A Therapeutic Perspective of HDAC8 in Different Diseases: An Overview of Selective Inhibitors
Abstract
:1. Introduction
2. HDAC8 Is a Class I HDAC Enzyme
- HDAC8 is an X-linked protein which acts independently, e.g., without forming any co-complexes for the activity;
- The L1 loop of HDAC8 is closest to the enzyme active site and undergoes conformational changes, differently depending on the substrate (Figure 2);
- L1 and L6 form a specific pocket which requires an “L” shape conformation for selective binding (Figure 2);
- HDAC8 presents a nuclear localization sequence between the catalytic domain of the enzyme and the serine binding motif found at the end of the catalytic domain [24].
2.1. HDAC8 Substrates
2.1.1. SMC3
2.1.2. p53
2.1.3. ERRα
2.1.4. inv(16)
2.1.5. CREB
3. Involvement of HDAC8 in Different Diseases
3.1. X-Linked Disorders
3.1.1. Cornelia de Lange Syndrome (CdLS)
3.1.2. Duchenne Muscular Dystrophy (DMD)
3.2. Aberrant Wound Healing
3.2.1. Pulmonary Fibrosis (PF)
3.2.2. Renal Fibrosis
3.2.3. Liver Fibrosis
3.2.4. Cardiac Fibrosis
3.2.5. Aberrant Wound Healing Associated with Diabetic Foot Ulcers (DFU)
3.3. Cancer
3.3.1. Haematological Malignancies
3.3.2. Solid Tumours
4. Neuropathological Disorders and Conditions
4.1. HDACis Drug Design
4.2. HDAC8 Drug Design
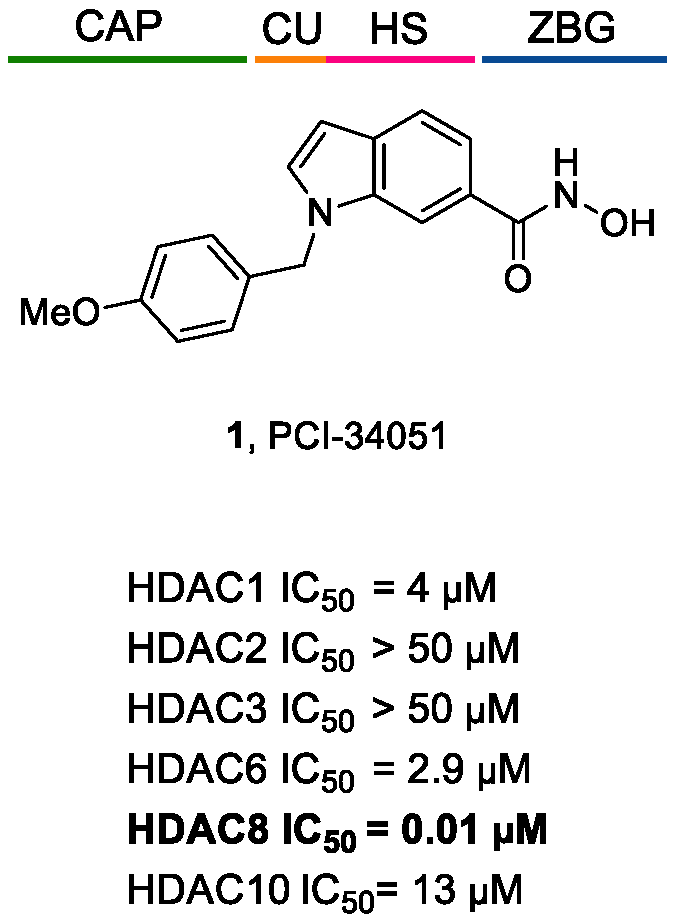
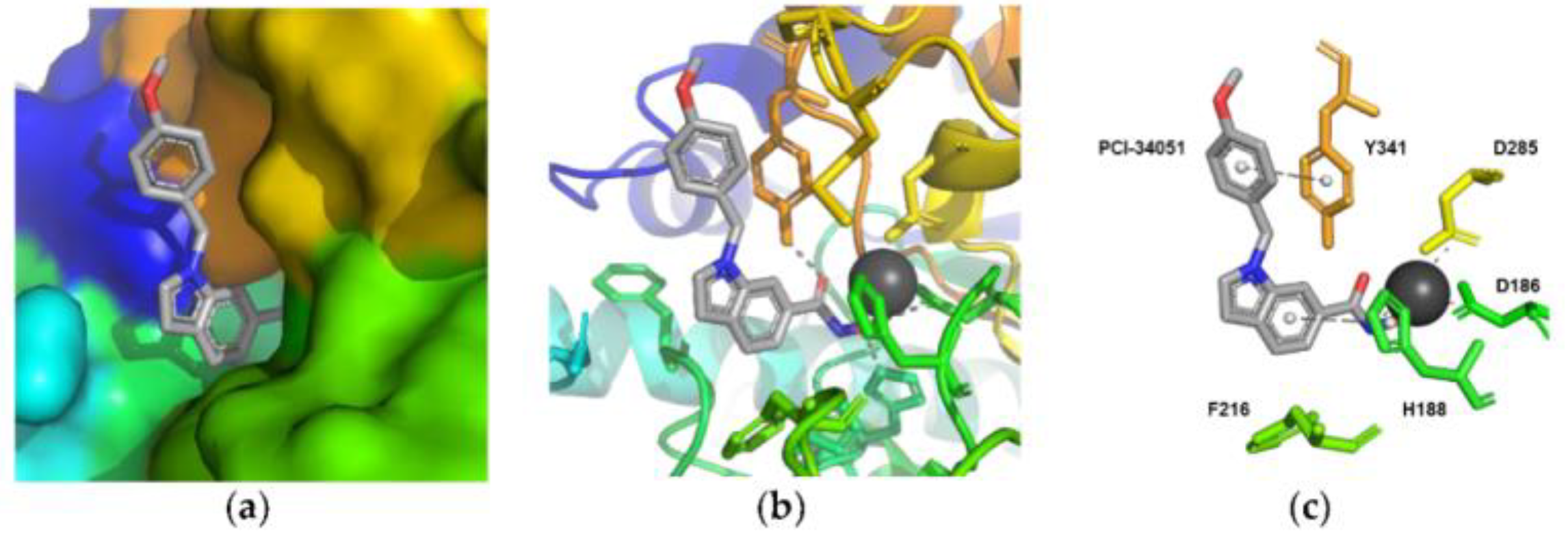
5. HDAC8 Inhibitors
5.1. HDAC8is Bearing Hydroxamic Acids as ZBG
5.1.1. Aromatic-Based Linkers
5.1.2. Flexible Aliphatic-Based Linkers
5.1.3. Alkenyl-Based Linkers
 | |||
|---|---|---|---|
| IC50 (μM) 2 | 40 R1 = H- | 41 R1 = Br- | 12 R1 = Ph- |
| HeLa HDACs 1 | >10 | >10 | >10 |
| hHDAC1 | ND | 4.5 ± 0.1 | 3.0 ± 0.2 |
| hHDAC2 | ND | >20 | >20 |
| hHDAC3 | ND | 4.8 ± 0.5 | 3.0 ± 0.1 |
| hHDAC4 | ND | >20 | >20 |
| hHDAC6 | ND | >20 | >20 |
| hHDAC8 | 0.724 ± 0.0001 | 0.0057 ± 0.0001 | 0.027 ± 0.003 |
| hHDAC10 | ND | >20 | >20 |
| hHDAC11 | ND | >20 | >20 |
5.1.4. Linkerless HDAC8 Inhibitors
5.2. HDAC8is Bearing Novel ZBGs
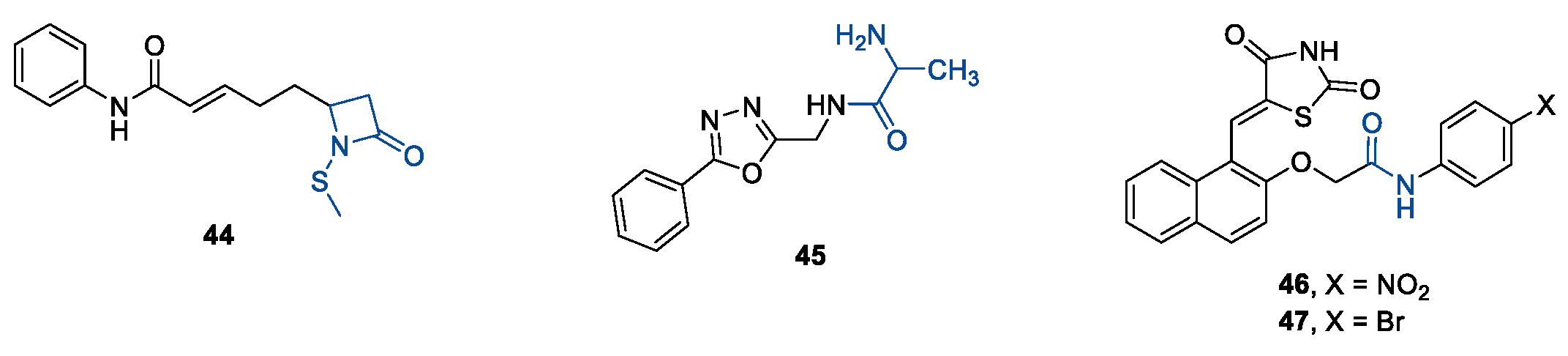
6. Multi-Target Pharmacological Tools Acting on HDAC8
6.1. Multi-Drug Combinations Targeting HDAC8
6.2. Polypharmacological Tools Targeting HDAC8
6.2.1. Selective HDAC6/8 Dual Inhibitors
6.2.2. Selective HDAC6/8/10 Multi-Target Inhibitors
6.2.3. HDAC1-3/8 Dual Inhibitors
6.2.4. Tubulin Polymerization and HDAC8 Dual Inhibitors (TP/HDAC8is)
6.2.5. MMP2/HDAC8 Dual Inhibitors
6.2.6. Bromodomain BRPF1 and HDAC8 Dual Inhibitors
6.2.7. Selective HDAC8-Degrading PROTACs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q.; Dai, Y.; Cai, Z.; Mou, L. HDAC Inhibitors: Novel Immunosuppressants for Allo- and Xeno- Transplantation. ChemistrySelect 2018, 3, 176–187. [Google Scholar] [CrossRef]
- Brindisi, M.; Saraswati, A.P.; Brogi, S.; Gemma, S.; Butini, S.; Campiani, G. Old but Gold: Tracking the New Guise of Histone Deacetylase 6 (HDAC6) Enzyme as a Biomarker and Therapeutic Target in Rare Diseases. J. Med. Chem. 2020, 63, 23–39. [Google Scholar] [CrossRef]
- Relitti, N.; Saraswati, A.P.; Chemi, G.; Brindisi, M.; Brogi, S.; Herp, D.; Schmidtkunz, K.; Saccoccia, F.; Ruberti, G.; Ulivieri, C.; et al. Novel quinolone-based potent and selective HDAC6 inhibitors: Synthesis, molecular modeling studies and biological investigation. Eur. J. Med. Chem. 2021, 212, 112998. [Google Scholar] [CrossRef]
- Saraswati, A.P.; Relitti, N.; Brindisi, M.; Osko, J.D.; Chemi, G.; Federico, S.; Grillo, A.; Brogi, S.; McCabe, N.H.; Turkington, R.C.; et al. Spiroindoline-Capped Selective HDAC6 Inhibitors: Design, Synthesis, Structural Analysis, and Biological Evaluation. ACS Med. Chem. Lett. 2020, 11, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Brindisi, M.; Senger, J.; Cavella, C.; Grillo, A.; Chemi, G.; Gemma, S.; Cucinella, D.M.; Lamponi, S.; Sarno, F.; Iside, C.; et al. Novel spiroindoline HDAC inhibitors: Synthesis, molecular modelling and biological studies. Eur. J. Med. Chem. 2018, 157, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Saccoccia, F.; Brindisi, M.; Gimmelli, R.; Relitti, N.; Guidi, A.; Saraswati, A.P.; Cavella, C.; Brogi, S.; Chemi, G.; Butini, S.; et al. Screening and Phenotypical Characterization of Schistosoma mansoni Histone Deacetylase 8 (SmHDAC8) Inhibitors as Multistage Antischistosomal Agents. ACS Infect. Dis. 2020, 6, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Oehme, I.; Deubzer, H.E.; Lodrini, M.; Milde, T.; Witt, O. Targeting of HDAC8 and investigational inhibitors in neuroblastoma. Expert Opin. Investig. Drugs 2009, 18, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Carullo, G.; Federico, S.; Relitti, N.; Gemma, S.; Butini, S.; Campiani, G. Retinitis Pigmentosa and Retinal Degenerations: Deciphering Pathways and Targets for Drug Discovery and Development. ACS Chem. Neurosci. 2020, 11, 2173–2191. [Google Scholar] [CrossRef]
- Castañeda, C.A.; Wolfson, N.A.; Leng, K.R.; Kuo, Y.-M.; Andrews, A.J.; Fierke, C.A. HDAC8 substrate selectivity is determined by long- and short-range interactions leading to enhanced reactivity for full-length histone substrates compared with peptides. J. Biol. Chem. 2017, 292, 21568–21577. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Melesina, J.; Kolbinger, F.; Oehme, I.; Senger, J.; Witt, O.; Sippl, W.; Jung, M. Targeting histone deacetylase 8 as a therapeutic approach to cancer and neurodegenerative diseases. Futur. Med. Chem. 2016, 8, 1609–1634. [Google Scholar] [CrossRef]
- Durst, K.L.; Lutterbach, B.; Kummalue, T.; Friedman, A.D.; Hiebert, S.W. The inv(16) Fusion Protein Associates with Corepressors via a Smooth Muscle Myosin Heavy-Chain Domain. Mol. Cell. Biol. 2003, 23, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Aramsangtienchai, P.; Spiegelman, N.; He, B.; Miller, S.P.; Dai, L.; Zhao, Y.; Lin, H. HDAC8 Catalyzes the Hydrolysis of Long Chain Fatty Acyl Lysine. ACS Chem. Biol. 2016, 11, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Porter, N.J.; Christianson, D.W. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr. Opin. Struct. Biol. 2019, 59, 9–18. [Google Scholar] [CrossRef]
- Somoza, J.R.; Skene, R.; Katz, B.A.; Mol, C.; Ho, J.D.; Jennings, A.J.; Luong, C.; Arvai, A.; Buggy, J.J.; Chi, E.; et al. Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases. Structure 2004, 12, 1325–1334. [Google Scholar] [CrossRef]
- Dowling, D.P.; Gantt, S.L.; Gattis, S.G.; Fierke, C.A.; Christianson, D.W. Structural Studies of Human Histone Deacetylase 8 and Its Site-Specific Variants Complexed with Substrate and Inhibitors. Biochemistry 2008, 47, 13554–13563. [Google Scholar] [CrossRef] [PubMed]
- Vannini, A.; Volpari, C.; Gallinari, P.; Jones, P.; Mattu, M.; Carfí, A.; De Francesco, R.; Steinkühler, C.; Di Marco, S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO Rep. 2007, 8, 879–884. [Google Scholar] [CrossRef]
- Porter, N.J.; Christianson, N.H.; Decroos, C.; Christianson, D.W. Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8. Biochemistry 2016, 55, 6718–6729. [Google Scholar] [CrossRef]
- Marek, M.; Shaik, T.B.; Heimburg, T.; Chakrabarti, A.; Lancelot, J.; Morales, E.R.; Da Veiga, C.; Kalinin, D.; Melesina, J.; Robaa, D.; et al. Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants. J. Med. Chem. 2018, 61, 10000–10016. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Ramos, J.; Luo, W.; Sirisawad, M.; Verner, E.; Buggy, J.J. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 2008, 22, 1026–1034. [Google Scholar] [CrossRef]
- Ho, T.C.S.; Chan, A.H.Y.; Ganesan, A. Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight. J. Med. Chem. 2020, 63, 12460–12484. [Google Scholar] [CrossRef]
- Federico, S.; Khan, T.; Fontana, A.; Brogi, S.; Benedetti, R.; Sarno, F.; Carullo, G.; Pezzotta, A.; Saraswati, A.P.; Passaro, E.; et al. Azetidin-2-one-based small molecules as dual hHDAC6/HDAC8 inhibitors: Investigation of their mechanism of action and impact of dual inhibition profile on cell viability. Eur. J. Med. Chem. 2022, 238, 114409. [Google Scholar] [CrossRef] [PubMed]
- Tabackman, A.A.; Frankson, R.; Marsan, E.S.; Perry, K.; Cole, K.E. Structure of ‘linkerless’ hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket. J. Struct. Biol. 2016, 195, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Campiani, G.; Cavella, C.; Osko, J.D.; Brindisi, M.; Relitti, N.; Brogi, S.; Saraswati, A.P.; Federico, S.; Chemi, G.; Maramai, S.; et al. Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors. J. Med. Chem. 2021, 64, 9960–9988. [Google Scholar] [CrossRef]
- Hassan, M.M.; Israelian, J.; Nawar, N.; Ganda, G.; Manaswiyoungkul, P.; Raouf, Y.S.; Armstrong, D.; Sedighi, A.; Olaoye, O.O.; Erdogan, F.; et al. Characterization of Conformationally Constrained Benzanilide Scaffolds for Potent and Selective HDAC8 Targeting. J. Med. Chem. 2020, 63, 8634–8648. [Google Scholar] [CrossRef]
- Riester, D.; Hildmann, C.; Grünewald, S.; Beckers, T.; Schwienhorst, A. Factors affecting the substrate specificity of histone deacetylases. Biochem. Biophys. Res. Commun. 2007, 357, 439–445. [Google Scholar] [CrossRef]
- Dose, A.; Liokatis, S.; Theillet, F.-X.; Selenko, P.; Schwarzer, D. NMR Profiling of Histone Deacetylase and Acetyl-transferase Activities in Real Time. ACS Chem. Biol. 2011, 6, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Oehme, I.; Witt, O.; Oliveira, G.; Sippl, W.; Romier, C.; Pierce, R.J.; Jung, M. HDAC8: A multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 2015, 36, 481–492. [Google Scholar] [CrossRef]
- Dasgupta, T.; Antony, J.; Braithwaite, A.W.; Horsfield, J.A. HDAC8 Inhibition Blocks SMC3 Deacetylation and Delays Cell Cycle Progression without Affecting Cohesin-dependent Transcription in MCF7 Cancer Cells. J. Biol. Chem. 2016, 291, 12761–12770. [Google Scholar] [CrossRef]
- Yan, W.; Liu, S.; Xu, E.; Zhang, J.; Zhang, Y.; Chen, X. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene 2013, 32, 599–609. [Google Scholar] [CrossRef]
- Wilson, B.J.; Tremblay, A.M.; Deblois, G.; Sylvain-Drolet, G.; Giguere, V. An Acetylation Switch Modulates the Transcriptional Activity of Estrogen-Related Receptor α. Mol. Endocrinol. 2010, 24, 1349–1358. [Google Scholar] [CrossRef] [Green Version]
- Gurard-Levin, Z.A.; Kim, J.; Mrksich, M. Combining Mass Spectrometry and Peptide Arrays to Profile the Specificities of Histone Deacetylases. ChemBioChem 2009, 10, 2159–2161. [Google Scholar] [CrossRef] [PubMed]
- Karolczak-Bayatti, M.; Sweeney, M.; Cheng, J.; Edey, L.; Robson, S.C.; Ulrich, S.M.; Treumann, A.; Taggart, M.J.; Europe-Finner, G.N. Acetylation of Heat Shock Protein 20 (Hsp20) Regulates Human Myometrial Activity. J. Biol. Chem. 2011, 286, 34346–34355. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, N.A.; Pitcairn, C.A.; Fierke, C.A. HDAC8 substrates: Histones and beyond. Biopolymers 2013, 99, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zang, J.; Ding, Q.; Inks, E.S.; Xu, W.; Chou, C.J.; Zhang, Y. Discovery of meta-sulfamoyl N-hydroxybenzamides as HDAC8 selective inhibitors. Eur. J. Med. Chem. 2018, 150, 282–291. [Google Scholar] [CrossRef]
- Negmeldin, A.T.; Pflum, M.K.H. The structural requirements of histone deacetylase inhibitors: SAHA analogs modified at the C5 position display dual HDAC6/8 selectivity. Bioorganic Med. Chem. Lett. 2017, 27, 3254–3258. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Wong, J.C.; Zhang, W.; Wang, Z.; Zhang, N.; Peng, Z.; Zhang, Z.; Rong, Y.; Li, S.; Zhang, M.; et al. Identification of a Novel Aminotetralin Class of HDAC6 and HDAC8 Selective Inhibitors. J. Med. Chem. 2014, 57, 8026–8034. [Google Scholar] [CrossRef]
- Chiu, C.-F.; Chin, H.-K.; Huang, W.-J.; Bai, L.-Y.; Huang, H.-Y.; Weng, J.-R. Induction of Apoptosis and Autophagy in Breast Cancer Cells by a Novel HDAC8 Inhibitor. Biomolecules 2019, 9, 824. [Google Scholar] [CrossRef]
- Gao, X.; Huang, Z.; Fan, Y.; Sun, Y.; Liu, H.; Wang, L.; Gu, X.-F.; Yu, Y. A Functional Mutation in HDAC8 Gene as Novel Diagnostic Marker for Cornelia De Lange Syndrome. Cell. Physiol. Biochem. 2018, 47, 2388–2395. [Google Scholar] [CrossRef]
- Du, J.; Li, W.; Liu, B.; Zhang, Y.; Yu, J.; Hou, X.; Fang, H. An in silico mechanistic insight into HDAC8 activation facilitates the discovery of new small-molecule activators. Bioorganic Med. Chem. 2020, 28, 115607. [Google Scholar] [CrossRef]
- Elangkovan, N.; Dickson, G. Gene Therapy for Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021, 8, S303–S316. [Google Scholar] [CrossRef]
- Sarogni, P.; Pallotta, M.M.; Musio, A. Cornelia de Lange syndrome: From molecular diagnosis to therapeutic approach. J. Med Genet. 2020, 57, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Ramos, F.J.; Puisac, B.; Baquero-Montoya, C.; Gil-Rodríguez, M.C.; Bueno, I.; Deardorff, M.A.; Hennekam, R.C.; Kaiser, F.J.; Krantz, I.D.; Musio, A.; et al. Clinical utility gene card for: Cornelia de Lange syndrome. Eur. J. Hum. Genet. 2015, 23, 1431. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, M.A.; Bando, M.; Nakato, R.; Watrin, E.; Itoh, T.; Minamino, M.; Saitoh, K.; Komata, M.; Katou, Y.; Clark, D.; et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 2012, 489, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, F.J.; Ansari, M.; Braunholz, D.; Gil-Rodríguez, M.C.; Decroos, C.; Wilde, J.J.; Fincher, C.T.; Kaur, M.; Bando, M.; Amor, D.J.; et al. Loss-of-function HDAC8 mutations cause a phenotypic spectrum of Cornelia de Lange syndrome-like features, ocular hypertelorism, large fontanelle and X-linked inheritance. Hum. Mol. Genet. 2014, 23, 2888–2900. [Google Scholar] [CrossRef]
- Kline, A.D.; Moss, J.; Selicorni, A.; Bisgaard, A.-M.; Deardorff, M.A.; Gillett, P.M.; Ishman, S.L.; Kerr, L.M.; Levin, A.V.; Mulder, P.A.; et al. Diagnosis and management of Cornelia de Lange syndrome: First international consensus statement. Nat. Rev. Genet. 2018, 19, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Mandal, T.; Balsubramanian, N.; Viaene, T.; Leedahl, T.; Sule, N.; Cook, G.; Srivastava, D. Histone deacetylase activators: N-acetylthioureas serve as highly potent and isozyme selective activators for human histone deacetylase-8 on a fluorescent substrate. Bioorganic Med. Chem. Lett. 2011, 21, 5920–5923. [Google Scholar] [CrossRef]
- Decroos, C.; Bowman, C.M.; Moser, J.-A.S.; Christianson, K.E.; Deardorff, M.A.; Christianson, D.W. Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders. ACS Chem. Biol. 2014, 9, 2157–2164. [Google Scholar] [CrossRef]
- Goemans, N.; Buyse, G. Current Treatment and Management of Dystrophinopathies. Curr. Treat. Options Neurol. 2014, 16, 287. [Google Scholar] [CrossRef]
- Duan, D.; Goemans, N.; Takeda, S.; Mercuri, E.; Aartsma-Rus, A. Duchenne Muscular Dystrophy. Nat. Rev. Dis. Primers 2021, 7, 13. [Google Scholar] [CrossRef]
- Minetti, G.C.; Colussi, C.; Adami, R.; Serra, C.; Mozzetta, C.; Parente, V.; Fortuni, S.; Straino, S.; Sampaolesi, M.; Di Padova, M.; et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat. Med. 2006, 12, 1147–1150. [Google Scholar] [CrossRef] [Green Version]
- Johnson, N.M.; Farr, G.H.; Maves, L. The HDAC Inhibitor TSA Ameliorates a Zebrafish Model of Duchenne Muscular Dystrophy. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Consalvi, S.; Mozzetta, C.; Bettica, P.; Germani, M.; Fiorentini, F.; Del Bene, F.; Rocchetti, M.; Leoni, F.; Monzani, V.; Mascagni, P.; et al. Preclinical Studies in the mdx Mouse Model of Duchenne Muscular Dystrophy with the Histone Deacetylase Inhibitor Givinostat. Mol. Med. 2013, 19, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Licandro, S.A.; Crippa, L.; Pomarico, R.; Perego, R.; Fossati, G.; Leoni, F.; Steinkühler, C. The pan HDAC inhibitor Givinostat improves muscle function and histological parameters in two Duchenne muscular dystrophy murine models expressing different haplotypes of the LTBP4 gene. Skelet. Muscle 2021, 11, 19. [Google Scholar] [CrossRef]
- Spreafico, M.; Cafora, M.; Bragato, C.; Capitanio, D.; Marasca, F.; Bodega, B.; De Palma, C.; Mora, M.; Gelfi, C.; Marozzi, A.; et al. Targeting HDAC8 to ameliorate skeletal muscle differentiation in Duchenne muscular dystrophy. Pharmacol. Res. 2021, 170, 105750. [Google Scholar] [CrossRef] [PubMed]
- Artlett, C.M. Inflammasomes in wound healing and fibrosis. J. Pathol. 2013, 229, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Pardali, E.; Sanchez-Duffhues, G.; Gomez-Puerto, M.C.; Dijke, P.T. TGF-β-Induced Endothelial-Mesenchymal Transition in Fibrotic Diseases. Int. J. Mol. Sci. 2017, 18, 2157. [Google Scholar] [CrossRef] [PubMed]
- Kendall, R.T.; Feghali-Bostwick, C.A. Fibroblasts in fibrosis: Novel roles and mediators. Front. Pharmacol. 2014, 5, 123. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chen, D.; Vaziri, N.D.; Guo, Y.; Zhao, Y. Small molecule inhibitors of epithelial-mesenchymal transition for the treatment of cancer and fibrosis. Med. Res. Rev. 2020, 40, 54–78. [Google Scholar] [CrossRef]
- Yoon, S.; Kang, G.; Eom, G.H. HDAC Inhibitors: Therapeutic Potential in Fibrosis-Associated Human Diseases. Int. J. Mol. Sci. 2019, 20, 1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, H.; Wu, X.-Q.; Zhang, D.-D.; Wang, Y.-N.; Guo, Y.; Li, P.; Xiong, Q.; Zhao, Y.-Y. Deciphering the cellular mechanisms underlying fibrosis-associated diseases and therapeutic avenues. Pharmacol. Res. 2021, 163, 105316. [Google Scholar] [CrossRef] [PubMed]
- Pang, M.; Zhuang, S. Histone Deacetylase: A Potential Therapeutic Target for Fibrotic Disorders. J. Pharmacol. Exp. Ther. 2010, 335, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. HDAC Inhibitors as Epigenetic Regulators of the Immune System: Impacts on Cancer Therapy and Inflammatory Diseases. BioMed Res. Int. 2016, 2016, 8797206. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Shan, B.; Klingsberg, R.C.; Qin, X.; Lasky, J.A. Abrogation of TGF-β1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am. J. Physiol. Cell. Mol. Physiol. 2009, 297, L864–L870. [Google Scholar] [CrossRef]
- Korfei, M.; Mahavadi, P.; Guenther, A. Targeting Histone Deacetylases in Idiopathic Pulmonary Fibrosis: A Future Therapeutic Option. Cells 2022, 11, 1626. [Google Scholar] [CrossRef]
- Mathai, S.K.; Schwartz, D.A. Translational research in pulmonary fibrosis. Transl. Res. 2019, 209, 1–13. [Google Scholar] [CrossRef]
- Chen, F.; Gao, Q.; Zhang, L.; Ding, Y.; Wang, H.; Cao, W. Inhibiting HDAC3 (Histone Deacetylase 3) Aberration and the Resultant Nrf2 (Nuclear Factor Erythroid-Derived 2-Related Factor-2) Repression Mitigates Pulmonary Fibrosis. Hypertension 2021, 78, e15–e25. [Google Scholar] [CrossRef]
- Saito, S.; Zhuang, Y.; Suzuki, T.; Ota, Y.; Bateman, M.E.; Alkhatib, A.; Morris, G.F.; Lasky, J.A. HDAC8 inhibition ameliorates pulmonary fibrosis. Am. J. Physiol. Cell. Mol. Physiol. 2019, 316, L175–L186. [Google Scholar] [CrossRef]
- François, H.; Chatziantoniou, C. Renal fibrosis: Recent translational aspects. Matrix Biol. 2018, 68–69, 318–332. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, X.-M.; Huang, X.-R.; Lan, H.Y. The preventive and therapeutic implication for renal fibrosis by targetting TGF-β/Smad3 signaling. Clin. Sci. 2018, 132, 1403–1415. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Xu, J.; Xu, Z.; Yang, B.; Luo, P.; He, Q. Defining therapeutic targets for renal fibrosis: Exploiting the biology of pathogenesis. Biomed. Pharmacother. 2021, 143, 112115. [Google Scholar] [CrossRef] [PubMed]
- Long, K.; Vaughn, Z.; McDaniels, M.D.; Joyasawal, S.; Przepiorski, A.; Parasky, E.; Sander, V.; Close, D.; Johnston, P.A.; Davidson, A.J.; et al. Validation of HDAC8 Inhibitors as Drug Discovery Starting Points to Treat Acute Kidney Injury. ACS Pharmacol. Transl. Sci. 2022, 5, 207–215. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, J.; Tolbert, E.; Zhao, T.C.; Bayliss, G.; Zhuang, S. Identification of histone deacetylase 8 as a novel therapeutic target for renal fibrosis. FASEB J. 2020, 34, 7295–7310. [Google Scholar] [CrossRef]
- Van Beneden, K.; Mannaerts, I.; Pauwels, M.; Branden, C.V.D.; van Grunsven, L.A. HDAC inhibitors in experimental liver and kidney fibrosis. Fibrogenesis Tissue Repair 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Park, J.H.; Jeon, J.Y.; Kim, S.Y.; Kim, J.M.; Lim, C.Y.; Lee, T.H.; Kim, H.K.; Lee, H.G.; Kwon, H.J.; et al. A new histone deacetylase inhibitor improves liver fibrosis inBDLrats through suppression of hepatic stellate cells. Br. J. Pharmacol. 2014, 171, 4820–4830. [Google Scholar] [CrossRef] [PubMed]
- Aher, J.S.; Khan, S.; Jain, S.; Tikoo, K.; Jena, G. Valproate ameliorates thioacetamide-induced fibrosis by hepatic stellate cell inactivation. Hum. Exp. Toxicol. 2015, 34, 44–55. [Google Scholar] [CrossRef]
- Lu, P.; Yan, M.; He, L.; Li, J.; Ji, Y.; Ji, J. Crosstalk between Epigenetic Modulations in Valproic Acid Deactivated Hepatic Stellate Cells: An Integrated Protein and miRNA Profiling Study. Int. J. Biol. Sci. 2019, 15, 93–104. [Google Scholar] [CrossRef]
- Lee, C.H.; Choi, Y.; Cho, H.; Bang, I.H.; Hao, L.; Lee, S.-O.; Jeon, R.; Bae, E.J.; Park, B.-H. Histone deacetylase 8 inhibition alleviates cholestatic liver injury and fibrosis. Biochem. Pharmacol. 2021, 183, 114312. [Google Scholar] [CrossRef]
- Lyu, X.; Hu, M.; Peng, J.; Zhang, X.; Sanders, Y.Y. HDAC inhibitors as antifibrotic drugs in cardiac and pulmonary fibrosis. Ther. Adv. Chronic Dis. 2019, 10, 2040622319862697. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Levin, M.D.; Petrenko, N.B.; Lu, M.M.; Wang, T.; Yuan, L.J.; Stout, A.L.; Epstein, J.A.; Patel, V.V. Histone-deacetylase inhibition reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy independent of angiotensin. J. Mol. Cell. Cardiol. 2008, 45, 715–723. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-H.; Seok, Y.M.; Song, M.-J.; Lee, H.-A.; Kurz, T.; Kim, I. Histone Deacetylase Inhibition Attenuates Cardiac Hypertrophy and Fibrosis through Acetylation of Mineralocorticoid Receptor in Spontaneously Hypertensive Rats. Mol. Pharmacol. 2015, 87, 782–791. [Google Scholar] [CrossRef]
- Zhao, T.; Kee, H.J.; Bai, L.; Kim, M.-K.; Kee, S.-J.; Jeong, M.H. Selective HDAC8 Inhibition Attenuates Isoproterenol-Induced Cardiac Hypertrophy and Fibrosis via p38 MAPK Pathway. Front. Pharmacol. 2021, 12, 677757. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Kee, H.J.; Kee, S.-J.; Jeong, M.H. Hdac8 Inhibitor Alleviates Transverse Aortic Constriction-Induced Heart Failure in Mice by Downregulating Ace1. Oxidative Med. Cell. Longev. 2022, 2022, 6227330. [Google Scholar] [CrossRef] [PubMed]
- Carullo, G.; Governa, P.; Leo, A.; Gallelli, L.; Citraro, R.; Cione, E.; Caroleo, M.C.; Biagi, M.; Aiello, F.; Manetti, F. Quercetin-3-Oleate Contributes to Skin Wound Healing Targeting FFA1/GPR40. ChemistrySelect 2019, 4, 8429–8433. [Google Scholar] [CrossRef]
- Carullo, G.; Sciubba, F.; Governa, P.; Mazzotta, S.; Frattaruolo, L.; Grillo, G.; Cappello, A.R.; Cravotto, G.; Di Cocco, M.E.; Aiello, F. Mantonico and Pecorello Grape Seed Extracts: Chemical Characterization and Evaluation of In Vitro Wound-Healing and Anti-Inflammatory Activities. Pharmaceuticals 2020, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Mazzotta, S.; Governa, P.; Borgonetti, V.; Marcolongo, P.; Nanni, C.; Gamberucci, A.; Manetti, F.; Pessina, F.; Carullo, G.; Brizzi, A.; et al. Pinocembrin and its linolenoyl ester derivative induce wound healing activity in HaCaT cell line potentially involving a GPR120/FFA4 mediated pathway. Bioorganic Chem. 2021, 108, 104657. [Google Scholar] [CrossRef]
- Fu, W.; Liang, D.; Wu, X.; Chen, H.; Hong, X.; Wang, J.; Zhu, T.; Zeng, T.; Lin, W.; Chen, S.; et al. Long noncoding RNA LINC01435 impedes diabetic wound healing by facilitating YY1-mediated HDAC8 expression. iScience 2022, 25, 104006. [Google Scholar] [CrossRef]
- Li, Y.; Sun, R.; Zou, J.; Ying, Y.; Luo, Z. Dual Roles of the AMP-Activated Protein Kinase Pathway in Angiogenesis. Cells 2019, 8, 752. [Google Scholar] [CrossRef]
- Begum, F.; Keni, R.; Ahuja, T.N.; Beegum, F.; Nandakumar, K.; Shenoy, R.R. Notch signaling: A possible therapeutic target and its role in diabetic foot ulcers. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102542. [Google Scholar] [CrossRef]
- Bhagat, T.D.; Zou, Y.; Huang, S.; Park, J.; Palmer, M.B.; Hu, C.; Li, W.; Shenoy, N.; Giricz, O.; Choudhary, G.; et al. Notch Pathway Is Activated via Genetic and Epigenetic Alterations and Is a Therapeutic Target in Clear Cell Renal Cancer. J. Biol. Chem. 2017, 292, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Adhikari, N.; Amin, S.A.; Jha, T. Histone deacetylase 8 (HDAC8) and its inhibitors with selectivity to other isoforms: An overview. Eur. J. Med. Chem. 2019, 164, 214–240. [Google Scholar] [CrossRef] [PubMed]
- Pantelaiou-Prokaki, G.; Mieczkowska, I.; Schmidt, G.E.; Fritzsche, S.; Prokakis, E.; Gallwas, J.; Wegwitz, F. HDAC8 suppresses the epithelial phenotype and promotes EMT in chemotherapy-treated basal-like breast cancer. Clin. Epigenetics 2022, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Spreafico, M.; Gruszka, A.M.; Valli, D.; Mazzola, M.; Deflorian, G.; Quintè, A.; Totaro, M.G.; Battaglia, C.; Alcalay, M.; Marozzi, A.; et al. HDAC8: A Promising Therapeutic Target for Acute Myeloid Leukemia. Front. Cell Dev. Biol. 2020, 8, 844. [Google Scholar] [CrossRef] [PubMed]
- Watters, J.M.; Wright, G.; Smith, M.A.; Shah, B.; Wright, K.L. Histone deacetylase 8 inhibition suppresses mantle cell lymphoma viability while preserving natural killer cell function. Biochem. Biophys. Res. Commun. 2021, 534, 773–779. [Google Scholar] [CrossRef]
- An, P.; Chen, F.; Li, Z.; Ling, Y.; Peng, Y.; Zhang, H.; Li, J.; Chen, Z.; Wang, H. HDAC8 promotes the dissemination of breast cancer cells via AKT/GSK-3β/Snail signals. Oncogene 2020, 39, 4956–4969. [Google Scholar] [CrossRef]
- Vanaja, G.R.; Ramulu, H.G.; Kalle, A.M. Overexpressed HDAC8 in cervical cancer cells shows functional redundancy of tubulin deacetylation with HDAC6. Cell Commun. Signal. 2018, 16, 20. [Google Scholar] [CrossRef]
- Zheng, H.; Zhao, W.; Yan, C.; Watson, C.C.; Massengill, M.; Xie, M.; Massengill, C.; Noyes, D.R.; Martinez, G.V.; Afzal, R.; et al. HDAC Inhibitors Enhance T-Cell Chemokine Expression and Augment Response to PD-1 Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2016, 22, 4119–4132. [Google Scholar] [CrossRef]
- Beg, A.A.; Gray, J.E. HDAC inhibitors with PD-1 blockade: A promising strategy for treatment of multiple cancer types? Epigenomics 2016, 8, 1015–1017. [Google Scholar] [CrossRef]
- Stone, M.L.; Chiappinelli, K.B.; Li, H.; Murphy, L.M.; Travers, M.E.; Topper, M.J.; Mathios, D.; Lim, M.; Shih, I.-M.; Wang, T.-L.; et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc. Natl. Acad. Sci. USA 2017, 114, E10981–E10990. [Google Scholar] [CrossRef] [Green Version]
- Briere, D.; Sudhakar, N.; Woods, D.M.; Hallin, J.; Engstrom, L.D.; Aranda, R.; Chiang, H.; Sodré, A.L.; Olson, P.; Weber, J.S.; et al. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol. Immunother. 2018, 67, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Knox, T.; Sahakian, E.; Banik, D.; Hadley, M.; Palmer, E.; Noonepalle, S.; Kim, J.; Powers, J.; Gracia-Hernandez, M.; Oliveira, V.; et al. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci. Rep. 2019, 9, 6136. [Google Scholar] [CrossRef]
- Maharaj, K.; Powers, J.J.; Mediavilla-Varela, M.; Achille, A.; Gamal, W.; Quayle, S.; Jones, S.S.; Sahakian, E.; Pinilla-Ibarz, J. HDAC6 Inhibition Alleviates CLL-Induced T-Cell Dysfunction and Enhances Immune Checkpoint Blockade Efficacy in the Eμ-TCL1 Model. Front. Immunol. 2020, 11, 590072. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Eden, C.; Perez, C.; Belshoff, A.; Hart, S.; Plaza-Rojas, L.; Delos Reyes, M.; Prajapati, K.; Voelkel-Johnson, C.; Henry, E.; et al. Inhibition of Histone Deacetylase (HDAC) Enhances Checkpoint Blockade Efficacy by Rendering Bladder Cancer Cells Visible for T Cell-Mediated Destruction. Front. Oncol. 2020, 10, 699. [Google Scholar] [CrossRef] [PubMed]
- Baretti, M.; Yarchoan, M. Epigenetic modifiers synergize with immune-checkpoint blockade to enhance long-lasting antitumor efficacy. J. Clin. Investig. 2021, 131, e151002. [Google Scholar] [CrossRef]
- Borcoman, E.; Kamal, M.; Marret, G.; Dupain, C.; Castel-Ajgal, Z.; Le Tourneau, C. HDAC Inhibition to Prime Immune Checkpoint Inhibitors. Cancers 2021, 14, 66. [Google Scholar] [CrossRef]
- Yang, W.; Feng, Y.; Zhou, J.; Cheung, O.K.-W.; Cao, J.; Wang, J.; Tang, W.; Tu, Y.; Xu, L.; Wu, F.; et al. A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci. Transl. Med. 2021, 13, eaaz6804. [Google Scholar] [CrossRef]
- Mormino, A.; Cocozza, G.; Fontemaggi, G.; Valente, S.; Esposito, V.; Santoro, A.; Bernardini, G.; Santoni, A.; Fazi, F.; Mai, A.; et al. Histone-deacetylase 8 drives the immune response and the growth of glioma. Glia 2021, 69, 2682–2698. [Google Scholar] [CrossRef]
- Santos-Barriopedro, I.; Li, Y.; Bahl, S.; Seto, E. HDAC8 Affects MGMT Levels in Glioblastoma Cell Lines via Interaction with the Proteasome Receptor ADRM1. Genes Cancer 2019, 10, 119. [Google Scholar] [CrossRef]
- Oehme, I.; Deubzer, H.E.; Wegener, D.; Pickert, D.; Linke, J.-P.; Hero, B.; Kopp-Schneider, A.; Westermann, F.; Ulrich, S.M.; von Deimling, A.; et al. Histone Deacetylase 8 in Neuroblastoma Tumorigenesis. Clin. Cancer Res. 2009, 15, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Rettig, I.; Koeneke, E.; Trippel, F.; Mueller, W.C.; Burhenne, J.; Kopp-Schneider, A.; Fabian, J.; Schober, A.; Fernekorn, U.; Von Deimling, A.; et al. Selective inhibition of HDAC8 decreases neuroblastoma growth in vitro and in vivo and enhances retinoic acid-mediated differentiation. Cell Death Dis. 2015, 6, e1657. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Najafi, S.; Stäble, S.; Fabian, J.; Koeneke, E.; Kolbinger, F.; Wrobel, J.K.; Meder, B.; Distel, M.; Heimburg, T.; et al. A kinome-wide RNAi screen identifies ALK as a target to sensitize neuroblastoma cells for HDAC8-inhibitor treatment. Cell Death Differ. 2018, 25, 2053–2070. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado-Tejedor, M.; González, M.P.; García-Muñoz, C.; Muruzabal, D.; García-Barroso, C.; Rabal, O.; Segura, V.; Sánchez-Arias, J.A.; Oyarzabal, J.; Garcia-Osta, A. Taking Advantage of the Selectivity of Histone Deacetylases and Phosphodiesterase Inhibitors to Design Better Therapeutic Strategies to Treat Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Stertz, L.; Fries, G.R.; de Aguiar, B.W.; Pfaffenseller, B.; Valvassori, S.S.; Gubert, C.; Ferreira, C.L.; Moretti, M.; Ceresér, K.M.; Kauer-Sant’Anna, M.; et al. Histone deacetylase activity and brain-derived neurotrophic factor (BDNF) levels in a pharmacological model of mania. Rev. Bras. de Psiquiatr. 2013, 36, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Koppel, I.; Timmusk, T. Differential regulation of Bdnf expression in cortical neurons by class-selective histone deacetylase inhibitors. Neuropharmacology 2013, 75, 106–115. [Google Scholar] [CrossRef]
- Boyault, C.; Zhang, Y.; Fritah, S.; Caron, C.; Gilquin, B.; Kwon, S.H.; Garrido, C.; Yao, T.-P.; Vourc’H, C.; Matthias, P.; et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007, 21, 2172–2181. [Google Scholar] [CrossRef]
- Kim, H.J.; Rowe, M.; Ren, M.; Hong, J.-S.; Chen, P.-S.; Chuang, D.-M. Histone Deacetylase Inhibitors Exhibit Anti-Inflammatory and Neuroprotective Effects in a Rat Permanent Ischemic Model of Stroke: Multiple Mechanisms of Action. J. Pharmacol. Exp. Ther. 2007, 321, 892–901. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Liu, L.; Wang, X. HDAC inhibitor trichostatin A-inhibited survival of dopaminergic neuronal cells. Neurosci. Lett. 2009, 467, 212–216. [Google Scholar] [CrossRef]
- Rouaux, C.; Panteleeva, I.; René, F.; De Aguilar, J.-L.G.; Echaniz-Laguna, A.; Dupuis, L.; Menger, Y.; Boutillier, A.-L.; Loeffler, J.-P. Sodium Valproate Exerts Neuroprotective Effects In Vivo through CREB-Binding Protein-Dependent Mechanisms But Does Not Improve Survival in an Amyotrophic Lateral Sclerosis Mouse Model. J. Neurosci. 2007, 27, 5535–5545. [Google Scholar] [CrossRef]
- Lillico, R.; Zhou, T.; Ahmad, T.K.; Stesco, N.; Gozda, K.; Truong, J.; Kong, J.; Lakowski, T.M.; Namaka, M. Increased Post-Translational Lysine Acetylation of Myelin Basic Protein Is Associated with Peak Neurological Disability in a Mouse Experimental Autoimmune Encephalomyelitis Model of Multiple Sclerosis. J. Proteome Res. 2018, 17, 55–62. [Google Scholar] [CrossRef]
- Camelo, S.; Iglesias, A.H.; Hwang, D.; Due, B.; Ryu, H.; Smith, K.; Gray, S.; Imitola, J.; Duran, G.; Assaf, B.; et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2005, 164, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, N.; Rao, P.; Burkhardt, S.; Sananbenesi, F.; Schlüter, O.M.; Bradke, F.; Lu, J.; Fischer, A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Broide, R.S.; Redwine, J.M.; Aftahi, N.; Young, W.; Bloom, F.E.; Winrow, C.J. Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci. 2007, 31, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.-L.; Yen, J.-L.; Kuo, Y.-C.; Kang, J.-J.; Cheng, Y.-W.; Huang, W.-J.; Hsiao, G. HDAC8 Inhibitor WK2-16 Therapeutically Targets Lipopolysaccharide-Induced Mouse Model of Neuroinflammation and Microglial Activation. Int. J. Mol. Sci. 2019, 20, 410. [Google Scholar] [CrossRef]
- Hendrix, S.; Sanchez, S.; Ventriglia, E.; Lemmens, S. HDAC8 Inhibition Reduces Lesional Iba-1+ Cell Infiltration after Spinal Cord Injury without Effects on Functional Recovery. Int. J. Mol. Sci. 2020, 21, 4539. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Morii, A.; Makanga, J.O.; Suzuki, T.; Miyata, N.; Inazu, T. HDAC8 regulates neural differentiation through embryoid body formation in P19 cells. Biochem. Biophys. Res. Commun. 2018, 498, 45–51. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Ma, Q.; Wu, H.-P.; Khamis, M.Y.; Li, Y.-H.; Ma, L.-Y.; Liu, H.-M. A Review of Progress in Histone Deacetylase 6 Inhibitors Research: Structural Specificity and Functional Diversity. J. Med. Chem. 2021, 64, 1362–1391. [Google Scholar] [CrossRef]
- Sangwan, R.; Rajan, R.; Mandal, P.K. HDAC as onco target: Reviewing the synthetic approaches with SAR study of their inhibitors. Eur. J. Med. Chem. 2018, 158, 620–706. [Google Scholar] [CrossRef]
- Bondarev, A.D.; Attwood, M.M.; Jonsson, J.; Chubarev, V.N.; Tarasov, V.V.; Schiöth, H.B. Recent developments of HDAC inhibitors: Emerging indications and novel molecules. Br. J. Clin. Pharmacol. 2021, 87, 4577–4597. [Google Scholar] [CrossRef]
- He, X.; Hui, Z.; Xu, L.; Bai, R.; Gao, Y.; Wang, Z.; Xie, T.; Ye, X.-Y. Medicinal chemistry updates of novel HDACs inhibitors (2020 to present). Eur. J. Med. Chem. 2022, 227, 113946. [Google Scholar] [CrossRef]
- Melesina, J.; Simoben, C.V.; Praetorius, L.; Bülbül, E.F.; Robaa, D.; Sippl, W. Strategies To Design Selective Histone Deacetylase Inhibitors. ChemMedChem 2021, 16, 1336–1359. [Google Scholar] [CrossRef]
- Gupta, P.; Reid, R.C.; Iyer, A.; Sweet, M.J.; Fairlie, D.P. Towards Isozyme-Selective HDAC Inhibitors For Interrogating Disease. Curr. Top. Med. Chem. 2012, 12, 1479–1499. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Bertrand, P. Inside HDACs with more selective HDAC inhibitors. Eur. J. Med. Chem. 2016, 121, 451–483. [Google Scholar] [CrossRef] [PubMed]
- Uba, A.I.; Weako, J.; Keskin, O.; Gürsoy, A.; Yelekçi, K. Examining the stability of binding modes of the co-crystallized inhibitors of human HDAC8 by molecular dynamics simulation. J. Biomol. Struct. Dyn. 2020, 38, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ying, J.B.; Wang, S.S.; He, D.; Zhu, H.; Zhang, C.; Tang, L.; Lin, R.; Zhang, Y. Exploring the binding mechanism of HDAC8 selective inhibitors: Lessons from the modification of Cap group. J. Cell. Biochem. 2020, 121, 3162–3172. [Google Scholar] [CrossRef]
- Melesina, J.; Robaa, D.; Pierce, R.J.; Romier, C.; Sippl, W. Homology modeling of parasite histone deacetylases to guide the structure-based design of selective inhibitors. J. Mol. Graph. Model. 2015, 62, 342–361. [Google Scholar] [CrossRef]
- Buggy, J.; Balasubramanian, S.; Steggerda, S. Uses of Selective Inhibitors of HDAC8 for Treatment of Inflammatory Conditions. U.S. Patent WO2008061160A1, 22 May 2008. Available online: https://worldwide.espacenet.com/patent/search/family/039402009/publication/WO2008061160A1?q=WO%202008%2F061160 (accessed on 25 August 2022).
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Tang, W.; Luo, T.; Greenberg, E.F.; Bradner, J.E.; Schreiber, S.L. Discovery of Histone Deacetylase 8 Selective Inhibitors. Bioorganic Med. Chem. Lett. 2011, 21, 2601–2605. [Google Scholar] [CrossRef]
- Olaoye, O.O.; Watson, P.R.; Nawar, N.; Geletu, M.; Sedighi, A.; Bukhari, S.; Raouf, Y.S.; Manaswiyoungkul, P.; Erdogan, F.; Abdeldayem, A.; et al. Unique Molecular Interaction with the Histone Deacetylase 6 Catalytic Tunnel: Crystallographic and Biological Characterization of a Model Chemotype. J. Med. Chem. 2021, 64, 2691–2704. [Google Scholar] [CrossRef]
- Suzuki, T.; Ota, Y.; Ri, M.; Bando, M.; Gotoh, A.; Itoh, Y.; Tsumoto, H.; Tatum, P.R.; Mizukami, T.; Nakagawa, H.; et al. Rapid Discovery of Highly Potent and Selective Inhibitors of Histone Deacetylase 8 Using Click Chemistry to Generate Candidate Libraries. J. Med. Chem. 2012, 55, 9562–9575. [Google Scholar] [CrossRef]
- Suzuki, T.; Muto, N.; Bando, M.; Itoh, Y.; Masaki, A.; Ri, M.; Ota, Y.; Nakagawa, H.; Iida, S.; Shirahige, K.; et al. Design, Synthesis, and Biological Activity of NCC149 Derivatives as Histone Deacetylase 8-Selective Inhibitors. ChemMedChem 2014, 9, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Heimburg, T.; Kolbinger, F.R.; Zeyen, P.; Ghazy, E.; Herp, D.; Schmidtkunz, K.; Melesina, J.; Shaik, T.B.; Erdmann, F.; Schmidt, M.; et al. Structure-Based Design and Biological Characterization of Selective Histone Deacetylase 8 (HDAC8) Inhibitors with Anti-Neuroblastoma Activity. J. Med. Chem. 2017, 60, 10188–10204. [Google Scholar] [CrossRef]
- Ingham, O.J.; Paranal, R.M.; Smith, W.B.; Escobar, R.A.; Yueh, H.; Snyder, T.; Porco, J.J.A.; Bradner, J.E.; Beeler, A.B. Development of a Potent and Selective HDAC8 Inhibitor. ACS Med. Chem. Lett. 2016, 7, 929–932. [Google Scholar] [CrossRef] [PubMed]
- Taha, T.Y.; Aboukhatwa, S.M.; Knopp, R.C.; Ikegaki, N.; Abdelkarim, H.; Neerasa, J.; Lu, Y.; Neelarapu, R.; Hanigan, T.W.; Thatcher, G.R.J.; et al. Design, Synthesis, and Biological Evaluation of Tetrahydroisoquinoline-Based Histone Deacetylase 8 Selective Inhibitors. ACS Med. Chem. Lett. 2017, 8, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Adhikari, N.; Amin, S.A.; Bobde, Y.; Ganesh, R.; Jha, T.; Ghosh, B. Design, synthesis, biological evaluation and molecular docking study of arylcarboxamido piperidine and piperazine-based hydroxamates as potential HDAC8 inhibitors with promising anticancer activity. Eur. J. Pharm. Sci. 2019, 138, 105046. [Google Scholar] [CrossRef]
- Kaul-Ghanekar, R.; Patil, M.; Choudhari, A.S.; Pandita, S.; Islam, A.; Raina, P. Cinnamaldehyde, cinnamic acid, and cinnamyl alcohol, the bioactives of Cinnamomum cassia exhibit HDAC8 inhibitory activity: An In vitro and In silico study. Pharmacogn. Mag. 2017, 13, S645–S651. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-J.; Wang, Y.-C.; Chao, S.-W.; Yang, C.-Y.; Chen, L.-C.; Lin, M.-H.; Hou, W.-C.; Chen, M.-Y.; Lee, T.-L.; Yang, P.; et al. Synthesis and Biological Evaluation ofortho-ArylN-Hydroxycinnamides as Potent Histone Deacetylase (HDAC) 8 Isoform-Selective Inhibitors. ChemMedChem 2012, 7, 1815–1824. [Google Scholar] [CrossRef]
- Kuo, Y.; Huang, W.; I-Chung, C. HDAC8 Inhibitors for Treating Cancer. U.S. Patent WO2015026935A2, 26 February 2015. Available online: https://worldwide.espacenet.com/patent/search/family/052484262/publication/WO2015026935A2?q=WO2015026935A2%20%281%29 (accessed on 25 August 2022).
- KrennHrubec, K.; Marshall, B.L.; Hedglin, M.; Verdin, E.; Ulrich, S.M. Design and evaluation of ‘Linkerless’ hydroxamic acids as selective HDAC8 inhibitors. Bioorganic Med. Chem. Lett. 2007, 17, 2874–2878. [Google Scholar] [CrossRef] [PubMed]
- Galletti, P.; Quintavalla, A.; Ventrici, C.; Giannini, G.; Cabri, W.; Penco, S.; Gallo, G.; Vincenti, S.; Giacomini, D. Azetidinones as Zinc-Binding Groups to Design Selective HDAC8 Inhibitors. ChemMedChem 2009, 4, 1991–2001. [Google Scholar] [CrossRef]
- Pidugu, V.R.; Yarla, N.S.; Bishayee, A.; Kalle, A.M.; Satya, A.K. Novel histone deacetylase 8-selective inhibitor 1,3,4-oxadiazole-alanine hybrid induces apoptosis in breast cancer cells. Apoptosis 2017, 22, 1394–1403. [Google Scholar] [CrossRef]
- Pidugu, V.R.; Yarla, N.S.; Pedada, S.R.; Kalle, A.M.; Satya, A.K. Design and synthesis of novel HDAC8 inhibitory 2,5-disubstituted-1,3,4-oxadiazoles containing glycine and alanine hybrids with anti cancer activity. Bioorganic Med. Chem. 2016, 24, 5611–5617. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, N.; Tilekar, K.; Jänsch, N.; Schweipert, M.; Hess, J.D.; Macias, L.H.; Mrowka, P.; Aguilera, R.J.; Choe, J.-Y.; Meyer-Almes, F.-J.; et al. Discovery of novel N-substituted thiazolidinediones (TZDs) as HDAC8 inhibitors: In-silico studies, synthesis, and biological evaluation. Bioorganic Chem. 2020, 100, 103934. [Google Scholar] [CrossRef]
- Tilekar, K.; Upadhyay, N.; Jänsch, N.; Schweipert, M.; Mrowka, P.; Meyer-Almes, F.; Ramaa, C. Discovery of 5-naphthylidene-2,4-thiazolidinedione derivatives as selective HDAC8 inhibitors and evaluation of their cytotoxic effects in leukemic cell lines. Bioorganic Chem. 2020, 95, 103522. [Google Scholar] [CrossRef] [PubMed]
- Kleinschek, A.; Meyners, C.; Digiorgio, E.; Brancolini, C.; Meyer-Almes, F. Potent and Selective Non-hydroxamate Histone Deacetylase 8 Inhibitors. ChemMedChem 2016, 11, 2598–2606. [Google Scholar] [CrossRef] [PubMed]
- Wolff, B.; Jänsch, N.; Sugiarto, W.O.; Frühschulz, S.; Lang, M.; Altintas, R.; Oehme, I.; Meyer-Almes, F.-J. Synthesis and structure activity relationship of 1, 3-benzo-thiazine-2-thiones as selective HDAC8 inhibitors. Eur. J. Med. Chem. 2019, 184, 111756. [Google Scholar] [CrossRef] [PubMed]
- Smalley, J.P.; Cowley, S.M.; Hodgkinson, J.T. Bifunctional HDAC Therapeutics: One Drug to Rule Them All? Molecules 2020, 25, 4394. [Google Scholar] [CrossRef]
- Papa, A.; Pasquini, S.; Contri, C.; Gemma, S.; Campiani, G.; Butini, S.; Varani, K.; Vincenzi, F. Polypharmacological Approaches for CNS Diseases: Focus on Endocannabinoid Degradation Inhibition. Cells 2022, 11, 471. [Google Scholar] [CrossRef]
- Olson, D.E.; Wagner, F.F.; Kaya, T.; Gale, J.P.; Aidoud, N.; Davoine, E.L.; Lazzaro, F.; Weïwer, M.; Zhang, Y.-L.; Holson, E.B. Discovery of the First Histone Deacetylase 6/8 Dual Inhibitors. J. Med. Chem. 2013, 56, 4816–4820. [Google Scholar] [CrossRef]
- Negmeldin, A.; Knoff, J.R.; Pflum, M.K.H. The structural requirements of histone deacetylase inhibitors: C4-modified SAHA analogs display dual HDAC6/HDAC8 selectivity. Eur. J. Med. Chem. 2018, 143, 1790–1806. [Google Scholar] [CrossRef]
- Wagner, F.F.; Olson, D.E.; Gale, J.P.; Kaya, T.; Weïwer, M.; Aidoud, N.; Thomas, M.; Davoine, E.L.; Lemercier, B.C.; Zhang, Y.-L.; et al. Potent and Selective Inhibition of Histone Deacetylase 6 (HDAC6) Does Not Require a Surface-Binding Motif. J. Med. Chem. 2013, 56, 1772–1776. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, J.; Jia, Y.; Wang, X.; Zhang, L.; Liu, C.; Fang, H.; Xu, W. Development of Tetrahydroisoquinoline-Based Hydroxamic Acid Derivatives: Potent Histone Deacetylase Inhibitors with Marked in Vitro and in Vivo Antitumor Activities. J. Med. Chem. 2011, 54, 2823–2838. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.A.; Ferreira-Silva, G.; Ferreira, A.C.S.; Fernandes, R.A.; Kwee, J.K.; Sant’Anna, C.M.R.; Ionta, M.; Fraga, C.A.M. Design, Synthesis, and Pharmacological Evaluation of Novel N-Acylhydrazone Derivatives as Potent Histone Deacetylase 6/8 Dual Inhibitors. J. Med. Chem. 2016, 59, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Negmeldin, A.T.; Padige, G.; Bieliauskas, A.V.; Pflum, M.K.H. Structural Requirements of HDAC Inhibitors: SAHA Analogues Modified at the C2 Position Display HDAC6/8 Selectivity. ACS Med. Chem. Lett. 2017, 8, 281–286. [Google Scholar] [CrossRef]
- Bieliauskas, A.V.; Weerasinghe, S.V.; Pflum, M.K.H. Structural requirements of HDAC inhibitors: SAHA analogs functionalized adjacent to the hydroxamic acid. Bioorganic Med. Chem. Lett. 2007, 17, 2216–2219. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.E.; Weerasinghe, S.V.; Pflum, M.K.H. The structural requirements of histone deacetylase inhibitors: Suberoylanilide hydroxamic acid analogs modified at the C3 position display isoform selectivity. Bioorganic Med. Chem. Lett. 2011, 21, 6139–6142. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.E.; Pflum, M.K.H. The structural requirements of histone deacetylase inhibitors: Suberoylanilide hydroxamic acid analogs modified at the C6 position. Bioorganic Med. Chem. Lett. 2012, 22, 7084–7086. [Google Scholar] [CrossRef]
- Debnath, S.; Debnath, T.; Bhaumik, S.; Majumdar, S.; Kalle, A.M.; Aparna, V. Discovery of novel potential selective HDAC8 inhibitors by combine ligand-based, structure-based virtual screening and in-vitro biological evaluation. Sci. Rep. 2019, 9, 17174. [Google Scholar] [CrossRef]
- Kolbinger, F.R.; Koeneke, E.; Ridinger, J.; Heimburg, T.; Müller, M.; Bayer, T.; Sippl, W.; Jung, M.; Gunkel, N.; Miller, A.K.; et al. The HDAC6/8/10 inhibitor TH34 induces DNA damage-mediated cell death in human high-grade neuroblastoma cell lines. Arch. Toxicol. 2018, 92, 2649–2664. [Google Scholar] [CrossRef]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzym. Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef]
- Dinda, B.; Dinda, S.; DasSharma, S.; Banik, R.; Chakraborty, A.; Dinda, M. Therapeutic potentials of baicalin and its aglycone, baicalein against inflammatory disorders. Eur. J. Med. Chem. 2017, 131, 68–80. [Google Scholar] [CrossRef]
- Tuli, H.S.; Aggarwal, V.; Kaur, J.; Aggarwal, D.; Parashar, G.; Parashar, N.C.; Tuorkey, M.; Kaur, G.; Savla, R.; Sak, K.; et al. Baicalein: A metabolite with promising antineoplastic activity. Life Sci. 2020, 259, 118183. [Google Scholar] [CrossRef] [PubMed]
- Sowndhararajan, K.; Deepa, P.; Kim, M.; Park, S.J.; Kim, S. Baicalein as a potent neuroprotective agent: A review. Biomed. Pharmacother. 2017, 95, 1021–1032. [Google Scholar] [CrossRef]
- Yu, X.; Li, H.; Hu, P.; Qing, Y.; Wang, X.; Zhu, M.; Wang, H.; Wang, Z.; Xu, J.; Guo, Q.; et al. Natural HDAC-1/8 inhibitor baicalein exerts therapeutic effect in CBF-AML. Clin. Transl. Med. 2020, 10, e154. [Google Scholar] [CrossRef] [PubMed]
- Pal-Bhadra, M.; Ramaiah, M.J.; Reddy, T.L.; Krishnan, A.; Pushpavalli, S.; Babu, K.S.; Tiwari, A.K.; Rao, J.M.; Yadav, J.S.; Bhadra, U. Plant HDAC inhibitor chrysin arrest cell growth and induce p21 WAF1 by altering chromatin of STAT response element in A375 cells. BMC Cancer 2012, 12, 180. [Google Scholar] [CrossRef]
- Sun, L.-P.; Chen, A.-L.; Hung, H.-C.; Chien, Y.-H.; Huang, J.-S.; Huang, C.-Y.; Chen, Y.-W.; Chen, C.-N. Chrysin: A Histone Deacetylase 8 Inhibitor with Anticancer Activity and a Suitable Candidate for the Standardization of Chinese Propolis. J. Agric. Food Chem. 2012, 60, 11748–11758. [Google Scholar] [CrossRef] [PubMed]
- Neelarapu, R.; Holzle, D.L.; Velaparthi, S.; Bai, H.; Brunsteiner, M.; Blond, S.Y.; Petukhov, P.A. Design, Synthesis, Docking, and Biological Evaluation of Novel Diazide-Containing Isoxazole- and Pyrazole-Based Histone Deacetylase Probes. J. Med. Chem. 2011, 54, 4350–4364. [Google Scholar] [CrossRef] [PubMed]
- Lamaa, D.; Lin, H.-P.; Zig, L.; Bauvais, C.; Bollot, G.; Bignon, J.; Levaique, H.; Pamlard, O.; Dubois, J.; Ouaissi, M.; et al. Design and Synthesis of Tubulin and Histone Deacetylase Inhibitor Based on iso-Combretastatin A-4. J. Med. Chem. 2018, 61, 6574–6591. [Google Scholar] [CrossRef]
- Hauguel, C.; Ducellier, S.; Provot, O.; Ibrahim, N.; Lamaa, D.; Balcerowiak, C.; Letribot, B.; Nascimento, M.; Blanchard, V.; Askenatzis, L.; et al. Design, synthesis and biological evaluation of quinoline-2-carbonitrile-based hydroxamic acids as dual tubulin polymerization and histone deacetylases inhibitors. Eur. J. Med. Chem. 2022, 240, 114573. [Google Scholar] [CrossRef]
- Zhong, Y.; Lu, Y.-T.; Sun, Y.; Shi, Z.-H.; Li, N.-G.; Tang, Y.-P.; Duan, J.-A. Recent opportunities in matrix metalloproteinase inhibitor drug design for cancer. Expert Opin. Drug Discov. 2018, 13, 75–87. [Google Scholar] [CrossRef]
- Park, C.G.; Park, S.Y.; Jun, J.A.; Jeong, K.J.; Heo, H.J.; Sohn, J.S.; Lee, H.Y.; Kang, J. Histone deacetylases 1, 6 and 8 are critical for invasion in breast cancer. Oncol. Rep. 2011, 25, 1677–1681. [Google Scholar] [CrossRef] [Green Version]
- Amin, S.A.; Adhikari, N.; Jha, T. Is Dual Inhibition of Metalloenzymes HDAC-8 and MMP-2 a Potential Pharmacological Target to Combat Hematological Malignancies? Pharmacol. Res. 2017, 122, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Halder, A.K.; Mallick, S.; Shikha, D.; Saha, A.; Saha, K.D.; Jha, T. Design of dual MMP-2/HDAC-8 inhibitors by pharmacophore mapping, molecular docking, synthesis and biological activity. RSC Adv. 2015, 5, 72373–72386. [Google Scholar] [CrossRef]
- Ghazy, E.; Zeyen, P.; Herp, D.; Hügle, M.; Schmidtkunz, K.; Erdmann, F.; Robaa, D.; Schmidt, M.; Morales, E.R.; Romier, C.; et al. Design, synthesis, and biological evaluation of dual targeting inhibitors of histone deacetylase 6/8 and bromodomain BRPF1. Eur. J. Med. Chem. 2020, 200, 112338. [Google Scholar] [CrossRef] [PubMed]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Chotitumnavee, J.; Yamashita, Y.; Takahashi, Y.; Takada, Y.; Iida, T.; Oba, M.; Itoh, Y.; Suzuki, T. Selective degradation of histone deacetylase 8 mediated by a proteolysis targeting chimera (PROTAC). Chem. Commun. 2022, 58, 4635–4638. [Google Scholar] [CrossRef]
- Sun, Z.; Deng, B.; Yang, Z.; Mai, R.; Huang, J.; Ma, Z.; Chen, T.; Chen, J. Discovery of pomalidomide-based PROTACs for selective degradation of histone deacetylase 8. Eur. J. Med. Chem. 2022, 239, 114544. [Google Scholar] [CrossRef]
- Darwish, S.; Ghazy, E.; Heimburg, T.; Herp, D.; Zeyen, P.; Salem-Altintas, R.; Ridinger, J.; Robaa, D.; Schmidtkunz, K.; Erdmann, F.; et al. Design, Synthesis and Biological Characterization of Histone Deacetylase 8 (HDAC8) Proteolysis Targeting Chimeras (PROTACs) with Anti-Neuroblastoma Activity. Int. J. Mol. Sci. 2022, 23, 7535. [Google Scholar] [CrossRef]





 | ||||
|---|---|---|---|---|
| Cpds | R1 | hHDAC2 IC50 (μM) | hHDAC3/NCoR1 IC50 (μM) | hHDAC8 IC50 (μM) |
| 14 | H- | 20 | 18 | 0.052 |
| 15 |  | 6.3 | 6.2 | 0.029 |
| 16 |  | 3.6 | 15 | 0.023 |
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cpds | R1 | X | Ar | hHDAC1 IC50 (μM) | hHDAC2 IC50 (μM) | hHDAC4 IC50 (μM) | hHDAC6 IC50 (μM) | hHDAC8 IC50 (μM) |
| 17 | Ph | -CH2 | 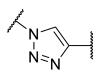 | 41 | 65 | 30 | 7.9 | 0.35 |
| 18 | Ph | -CH2CH2 |  | >100 | 76 | >100 | 3.2 | 0.18 |
| 13 | Ph | -SCH2 | 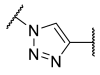 | 38 | >100 | 44 | 2.4 | 0.070 |
| 19 | 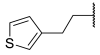 | -CH2CH2 | 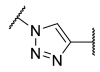 | >100 | >100 | >100 | 1.1 | 0.10 |
| 20 | Ph | -SCH2 | 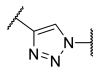 | >100 | >100 | >100 | 2.2 | 0.053 |
| 7 | Ph | -SCH2 |  | >100 | >100 | >100 | 14 | 0.15 |
 | ||||||
|---|---|---|---|---|---|---|
| Cpds | R1 | X | Y | hHDAC1 IC50 (μM) 1 | hHDAC6 IC50 (μM) 1 | hHDAC8 IC50 (μM) 1 |
| 21 | Cl | -NH | Me | 18.7 ± 2.5 | 14.4 ± 2.4 | 0.035 ± 0.04 |
| 22 | H | -NH | -OMe | 14.5 ± 1.4 | 5.1 ± 0.8 | 0.069 ± 0.017 |
| 23 | H | -O | -OMe | 12.1 ± 5.7 | 2.9 ± 0.3 | 0.027 ± 0.03 |
 | ||||
|---|---|---|---|---|
| Cpds | R1 | hHDAC2 IC50 (μM) 1 | hHDAC6 IC50 (μM) 1 | hHDAC8 IC50 (μM) 1 |
| 24 |  | 14.5 ± 2.4 | 1.5 ± 0.3 | 0.050 ± 0.010 |
| 25 | 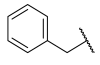 | 47.1 ± 7.0 | 2.6 ± 0.5 | 0.080 ± 0.020 |
| 26 |  | 11.2 ± 1.8 | 1.8 ± 0.3 | 0.060 ± 0.010 |
 | |||||||
|---|---|---|---|---|---|---|---|
| Cpds | R1 | R2 | X | hHDAC3 IC50 (μM) | hHDAC6 IC50 (μM) | hHDAC8 IC50 (μM) | hHDAC11 IC50 (μM) |
| 27 | 3-tBu | H |  | >1 | 0.066 | 0.0337 | >1 |
| 28 | 3-tBu | 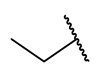 | 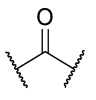 | >1 | >1 | 0.0835 | >1 |
| 29 | 3-tBu | 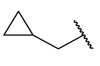 | 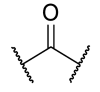 | >1 | >1 | 0.0660 | >1 |
| 30 | 3,5-CF3 |  | 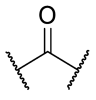 | >1 | >1 | 0.0234 | >1 |
| 31 | 3-OCF3 | 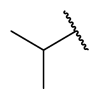 |  | >1 | >1 | 0.0655 | >1 |
 | |||||
|---|---|---|---|---|---|
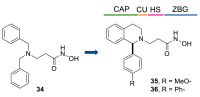
| Cpds | hHDAC1 IC50 (μM) 1 | hHDAC2 IC50 (μM) 1 | hHDAC3 IC50 (μM) 1 | hHDAC6 (% Inhibition) 2 | hHDAC8 IC50 (μM) 1 |
| 34 | 33 ± 1.1 | 2.7 ± 0.67 | 52 ± 3.0 | 97 ± 0.050 | 1.4 ± 0.41 |
| 35 | 27 ± 3.7 | >100 | >100 | 21 ± 0.040 | 0.082 ± 0.019 |
| 36 | 7.3 ± 0.48 | 47 ± 17 | 38 ± 2.2 | 39 ± 1.2 | 0.055 ± 0.014 |
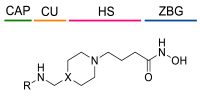 | |||||
|---|---|---|---|---|---|
| Cpds | R | X | HeLa HDACs IC50 (μM) 1 | hHDAC3/NCoR1 (% Inhibition) 2 | hHDAC8 IC50 (μM) |
| 37 |  | -CH- | 3.57 ± 2.19 | 4.21 | 3.14 ± 1.01 |
| 38 | 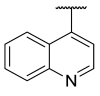 | -N- | 3.65 ± 2.39 | 4.42 | 1.74 ± 0.81 |
| 39 | 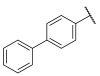 | -N- | 4.99 ± 1.82 | 5.35 | 4.29 ± 1.42 |
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cpds | R1 | R2 | X | n | hHDAC1 IC50 (μM) | hHDAC2 IC50 (μM) | hHDAC3 IC50 (μM) | hHDAC6 IC50 (μM) | hHDAC8 IC50 (μM) |
| 48 | H- | H- | -N- | 3 | 3.6 ± 0.8 | 32 ± 15 | >50 | 6.7 ± 0.8 | 0.011 ± 0.001 |
| 49 | H- | F- | -N- | 3 | 35 ± 3 | >50 | >50 | 5.2 ± 1.1 | 0.017 ± 0.0001 |
| 50 | Br- | H- | -S- | 2 | >50 | >50 | >50 | >50 | 0.260 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontana, A.; Cursaro, I.; Carullo, G.; Gemma, S.; Butini, S.; Campiani, G. A Therapeutic Perspective of HDAC8 in Different Diseases: An Overview of Selective Inhibitors. Int. J. Mol. Sci. 2022, 23, 10014. https://doi.org/10.3390/ijms231710014
Fontana A, Cursaro I, Carullo G, Gemma S, Butini S, Campiani G. A Therapeutic Perspective of HDAC8 in Different Diseases: An Overview of Selective Inhibitors. International Journal of Molecular Sciences. 2022; 23(17):10014. https://doi.org/10.3390/ijms231710014
Chicago/Turabian StyleFontana, Anna, Ilaria Cursaro, Gabriele Carullo, Sandra Gemma, Stefania Butini, and Giuseppe Campiani. 2022. "A Therapeutic Perspective of HDAC8 in Different Diseases: An Overview of Selective Inhibitors" International Journal of Molecular Sciences 23, no. 17: 10014. https://doi.org/10.3390/ijms231710014
APA StyleFontana, A., Cursaro, I., Carullo, G., Gemma, S., Butini, S., & Campiani, G. (2022). A Therapeutic Perspective of HDAC8 in Different Diseases: An Overview of Selective Inhibitors. International Journal of Molecular Sciences, 23(17), 10014. https://doi.org/10.3390/ijms231710014













