulating the transcription of NHX-type Na+ (K+)/H+ antiporters. Front. Plant Sci. 2020, 11, 38. [Google Scholar] [CrossRef]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitricoxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Rademacher, W. Growth Retardants: Effects on Gibberellin Biosynthesis and Other Metabolic Pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 501–531. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Qin, H.; Zhou, S.R.; Wei, P.C.; Zhang, H.W.; Zhou, Y.; Miao, Y.C.; Huang, R.F. The ubiquitin-binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 2020, 32, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Tanveera, M.; Shahzada, B.; Sharmac, A.; Biju, S.; Bhardwajc, R. 24-Epibrassinolide; an active brassinolide and its role in salt stress tolerance in plants: A review. Plant Physiol. Biochem. 2018, 130, 69–79. [Google Scholar] [CrossRef]
- Zhu, T.; Deng, X.G.; Zhou, X.; Zhu, L.S.; Zou, L.J.; Li, P.X.; Zhang, D.W.; Lin, H.H. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016, 6, 35392. [Google Scholar] [CrossRef] [Green Version]
- Katsuhara, M.; Kawasaki, T. Salt stress induced nuclear and DNA degradation in Meristematic cells of barley roots. Plant Cell Physiol. 1996, 37, 169–173. [Google Scholar] [CrossRef] [Green Version]
- Daud, M.K.; Sun, Y.Q.; Dawood, M.; Hayat, Y.; Variath, M.T.; Wu, Y.X.; Raziuddin; Mishkat, U.; Salahuddi; Najeeb, U.; et al. Cadmium-induced functional and ultrastructural alterations in roots of two transgenic cotton cultivars. J. Hazard. Mater. 2009, 161, 463–473. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and its relationship to plant hormones. Ann. Bot. 2018, 121, 195–207. [Google Scholar] [CrossRef] [Green Version]
- Huangfu, L.X.; Zhang, Z.H.; Zhou, Y.; Zhang, E.Y.; Chen, R.J.; Fang, H.M.; Li, P.C.; Xu, Y.; Yao, Y.L.; Zhu, M.Y.; et al. Integrated physiological, metabolomic and transcriptomic analyses provide insights into the roles of exogenous melatonin in promoting rice seed germination under salt stress. Plant Growth Regul. 2021, 95, 19–31. [Google Scholar] [CrossRef]
- Jiang, D.; Lu, B.; Liu, L.T.; Duan, W.J.; Chen, L.; Li, J.; Zhang, K.; Sun, H.C.; Zhang, Y.J.; Dong, H.Z.; et al. Exogenous melatonin improves salt stress adaptation of cotton seedlings by regulating active oxygen metabolism. Peer J. 2020, 8, e10486. [Google Scholar] [CrossRef]
- Mu, X.W.; Yan, M.W.; De, Z.C.; Gu, Y.C. Changes of free radicals and digestive enzymes in saliva in cases with deficiency in spleen-yin syndrome. J. Biomed. Res. 2010, 24, 250–255. [Google Scholar]
- Hong, X.L.; Yu, X.; Ling, L.C.; Yan, X.; Li, C.; Shi, H.Y.; Wang, J.W.; Ye, Y.H. Cerebroside C increases tolerance to chilling injury and alters lipid composition in wheat roots. PLoS ONE 2013, 8, e73380. [Google Scholar]
- Zhang, J.Q.; Shen, M.; Zhu, C.C.; Yu, F.X.; Liu, Z.Q.; Ally, N.; Sun, S.C.; Li, K.; Liu, H.L. 3-Nitropropionic acid induces ovarian oxidative stress and impairs follicle in mouse. PLoS ONE 2014, 9, e86589. [Google Scholar] [CrossRef]
- Loh, K.P.; Qi, J.; Tan, B.K.H.; Liu, X.H.; Wei, B.G.; Zhu, Y.Z. Leonurine Protects Middle Cerebral Artery Occluded Rats Through Antioxidant Effect and Regulation of Mitochondrial Function. Stroke 2010, 41, 2661–2668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemensson-Lindell, A. Triphenyltetrazolium chloride as an indicator of fine-root vitality and environmental stress in coniferous forest stands: Applications and limitations. Plant Soil 1994, 159, 297–300. [Google Scholar] [CrossRef]
- Bai, T.H.; Li, C.Y.; Ma, F.W.; Shu, H.R. Physiological responses and analysis of tolerance of apple rootstocks to root-zone hypoxia stress. Sci. Agric. Sin. 2008, 41, 4140–4148. [Google Scholar]
- Monica, R.C.; Lucia, G.; Muccifora, B.S.; Bottega, S.; Spanò, C. Root responses to different types of TiO2 nanoparticles and bulk counterpart in plant model system Vicia faba L. Environ. Exp. Bot. 2016, 130, 11–21. [Google Scholar]
- Guo, J.Y.; Shi, G.Y.; Guo, X.Y.; Zhang, L.W.; Xu, W.Y.; Wang, Y.M.; Su, Z.; Hua, J.P. Transcriptome analysis reveals that distinct metabolic pathways operate in salt-tolerant and salt-sensitive upland cotton varieties subjected to salinity stress. Plant Sci. 2015, 238, 33–45. [Google Scholar] [CrossRef] [PubMed]




| Treatments | CK | MT | S | MS |
|---|---|---|---|---|
| For 24 h | 0 μmol∙L−1 melatonin (distilled water) | 10 μmol∙L−1 melatonin | 0 μmol∙L−1 melatonin (distilled water) | 10 μmol∙L−1 melatonin |
| Two-true-leaf stage | 0 μmol∙L−1 NaCl | 0 μmol∙L−1 NaCl | 150 mmol∙L−1 NaCl | 150 mmol∙L−1 NaCl |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, W.; Lu, B.; Liu, L.; Meng, Y.; Ma, X.; Li, J.; Zhang, K.; Sun, H.; Zhang, Y.; Dong, H.; et al. Effects of Exogenous Melatonin on Root Physiology, Transcriptome and Metabolome of Cotton Seedlings under Salt Stress. Int. J. Mol. Sci. 2022, 23, 9456. https://doi.org/10.3390/ijms23169456
Duan W, Lu B, Liu L, Meng Y, Ma X, Li J, Zhang K, Sun H, Zhang Y, Dong H, et al. Effects of Exogenous Melatonin on Root Physiology, Transcriptome and Metabolome of Cotton Seedlings under Salt Stress. International Journal of Molecular Sciences. 2022; 23(16):9456. https://doi.org/10.3390/ijms23169456
Chicago/Turabian StyleDuan, Wenjing, Bin Lu, Liantao Liu, Yanjun Meng, Xinying Ma, Jin Li, Ke Zhang, Hongchun Sun, Yongjiang Zhang, Hezhong Dong, and et al. 2022. "Effects of Exogenous Melatonin on Root Physiology, Transcriptome and Metabolome of Cotton Seedlings under Salt Stress" International Journal of Molecular Sciences 23, no. 16: 9456. https://doi.org/10.3390/ijms23169456






