Phenotypic and Transcriptional Changes of Pulmonary Immune Responses in Dogs Following Canine Distemper Virus Infection
Abstract
1. Introduction
2. Results
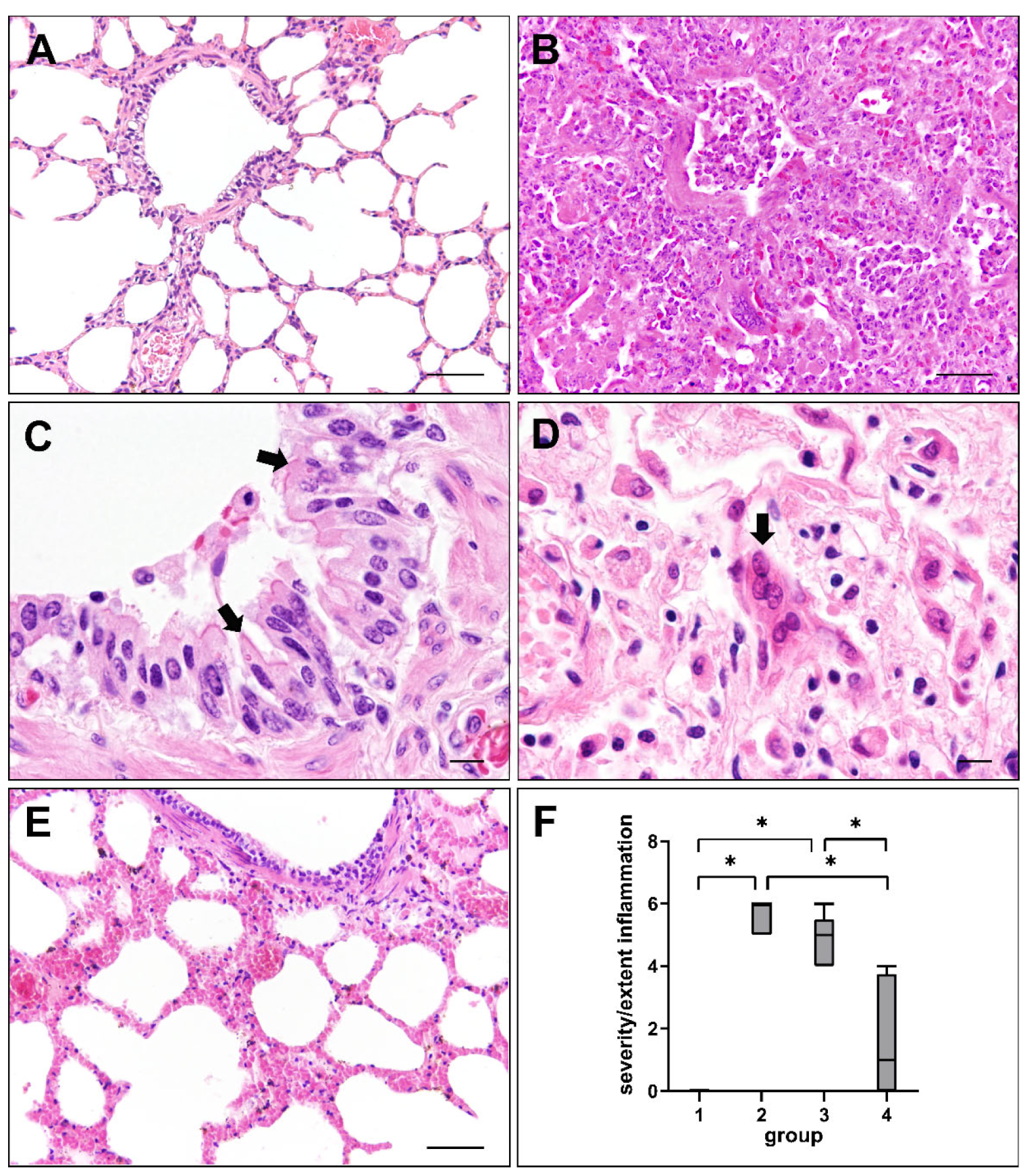
2.1. Histopathologic Changes in Canine Distemper Virus-Infected Lungs
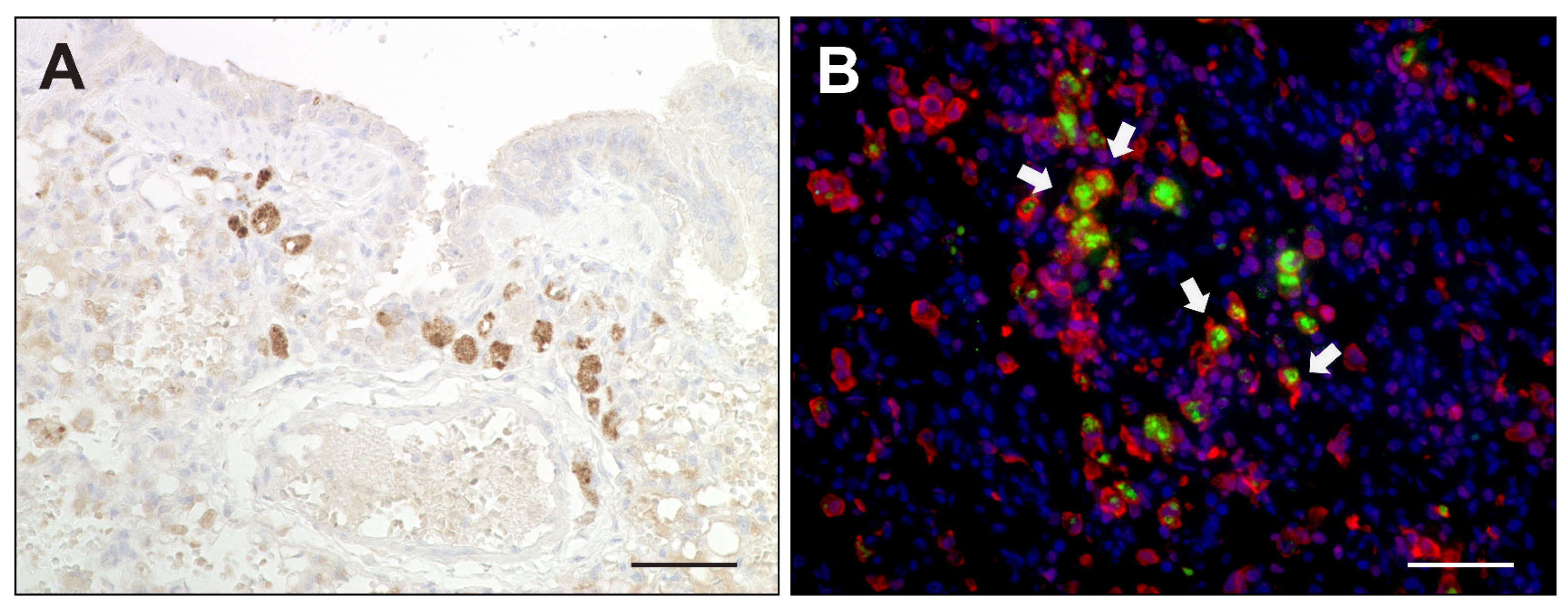
2.2. Virus Loads and Cell Tropism
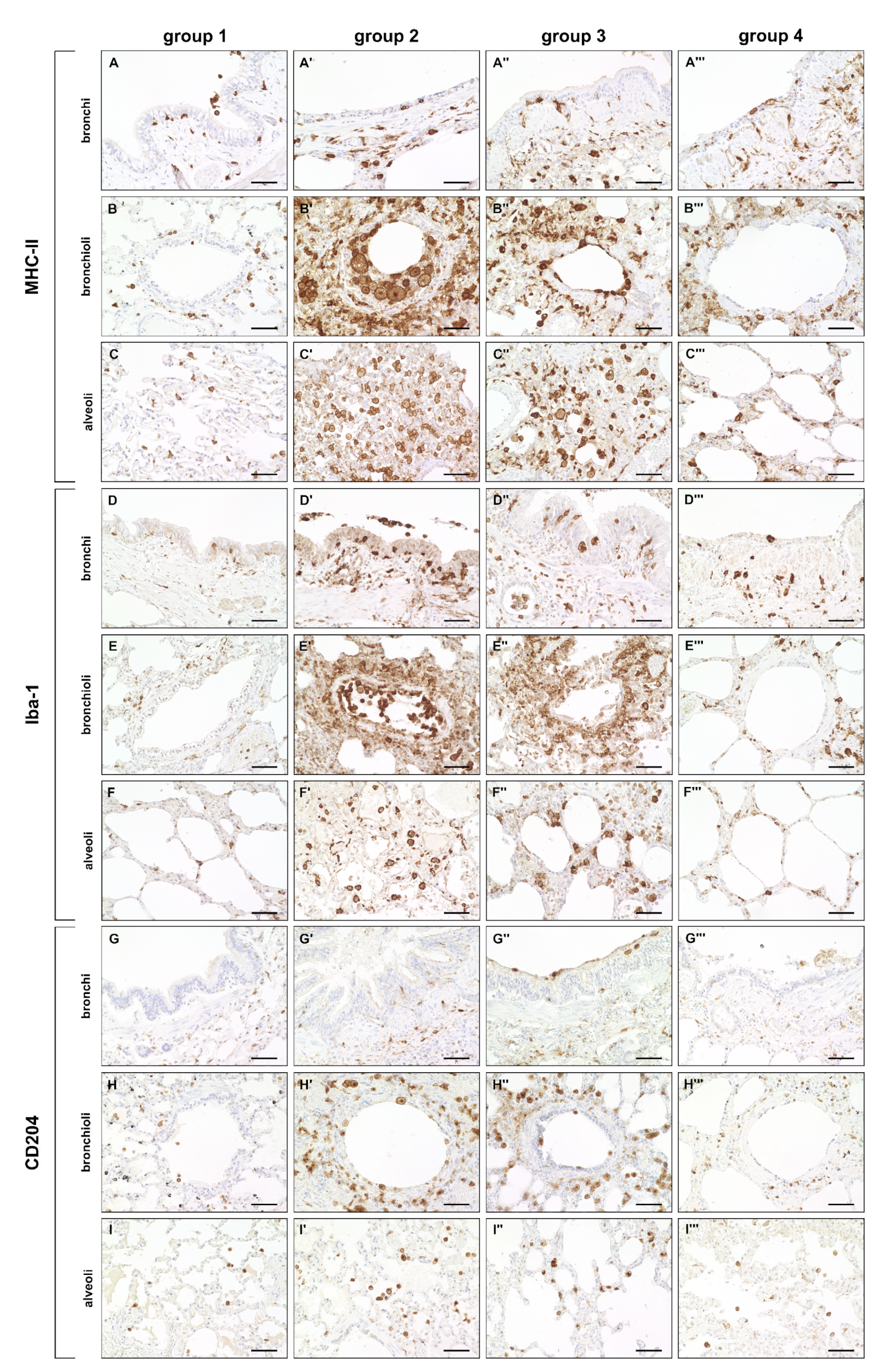
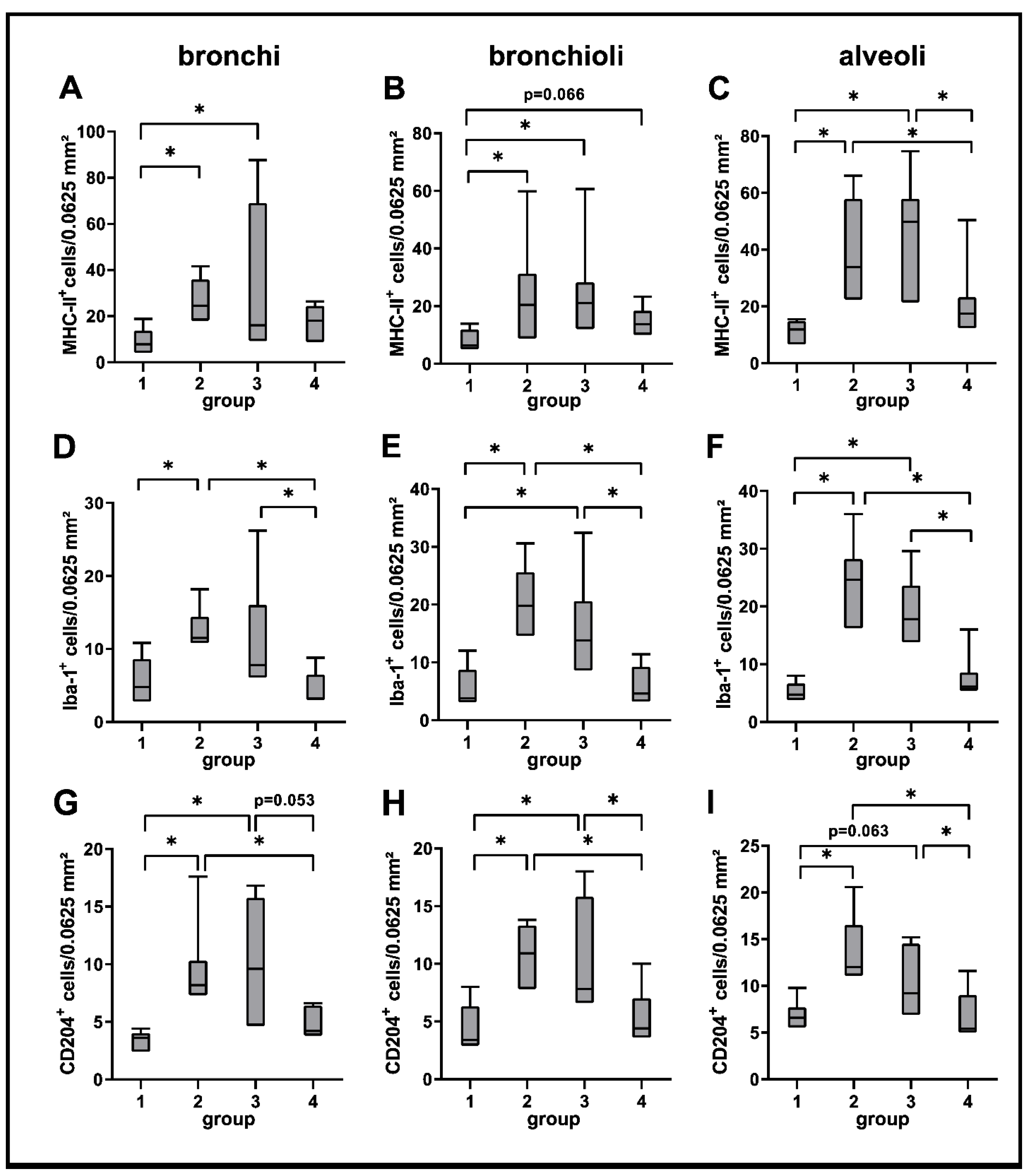
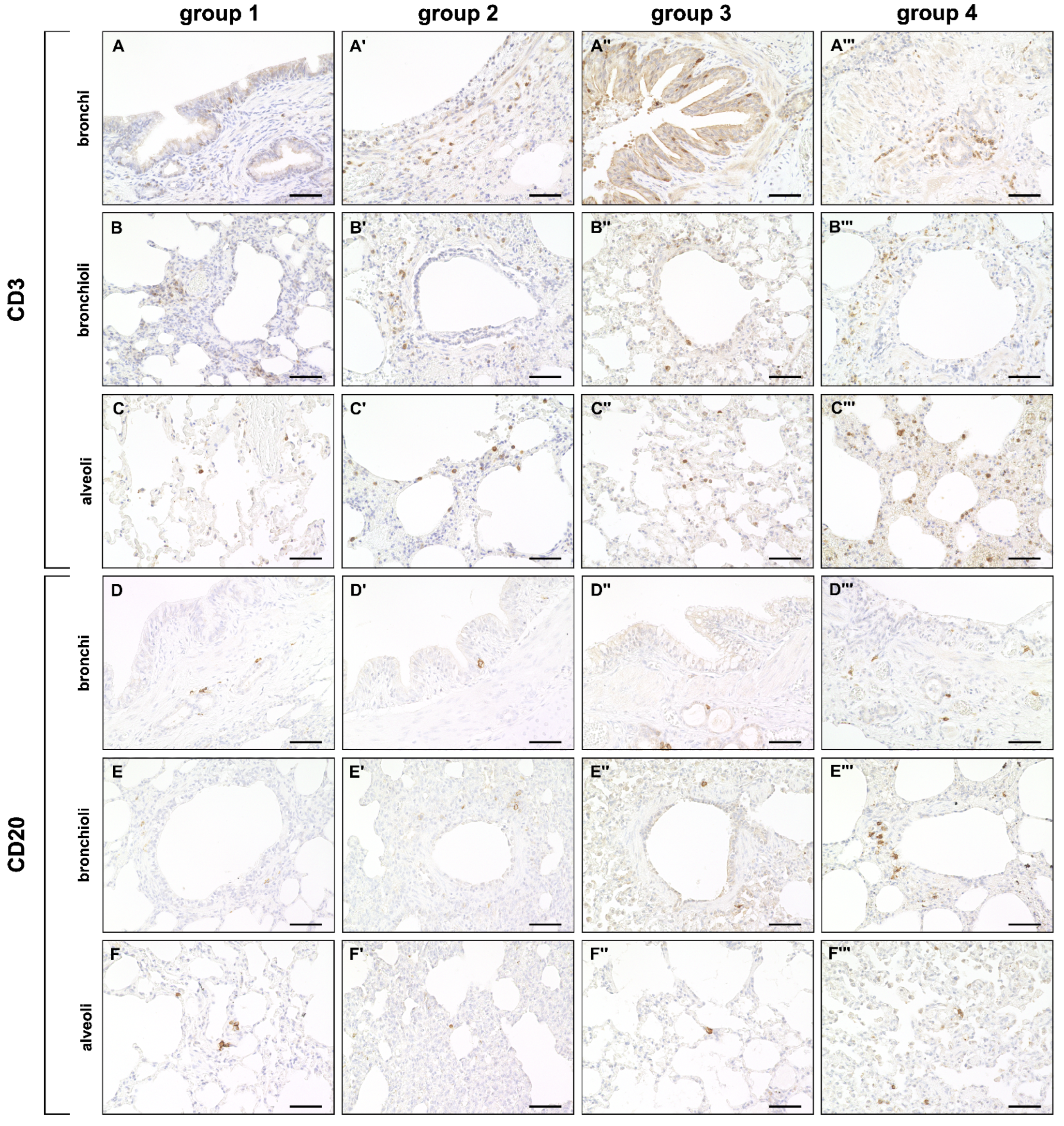
2.3. Phenotyping of Pulmonary Immune Responses
2.3.1. Innate Immune Cell Response
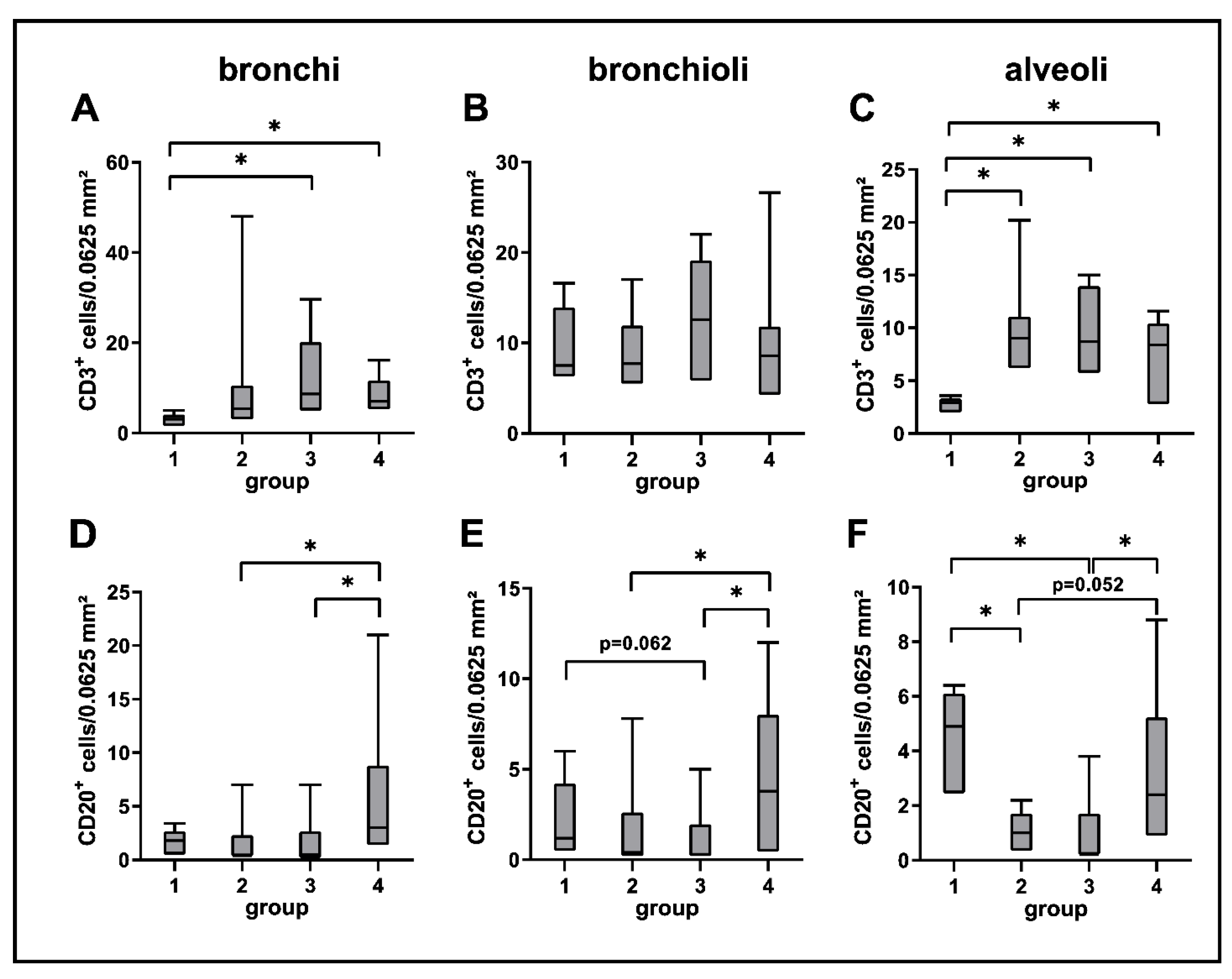
2.3.2. Adaptive Immune Cell Response
2.4. Global Transcriptome Analysis of Canine Distemper Virus-Infected Lung Tissue
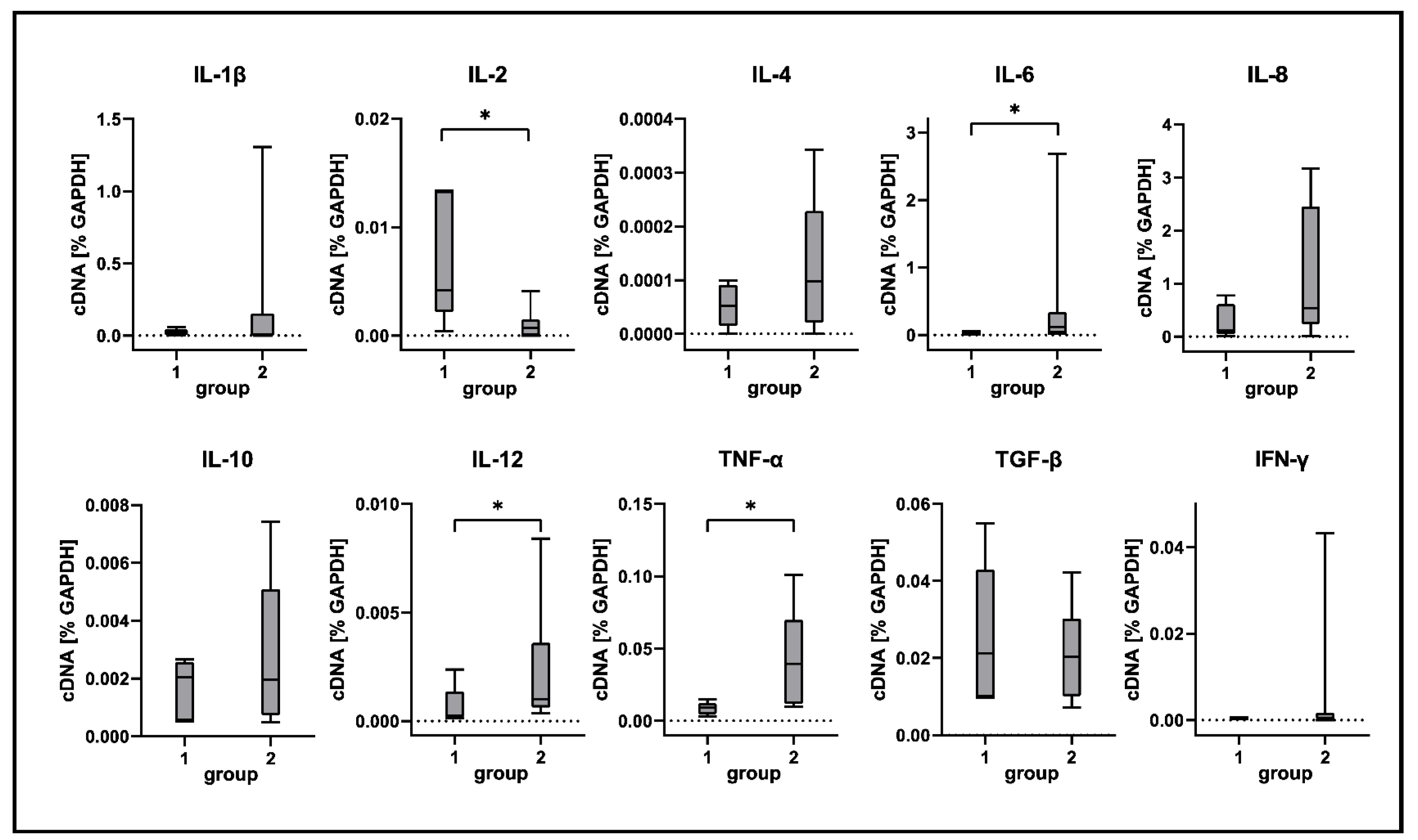
2.5. Cytokine Expression Analyses
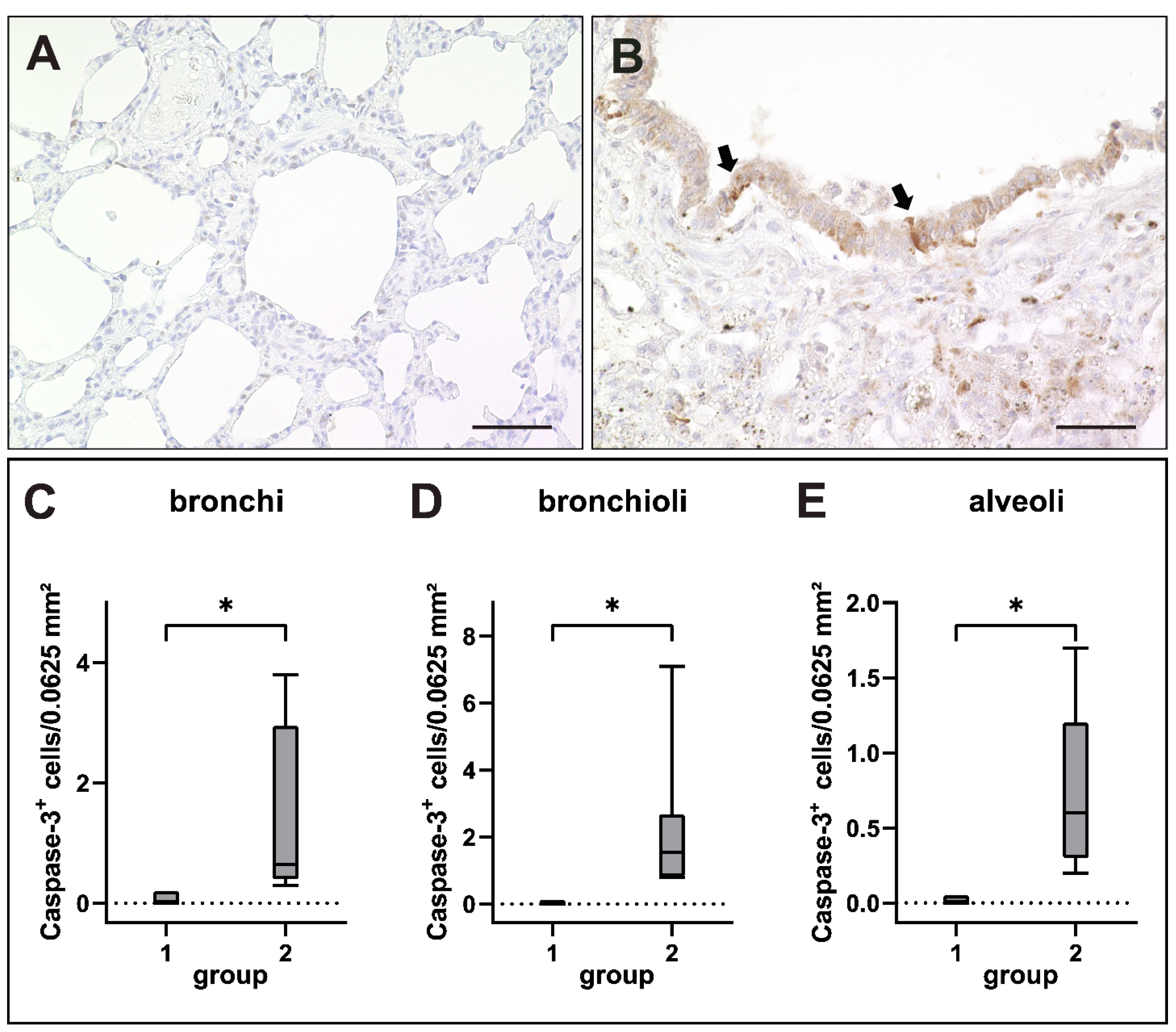
2.6. Apoptosis Induction in Canine Distemper Virus-Infected Lungs
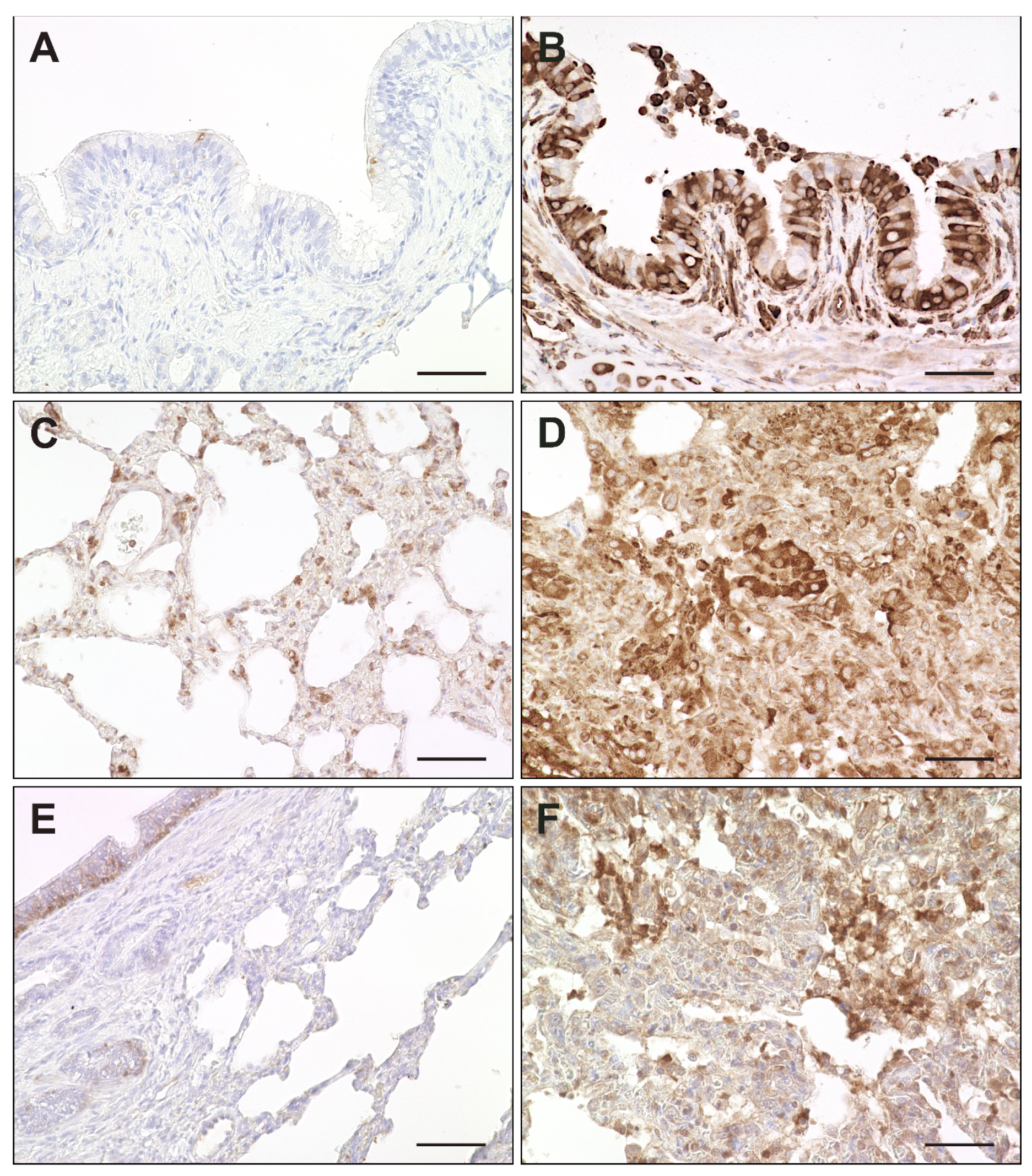
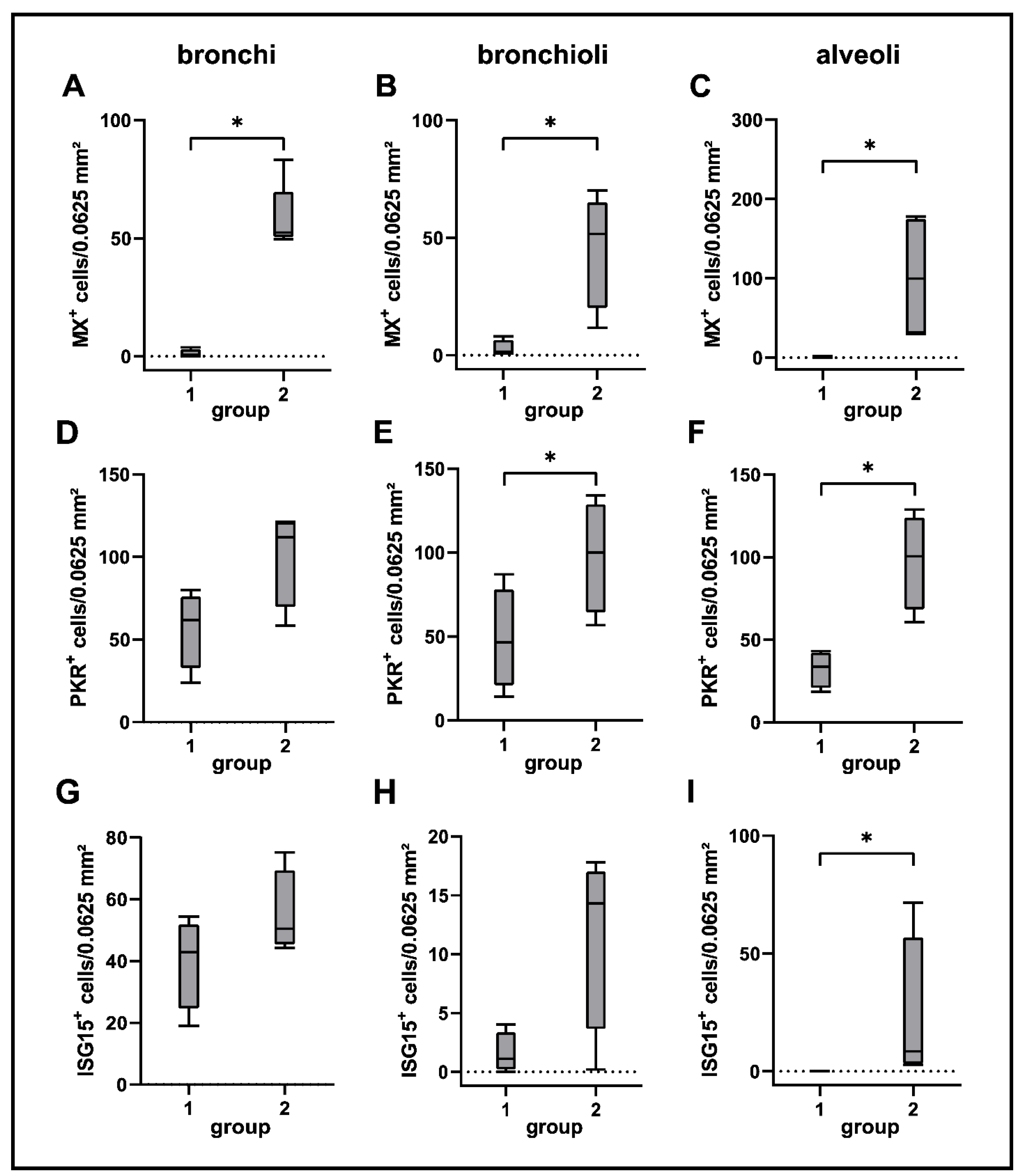
2.7. Interferon-Related Genes
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Animals, Tissue Samples and Processing
4.3. Classification of Disease Phases
4.4. Histological Scoring of Lung Lesions
4.5. Immunohistochemistry
4.6. Immunofluorescence Double Labeling
4.7. Molecular Investigations
4.7.1. RNA Isolation
4.7.2. RNA Sequencing Analysis
4.7.3. Reverse Transcription
4.7.4. Primers and Plasmids
4.7.5. Generation of Standard Dilutions
4.7.6. Reverse Transcription Quantitative PCR (RT-qPCR) Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sato, H.; Yoneda, M.; Honda, T.; Kai, C. Morbillivirus receptors and tropism: Multiple pathways for infection. Front. Microbiol. 2012, 3, 75. [Google Scholar] [CrossRef]
- Arruda, B.; Shen, H.; Zheng, Y.; Li, G. Novel Morbillivirus as Putative Cause of Fetal Death and Encephalitis among Swine. Emerg. Infect. Dis. 2021, 27, 1858–1866. [Google Scholar] [CrossRef]
- Beineke, A.; Baumgärtner, W.; Wohlsein, P. Cross-species transmission of canine distemper virus—An update. One Health 2015, 1, 49–59. [Google Scholar] [CrossRef]
- Appel, M.J. Distemper pathogenesis in dogs. J. Am. Vet. Med. Assoc. 1970, 156, 1681–1684. [Google Scholar]
- Barrett, T. Morbillivirus infections, with special emphasis on morbilliviruses of carnivores. Vet. Microbiol. 1999, 69, 3–13. [Google Scholar] [CrossRef]
- Deem, S.L.; Spelman, L.H.; Yates, R.A.; Montali, R.J. Canine distemper in terrestrial carnivores: A review. J. Zoo Wildl. Med. 2000, 31, 441–451. [Google Scholar] [CrossRef]
- Harder, T.C.; Osterhaus, A.D. Canine distemper virus—A morbillivirus in search of new hosts? Trends Microbiol. 1997, 5, 120–124. [Google Scholar] [CrossRef]
- Feng, N.; Yu, Y.; Wang, T.; Wilker, P.; Wang, J.; Li, Y.; Sun, Z.; Gao, Y.; Xia, X. Fatal canine distemper virus infection of giant pandas in China. Sci. Rep. 2016, 6, 27518. [Google Scholar] [CrossRef]
- Chinnadurai, S.K.; Kinsel, M.J.; Adkesson, M.J.; Terio, K. Canine Distemper in a Vaccinated Snow Leopard (Panthera uncia). J. Zoo Wildl Med. 2017, 48, 1200–1203. [Google Scholar] [CrossRef]
- Choi, Y.K.; Simon, M.A.; Kim, D.Y.; Yoon, B.I.; Kwon, S.W.; Lee, K.W.; Seo, I.B.; Kim, D.Y. Fatal measles virus infection in Japanese macaques (Macaca fuscata). Vet. Pathol. 1999, 36, 594–600. [Google Scholar] [CrossRef]
- Kennedy, S.; Kuiken, T.; Jepson, P.D.; Deaville, R.; Forsyth, M.; Barrett, T.; van de Bildt, M.W.; Osterhaus, A.D.; Eybatov, T.; Duck, C.; et al. Mass die-Off of Caspian seals caused by canine distemper virus. Emerg. Infect. Dis. 2000, 6, 637–639. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.; Sulikhan, N.; Uphyrkina, O.; Goncharuk, M.; Kerley, L.; Castro, E.H.; Reeve, R.; Seimon, T.; McAloose, D.; Seryodkin, I.V.; et al. Distemper, extinction, and vaccination of the Amur tiger. Proc. Natl. Acad. Sci. USA 2020, 117, 31954–31962. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, O.V.; Botelho, C.V.; Ferreira, C.G.; Scherer, P.O.; Soares-Martins, J.A.; Almeida, M.R.; Silva Junior, A. Immunopathogenic and neurological mechanisms of canine distemper virus. Adv. Virol. 2012, 2012, 163860. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Elia, G.; Buonavoglia, C. Canine distemper virus. Vet. Clin. North Am. Small Anim. Pract. 2008, 38, 787–797. [Google Scholar] [CrossRef]
- Pardo, M.C.; Bauman, J.E.; Mackowiak, M. Protection of dogs against canine distemper by vaccination with a canarypox virus recombinant expressing canine distemper virus fusion and hemagglutinin glycoproteins. Am. J. Vet. Res. 1997, 58, 833–836. [Google Scholar]
- Diallo, A. Morbillivirus group: Genome organisation and proteins. Vet. Microbiol. 1990, 23, 155–163. [Google Scholar] [CrossRef]
- Hall, W.W.; Lamb, R.A.; Choppin, P.W. The polypeptides of canine distemper virus: Synthesis in infected cells and relatedness to the polypeptides of other morbilliviruses. Virology 1980, 100, 433–449. [Google Scholar] [CrossRef]
- Örvell, C. Structural polypeptides of canine distemper virus. Arch. Virol. 1980, 66, 193–206. [Google Scholar] [CrossRef]
- Tatsuo, H.; Ono, N.; Yanagi, Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J. Virol. 2001, 75, 5842–5850. [Google Scholar] [CrossRef]
- Sawatsky, B.; Cattaneo, R.; von Messling, V. Canine Distemper Virus Spread and Transmission to Naive Ferrets: Selective Pressure on Signaling Lymphocyte Activation Molecule-Dependent Entry. J. Virol. 2018, 92, e00669-18. [Google Scholar] [CrossRef]
- De Vries, R.D.; Ludlow, M.; de Jong, A.; Rennick, L.J.; Verburgh, R.J.; van Amerongen, G.; van Riel, D.; van Run, P.; Herfst, S.; Kuiken, T.; et al. Delineating morbillivirus entry, dissemination and airborne transmission by studying in vivo competition of multicolor canine distemper viruses in ferrets. PLoS Pathog. 2017, 13, e1006371. [Google Scholar] [CrossRef] [PubMed]
- Lemon, K.; de Vries, R.D.; Mesman, A.W.; McQuaid, S.; van Amerongen, G.; Yuksel, S.; Ludlow, M.; Rennick, L.J.; Kuiken, T.; Rima, B.K.; et al. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog. 2011, 7, e1001263. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.S.; Frenzke, M.; Leonard, V.H.; Welstead, G.G.; Richardson, C.D.; Cattaneo, R. Measles virus infection of alveolar macrophages and dendritic cells precedes spread to lymphatic organs in transgenic mice expressing human signaling lymphocytic activation molecule (SLAM, CD150). J. Virol. 2010, 84, 3033–3042. [Google Scholar] [CrossRef]
- Von Messling, V.; Milosevic, D.; Cattaneo, R. Tropism illuminated: Lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc. Natl. Acad. Sci. USA 2004, 101, 14216–14221. [Google Scholar] [CrossRef]
- Iwatsuki, K.; Okita, M.; Ochikubo, F.; Gemma, T.; Shin, Y.S.; Miyashita, N.; Mikami, T.; Kai, C. Immunohistochemical analysis of the lymphoid organs of dogs naturally infected with canine distemper virus. J. Comp. Pathol. 1995, 113, 185–190. [Google Scholar] [CrossRef]
- Krakowka, S.; Higgins, R.J.; Koestner, A. Canine distemper virus: Review of structural and functional modulations in lymphoid tissues. Am. J. Vet. Res. 1980, 41, 284–292. [Google Scholar] [PubMed]
- Sawatsky, B.; Wong, X.X.; Hinkelmann, S.; Cattaneo, R.; von Messling, V. Canine distemper virus epithelial cell infection is required for clinical disease but not for immunosuppression. J. Virol. 2012, 86, 3658–3666. [Google Scholar] [CrossRef]
- Von Messling, V.; Springfeld, C.; Devaux, P.; Cattaneo, R. A ferret model of canine distemper virus virulence and immunosuppression. J. Virol. 2003, 77, 12579–12591. [Google Scholar] [CrossRef] [PubMed]
- Frisk, A.L.; Konig, M.; Moritz, A.; Baumgärtner, W. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J. Clin. Microbiol. 1999, 37, 3634–3643. [Google Scholar] [CrossRef]
- Ludlow, M.; Rennick, L.J.; Nambulli, S.; de Swart, R.L.; Duprex, W.P. Using the ferret model to study morbillivirus entry, spread, transmission and cross-species infection. Curr. Opin. Virol. 2014, 4, 15–23. [Google Scholar] [CrossRef]
- Pratakpiriya, W.; Seki, F.; Otsuki, N.; Sakai, K.; Fukuhara, H.; Katamoto, H.; Hirai, T.; Maenaka, K.; Techangamsuwan, S.; Lan, N.T.; et al. Nectin4 is an epithelial cell receptor for canine distemper virus and involved in neurovirulence. J. Virol. 2012, 86, 10207–10210. [Google Scholar] [CrossRef] [PubMed]
- Ducatelle, R.; Maenhout, D.; Coussement, W.; Hoorens, J. Dual adenovirus and distemper virus pneumonia in a dog. Vet. Q. 1982, 4, 84–88. [Google Scholar] [CrossRef]
- Miry, C.; Ducatelle, R.; Thoonen, H.; Hoorens, J. Immunoperoxidase study of canine distemper virus pneumonia. Res. Vet. Sci. 1983, 34, 145–148. [Google Scholar] [CrossRef]
- Damián, M.; Morales, E.; Salas, G.; Trigo, F.J. Immunohistochemical detection of antigens of distemper, adenovirus and parainfluenza viruses in domestic dogs with pneumonia. J. Comp. Pathol. 2005, 133, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Appel, M.J.; Summers, B.A. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet. Microbiol. 1995, 44, 187–191. [Google Scholar] [CrossRef]
- Summers, B.A.; Greisen, H.A.; Appel, M.J. Canine distemper encephalomyelitis: Variation with virus strain. J. Comp. Pathol. 1984, 94, 65–75. [Google Scholar] [CrossRef]
- Krakowka, S.; Koestner, A. Age-related susceptibility to infection with canine distemper virus in gnotobiotic dogs. J. Infect. Dis. 1976, 134, 629–632. [Google Scholar] [CrossRef]
- Krakowka, S.; Olsen, R.; Confer, A.; Koestner, A.; McCullough, B. Serologic response to canine distemper viral antigens in gnotobiotic dogs infected with canine distemper virus. J. Infect. Dis. 1975, 132, 384–392. [Google Scholar] [CrossRef]
- Rima, B.K.; Duffy, N.; Mitchell, W.J.; Summers, B.A.; Appel, M.J. Correlation between humoral immune responses and presence of virus in the CNS in dogs experimentally infected with canine distemper virus. Arch. Virol. 1991, 121, 1–8. [Google Scholar] [CrossRef]
- Axthelm, M.K.; Krakowka, S. Experimental old dog encephalitis (ODE) in a gnotobiotic dog. Vet. Pathol. 1998, 35, 527–534. [Google Scholar] [CrossRef]
- Markus, S.; Failing, K.; Baumgärtner, W. Increased expression of pro-inflammatory cytokines and lack of up-regulation of anti-inflammatory cytokines in early distemper CNS lesions. J. Neuroimmunol. 2002, 125, 30–41. [Google Scholar] [CrossRef]
- Toews, G.B. Cytokines and the lung. Eur. Respir. J. 2001, 34, 3s–17s. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. The cytokine network in asthma and chronic obstructive pulmonary disease. J. Clin. Investig. 2008, 118, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Belardelli, F. Role of interferons and other cytokines in the regulation of the immune response. APMIS 1995, 103, 161–179. [Google Scholar] [CrossRef]
- Rothwell, N.J. Sixteenth Gaddum Memorial Lecture December 1996. Neuroimmune interactions: The role of cytokines. Br. J. Pharmacol. 1997, 121, 841–847. [Google Scholar] [CrossRef][Green Version]
- Rath, P.C.; Aggarwal, B.B. TNF-induced signaling in apoptosis. J. Clin. Immunol. 1999, 19, 350–364. [Google Scholar] [CrossRef]
- Guttman, J.A.; Finlay, B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta 2009, 1788, 832–841. [Google Scholar] [CrossRef]
- Lloyd, C.M.; Marsland, B.J. Lung Homeostasis: Influence of Age, Microbes, and the Immune System. Immunity 2017, 46, 549–561. [Google Scholar] [CrossRef]
- Iwasaki, A.; Foxman, E.F.; Molony, R.D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 2017, 17, 7–20. [Google Scholar] [CrossRef]
- Stark, G.R.; Kerr, I.M.; Williams, B.R.; Silverman, R.H.; Schreiber, R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998, 67, 227–264. [Google Scholar] [CrossRef]
- Alldinger, S.; Wünschmann, A.; Baumgärtner, W.; Voss, C.; Kremmer, E. Up-regulation of major histocompatibility complex class II antigen expression in the central nervous system of dogs with spontaneous canine distemper virus encephalitis. Acta Neuropathol. 1996, 92, 273–280. [Google Scholar] [CrossRef]
- Baumgärtner, W.; Alldinger, S. The Pathogenesis of Canine Distemper Virus Induced Demyelination. In Experimental Models of Multiple Sclerosis; Lavi, E., Constantinescu, C.S., Eds.; Springer US: Boston, MA, USA, 2005; pp. 871–887. [Google Scholar]
- Beineke, A.; Markus, S.; Borlak, J.; Thum, T.; Baumgärtner, W. Increase of pro-inflammatory cytokine expression in non-demyelinating early cerebral lesions in nervous canine distemper. Viral Immunol. 2008, 21, 401–410. [Google Scholar] [CrossRef]
- Tipold, A.; Moore, P.; Zurbriggen, A.; Burgener, I.; Barben, G.; Vandevelde, M. Early T cell response in the central nervous system in canine distemper virus infection. Acta Neuropathol. 1999, 97, 45–56. [Google Scholar] [CrossRef]
- Klotz, D.; Gerhauser, I. Interferon-Stimulated Genes-Mediators of the Innate Immune Response during Canine Distemper Virus Infection. Int. J. Mol. Sci. 2019, 20, 1620. [Google Scholar] [CrossRef]
- Qeska, V.; Barthel, Y.; Iseringhausen, M.; Tipold, A.; Stein, V.M.; Khan, M.A.; Baumgärtner, W.; Beineke, A. Dynamic changes of Foxp3+ regulatory T cells in spleen and brain of canine distemper virus-infected dogs. Vet. Immunol. Immunopathol. 2013, 156, 215–222. [Google Scholar] [CrossRef]
- Wünschmann, A.; Alldinger, S.; Kremmer, E.; Baumgärtner, W. Identification of CD4+ and CD8+ T cell subsets and B cells in the brain of dogs with spontaneous acute, subacute-, and chronic-demyelinating distemper encephalitis. Vet. Immunol. Immunopathol. 1999, 67, 101–116. [Google Scholar] [CrossRef]
- Beineke, A.; Puff, C.; Seehusen, F.; Baumgärtner, W. Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet. Immunol. Immunopathol. 2009, 127, 1–18. [Google Scholar] [CrossRef]
- Moussallem, T.M.; Guedes, F.; Fernandes, E.R.; Pagliari, C.; Lancellotti, C.L.; de Andrade, H.F., Jr.; Duarte, M.I. Lung involvement in childhood measles: Severe immune dysfunction revealed by quantitative immunohistochemistry. Hum. Pathol. 2007, 38, 1239–1247. [Google Scholar] [CrossRef]
- Díaz-Delgado, J.; Groch, K.R.; Ressio, R.; Riskallah, I.P.J.; Sierra, E.; Sacchini, S.; Quesada-Canales, Ó.; Arbelo, M.; Fernández, A.; Santos-Neto, E.; et al. Comparative Immunopathology of Cetacean morbillivirus Infection in Free-Ranging Dolphins from Western Mediterranean, Northeast-Central, and Southwestern Atlantic. Front. Immunol. 2019, 10, 485. [Google Scholar] [CrossRef]
- Demonbreun, W.A. The Histopathology of Natural and Experimental Canine Distemper. Am. J. Pathol. 1937, 13, 187–212. [Google Scholar]
- Coffin, D.L.; Liu, C. Studies of canine distemper infection by means of fluorescein-labeled antibody. II. The pathology and diagnosis of the naturally occurring disease in dogs and the antigenic nature of the inclusion body. Virology 1957, 3, 132–145. [Google Scholar] [CrossRef]
- Look, D.C.; Walter, M.J.; Williamson, M.R.; Pang, L.; You, Y.; Sreshta, J.N.; Johnson, J.E.; Zander, D.S.; Brody, S.L. Effects of paramyxoviral infection on airway epithelial cell Foxj1 expression, ciliogenesis, and mucociliary function. Am. J. Pathol. 2001, 159, 2055–2069. [Google Scholar] [CrossRef]
- Tilley, A.E.; Walters, M.S.; Shaykhiev, R.; Crystal, R.G. Cilia dysfunction in lung disease. Annu. Rev. Physiol. 2015, 77, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Kantar, A.; Oggiano, N.; Giorgi, P.L.; Braga, P.C.; Fiorini, R. Polymorphonuclear leukocyte-generated oxygen metabolites decrease beat frequency of human respiratory cilia. Lung 1994, 172, 215–222. [Google Scholar] [CrossRef]
- Klemens, J.; Ciurkiewicz, M.; Chludzinski, E.; Iseringhausen, M.; Klotz, D.; Pfankuche, V.M.; Ulrich, R.; Herder, V.; Puff, C.; Baumgärtner, W.; et al. Neurotoxic potential of reactive astrocytes in canine distemper demyelinating leukoencephalitis. Sci. Rep. 2019, 9, 11689. [Google Scholar] [CrossRef]
- Gaedke, K.; Zurbriggen, A.; Baumgärtner, W. Lack of correlation between virus nucleoprotein and mRNA expression and the inflammatory response in demyelinating distemper encephalitis indicates a biphasic disease process. Eur. J. Vet. Pathol. 1999, 5, 9–20. [Google Scholar]
- Higgins, R.J.; Krakowka, S.G.; Metzler, A.E.; Koestner, A. Experimental canine distemper encephalomyelitis in neonatal gnotobiotic dogs. A sequential ultrastructural study. Acta Neuropathol. 1982, 57, 287–295. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Shenoy, A.T.; De Ana, C.L.; Arafa, E.I.; Salwig, I.; Barker, K.A.; Korkmaz, F.T.; Ramanujan, A.; Etesami, N.S.; Soucy, A.M.; Martin, I.M.C.; et al. Antigen presentation by lung epithelial cells directs CD4+TRM cell function and regulates barrier immunity. Nat. Commun. 2021, 12, 5834. [Google Scholar] [CrossRef]
- Kawasaki, T.; Ikegawa, M.; Kawai, T. Antigen Presentation in the Lung. Front. Immunol. 2022, 13, 860915. [Google Scholar] [CrossRef]
- Toulmin, S.A.; Bhadiadra, C.; Paris, A.J.; Lin, J.H.; Katzen, J.; Basil, M.C.; Morrisey, E.E.; Worthen, G.S.; Eisenlohr, L.C. Type II alveolar cell MHCII improves respiratory viral disease outcomes while exhibiting limited antigen presentation. Nat. Commun. 2021, 12, 3993. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.C.; Milne, D.S.; Wilkes, J.; Dark, J.H.; Tetley, T.D.; Kirby, J.A. Constitutive expression of MHC and adhesion molecules by alveolar epithelial cells (type II pneumocytes) isolated from human lung and comparison with immunocytochemical findings. J. Cell Sci. 1994, 107, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Glanville, A.R.; Tazelaar, H.D.; Theodore, J.; Imoto, E.; Rouse, R.V.; Baldwin, J.C.; Robin, E.D. The distribution of MHC class I and II antigens on bronchial epithelium. Am. Rev. Respir. Dis. 1989, 139, 330–334. [Google Scholar] [CrossRef]
- Stein, V.M.; Czub, M.; Schreiner, N.; Moore, P.F.; Vandevelde, M.; Zurbriggen, A.; Tipold, A. Microglial cell activation in demyelinating canine distemper lesions. J. Neuroimmunol. 2004, 153, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Manchester, M.; Eto, D.S.; Oldstone, M.B. Characterization of the inflammatory response during acute measles encephalitis in NSE-CD46 transgenic mice. J. Neuroimmunol. 1999, 96, 207–217. [Google Scholar] [CrossRef]
- Moss, W.J.; Ryon, J.J.; Monze, M.; Griffin, D.E. Differential regulation of interleukin (IL)-4, IL-5, and IL-10 during measles in Zambian children. J. Infect. Dis. 2002, 186, 879–887. [Google Scholar] [CrossRef]
- McCullough, B.; Krakowka, S.; Koestner, A. Experimental canine distemper virus-induced lymphoid depletion. Am. J. Pathol. 1974, 74, 155–170. [Google Scholar]
- Moro, L.; de Sousa Martins, A.; de Moraes Alves, C.; de Araujo Santos, F.G.; dos Santos Nunes, J.E.; Carneiro, R.A.; Carvalho, R.; Vasconcelos, A.C. Apoptosis in canine distemper. Arch. Virol. 2003, 148, 153–164. [Google Scholar] [CrossRef]
- Kumagai, K.; Yamaguchi, R.; Uchida, K.; Tateyama, S. Lymphoid apoptosis in acute canine distemper. J. Vet. Med. Sci. 2004, 66, 175–181. [Google Scholar] [CrossRef]
- De Vries, R.D.; McQuaid, S.; van Amerongen, G.; Yüksel, S.; Verburgh, R.J.; Osterhaus, A.D.; Duprex, W.P.; de Swart, R.L. Measles immune suppression: Lessons from the macaque model. PLoS Pathog. 2012, 8, e1002885. [Google Scholar] [CrossRef]
- Wünschmann, A.; Kremmer, E.; Baumgärtner, W. Phenotypical characterization of T and B cell areas in lymphoid tissues of dogs with spontaneous distemper. Vet. Immunol. Immunopathol. 2000, 73, 83–98. [Google Scholar] [CrossRef]
- Hirsch, R.L.; Griffin, D.E.; Johnson, R.T.; Cooper, S.J.; de Soriano, I.L.; Roedenbeck, S.; Vaisberg, A. Cellular immune responses during complicated and uncomplicated measles virus infections of man. Clin. Immunol. Immunopathol. 1984, 31, 1–12. [Google Scholar] [CrossRef]
- Ward, B.J.; Johnson, R.T.; Vaisberg, A.; Jauregui, E.; Griffin, D.E. Cytokine production in vitro and the lymphoproliferative defect of natural measles virus infection. Clin. Immunol. Immunopathol. 1991, 61, 236–248. [Google Scholar] [CrossRef]
- Moore, B.B.; Moore, T.A.; Toews, G.B. Role of T- and B-lymphocytes in pulmonary host defences. Eur. Respir. J. 2001, 18, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef]
- Pandey, P.; Karupiah, G. Targeting tumour necrosis factor to ameliorate viral pneumonia. FEBS J. 2022, 289, 883–900. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Hoidal, J.R.; Mukherjee, T.K. Role of TNFalpha in pulmonary pathophysiology. Respir. Res. 2006, 7, 125. [Google Scholar] [CrossRef]
- Seo, S.H.; Webster, R.G. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 2002, 76, 1071–1076. [Google Scholar] [CrossRef]
- Wong, G.H.; Goeddel, D.V. Tumour necrosis factors alpha and beta inhibit virus replication and synergize with interferons. Nature 1986, 323, 819–822. [Google Scholar] [CrossRef]
- Van Campen, H. Influenza A virus replication is inhibited by tumor necrosis factor-alpha in vitro. Arch. Virol. 1994, 136, 439–446. [Google Scholar] [CrossRef]
- Czikora, I.; Alli, A.; Bao, H.F.; Kaftan, D.; Sridhar, S.; Apell, H.J.; Gorshkov, B.; White, R.; Zimmermann, A.; Wendel, A.; et al. A novel tumor necrosis factor-mediated mechanism of direct epithelial sodium channel activation. Am. J. Respir. Crit. Care Med. 2014, 190, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Braun, C.; Hamacher, J.; Morel, D.R.; Wendel, A.; Lucas, R. Dichotomal role of TNF in experimental pulmonary edema reabsorption. J. Immunol. 2005, 175, 3402–3408. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, T.; Yamagata, Y.; Nishimoto, T.; Hirano, T.; Nakanishi, M.; Minakata, Y.; Ichinose, M.; Dagenais, A.; Berthiaume, Y. The regulation of amiloride-sensitive epithelial sodium channels by tumor necrosis factor-alpha in injured lungs and alveolar type II cells. Respir. Physiol. Neurobiol. 2009, 166, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Hamacher, J.; Hadizamani, Y.; Borgmann, M.; Mohaupt, M.; Männel, D.N.; Moehrlen, U.; Lucas, R.; Stammberger, U. Cytokine-Ion Channel Interactions in Pulmonary Inflammation. Front. Immunol. 2017, 8, 1644. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, A.; Fréchette, R.; Yamagata, Y.; Yamagata, T.; Carmel, J.F.; Clermont, M.E.; Brochiero, E.; Massé, C.; Berthiaume, Y. Downregulation of ENaC activity and expression by TNF-alpha in alveolar epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2004, 286, L301–L311. [Google Scholar] [CrossRef]
- Patel, B.V.; Wilson, M.R.; O’Dea, K.P.; Takata, M. TNF-induced death signaling triggers alveolar epithelial dysfunction in acute lung injury. J. Immunol. 2013, 190, 4274–4282. [Google Scholar] [CrossRef]
- Lucas, R.; Hadizamani, Y.; Enkhbaatar, P.; Csanyi, G.; Caldwell, R.W.; Hundsberger, H.; Sridhar, S.; Lever, A.A.; Hudel, M.; Ash, D.; et al. Dichotomous Role of Tumor Necrosis Factor in Pulmonary Barrier Function and Alveolar Fluid Clearance. Front. Physiol. 2021, 12, 793251. [Google Scholar] [CrossRef]
- Faggioni, R.; Gatti, S.; Demitri, M.T.; Delgado, R.; Echtenacher, B.; Gnocchi, P.; Heremans, H.; Ghezzi, P. Role of xanthine oxidase and reactive oxygen intermediates in LPS- and TNF-induced pulmonary edema. J. Lab. Clin. Med. 1994, 123, 394–399. [Google Scholar]
- Hamacher, J.; Lucas, R.; Lijnen, H.R.; Buschke, S.; Dunant, Y.; Wendel, A.; Grau, G.E.; Suter, P.M.; Ricou, B. Tumor necrosis factor-alpha and angiostatin are mediators of endothelial cytotoxicity in bronchoalveolar lavages of patients with acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2002, 166, 651–656. [Google Scholar] [CrossRef]
- Ferro, T.J.; Hocking, D.C.; Johnson, A. Tumor necrosis factor-alpha alters pulmonary vasoreactivity via neutrophil-derived oxidants. Am. J. Physiol. 1993, 265, L462–L471. [Google Scholar] [CrossRef]
- Petrache, I.; Birukova, A.; Ramirez, S.I.; Garcia, J.G.; Verin, A.D. The role of the microtubules in tumor necrosis factor-alpha-induced endothelial cell permeability. Am. J. Respir. Cell Mol. Biol. 2003, 28, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, E.; Cuzzocrea, S. Role of TNF-alpha in lung tight junction alteration in mouse model of acute lung inflammation. Respir. Res. 2007, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.R.; Kim, R.K.; Pober, J.S.; Kluger, M.S. Tumor necrosis factor disrupts claudin-5 endothelial tight junction barriers in two distinct NF-κB-dependent phases. PLoS ONE 2015, 10, e0120075. [Google Scholar] [CrossRef] [PubMed]
- Ishii, Y.; Partridge, C.A.; Del Vecchio, P.J.; Malik, A.B. Tumor necrosis factor-alpha-mediated decrease in glutathione increases the sensitivity of pulmonary vascular endothelial cells to H2O2. J. Clin. Investig. 1992, 89, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Gröne, A.; Alldinger, S.; Baumgärtner, W. Interleukin-1beta, -6, -12 and tumor necrosis factor-alpha expression in brains of dogs with canine distemper virus infection. J. Neuroimmunol. 2000, 110, 20–30. [Google Scholar] [CrossRef]
- Schobesberger, M.; Summerfield, A.; Doherr, M.G.; Zurbriggen, A.; Griot, C. Canine distemper virus-induced depletion of uninfected lymphocytes is associated with apoptosis. Vet. Immunol. Immunopathol. 2005, 104, 33–44. [Google Scholar] [CrossRef]
- Schneider-Schaulies, J.; Schneider-Schaulies, S.; Ter Meulen, V. Differential induction of cytokines by primary and persistent measles virus infections in human glial cells. Virology 1993, 195, 219–228. [Google Scholar] [CrossRef]
- Plaza, J.A.; Nuovo, G.J. Histologic and molecular correlates of fatal measles infection in children. Diagn. Mol. Pathol. 2005, 14, 97–102. [Google Scholar] [CrossRef]
- Manetti, R.; Parronchi, P.; Giudizi, M.G.; Piccinni, M.P.; Maggi, E.; Trinchieri, G.; Romagnani, S. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J. Exp. Med. 1993, 177, 1199–1204. [Google Scholar] [CrossRef]
- Frisk, A.L.; Baumgärtner, W.; Gröne, A. Dominating interleukin-10 mRNA expression induction in cerebrospinal fluid cells of dogs with natural canine distemper virus induced demyelinating and non-demyelinating CNS lesions. J. Neuroimmunol. 1999, 97, 102–109. [Google Scholar] [CrossRef]
- Williams, M.A.; Tyznik, A.J.; Bevan, M.J. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature 2006, 441, 890–893. [Google Scholar] [CrossRef]
- Dooms, H.; Kahn, E.; Knoechel, B.; Abbas, A.K. IL-2 induces a competitive survival advantage in T lymphocytes. J. Immunol. 2004, 172, 5973–5979. [Google Scholar] [CrossRef] [PubMed]
- Furtado, G.C.; de Lafaille, M.A.C.; Kutchukhidze, N.; Lafaille, J.J. Interleukin 2 signaling is required for CD4+ regulatory T cell function. J. Exp. Med. 2002, 196, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Sadlack, B.; Löhler, J.; Schorle, H.; Klebb, G.; Haber, H.; Sickel, E.; Noelle, R.J.; Horak, I. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur. J. Immunol. 1995, 25, 3053–3059. [Google Scholar] [CrossRef] [PubMed]
- Latchman, Y.E.; Liang, S.C.; Wu, Y.; Chernova, T.; Sobel, R.A.; Klemm, M.; Kuchroo, V.K.; Freeman, G.J.; Sharpe, A.H. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA 2004, 101, 10691–10696. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef]
- Veinalde, R.; Pidelaserra-Martí, G.; Moulin, C.; Jeworowski, L.M.; Küther, L.; Buchholz, C.J.; Jäger, D.; Ungerechts, G.; Engeland, C.E. Oncolytic measles vaccines encoding PD-1 and PD-L1 checkpoint blocking antibodies to increase tumor-specific T cell memory. Mol. Ther. Oncolytics 2022, 24, 43–58. [Google Scholar] [CrossRef]
- Balkhi, M.Y.; Ma, Q.; Ahmad, S.; Junghans, R.P. T cell exhaustion and Interleukin 2 downregulation. Cytokine 2015, 71, 339–347. [Google Scholar] [CrossRef]
- West, E.E.; Jin, H.T.; Rasheed, A.U.; Penaloza-Macmaster, P.; Ha, S.J.; Tan, W.G.; Youngblood, B.; Freeman, G.J.; Smith, K.A.; Ahmed, R. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J. Clin. Investig. 2013, 123, 2604–2615. [Google Scholar] [CrossRef]
- Bogdan, C.; Paik, J.; Vodovotz, Y.; Nathan, C. Contrasting mechanisms for suppression of macrophage cytokine release by transforming growth factor-beta and interleukin-10. J. Biol. Chem. 1992, 267, 23301–23308. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Merrill, J.E.; Zimmerman, R.P. Natural and induced cytotoxicity of oligodendrocytes by microglia is inhibitable by TGF beta. Glia 1991, 4, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, W.; Liu, Z.; Liu, S.; Liang, X. Virus Infection and Death Receptor-Mediated Apoptosis. Viruses 2017, 9, 316. [Google Scholar] [CrossRef] [PubMed]
- Orzalli, M.H.; Kagan, J.C. Apoptosis and Necroptosis as Host Defense Strategies to Prevent Viral Infection. Trends Cell Biol. 2017, 27, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; Kobune, F.; Sato, T.A.; Kohama, T.; Takeuchi, Y.; Abe, T.; Takayama, N.; Tsuchiya, T.; Tashiro, M. Extensive lymphopenia due to apoptosis of uninfected lymphocytes in acute measles patients. Arch. Virol. 2000, 145, 905–920. [Google Scholar] [CrossRef]
- Pillet, S.; von Messling, V. Canine distemper virus selectively inhibits apoptosis progression in infected immune cells. J. Virol. 2009, 83, 6279–6287. [Google Scholar] [CrossRef]
- Kajita, M.; Katayama, H.; Murata, T.; Kai, C.; Hori, M.; Ozaki, H. Canine distemper virus induces apoptosis through caspase-3 and -8 activation in vero cells. J. Vet. Med. B Infect. Dis Vet. Public Health 2006, 53, 273–277. [Google Scholar] [CrossRef]
- Vuorinen, T.; Peri, P.; Vainionpää, R. Measles virus induces apoptosis in uninfected bystander T cells and leads to granzyme B and caspase activation in peripheral blood mononuclear cell cultures. Eur. J. Clin. Investig. 2003, 33, 434–442. [Google Scholar] [CrossRef]
- Esolen, L.M.; Park, S.W.; Hardwick, J.M.; Griffin, D.E. Apoptosis as a cause of death in measles virus-infected cells. J. Virol. 1995, 69, 3955–3958. [Google Scholar] [CrossRef]
- Klotz, D.; Baumgärtner, W.; Gerhauser, I. Type I interferons in the pathogenesis and treatment of canine diseases. Vet. Immunol. Immunopathol. 2017, 191, 80–93. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Schaulies, S.; Schneider-Schaulies, J.; Schuster, A.; Bayer, M.; Pavlovic, J.; ter Meulen, V. Cell type-specific MxA-mediated inhibition of measles virus transcription in human brain cells. J. Virol. 1994, 68, 6910–6917. [Google Scholar] [CrossRef] [PubMed]
- Schnorr, J.J.; Schneider-Schaulies, S.; Simon-Jödicke, A.; Pavlovic, J.; Horisberger, M.A.; ter Meulen, V. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J. Virol. 1993, 67, 4760–4768. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Kochs, G. Mx genes: Host determinants controlling influenza virus infection and trans-species transmission. Hum. Genet. 2020, 139, 695–705. [Google Scholar] [CrossRef]
- Lee, S.; Ishitsuka, A.; Noguchi, M.; Hirohama, M.; Fujiyasu, Y.; Petric, P.P.; Schwemmle, M.; Staeheli, P.; Nagata, K.; Kawaguchi, A. Influenza restriction factor MxA functions as inflammasome sensor in the respiratory epithelium. Sci. Immunol. 2019, 4, eaau4643. [Google Scholar] [CrossRef]
- Porter, B.F.; Ambrus, A.; Storts, R.W. Immunohistochemical evaluation of mx protein expression in canine encephalitides. Vet. Pathol. 2006, 43, 981–987. [Google Scholar] [CrossRef]
- Braun, B.A.; Marcovitz, A.; Camp, J.G.; Jia, R.; Bejerano, G. Mx1 and Mx2 key antiviral proteins are surprisingly lost in toothed whales. Proc. Natl. Acad. Sci. USA 2015, 112, 8036–8040. [Google Scholar] [CrossRef]
- Takeuchi, T.; Kobayashi, T.; Tamura, S.; Yokosawa, H. Negative regulation of protein phosphatase 2Cbeta by ISG15 conjugation. FEBS Lett. 2006, 580, 4521–4526. [Google Scholar] [CrossRef]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Dzimianski, J.V.; Scholte, F.E.M.; Bergeron, É.; Pegan, S.D. ISG15: It’s Complicated. J. Mol. Biol. 2019, 431, 4203–4216. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, B.J.; Ahn, H.S.; Go, H.J.; Kim, D.Y.; Kim, J.H.; Lee, J.B.; Park, S.Y.; Song, C.S.; Lee, S.W.; et al. Canine interferon lambda 3 expressed using an adenoviral vector effectively induces antiviral activity against canine influenza virus. Virus Res. 2021, 296, 198342. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, C.K.; Li, Z.; George, C.X.; Samuel, C.E. Protein kinase PKR and RNA adenosine deaminase ADAR1: New roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 2011, 23, 573–582. [Google Scholar] [CrossRef]
- García, M.A.; Meurs, E.F.; Esteban, M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie 2007, 89, 799–811. [Google Scholar] [CrossRef]
- Lee, S.B.; Esteban, M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 1994, 199, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.H. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007, 81, 12720–12729. [Google Scholar] [CrossRef]
- Drappier, M.; Michiels, T. Inhibition of the OAS/RNase L pathway by viruses. Curr. Opin. Virol. 2015, 15, 19–26. [Google Scholar] [CrossRef]
- Malathi, K.; Saito, T.; Crochet, N.; Barton, D.J.; Gale, M., Jr.; Silverman, R.H. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 2010, 16, 2108–2119. [Google Scholar] [CrossRef]
- Malathi, K.; Siddiqui, M.A.; Dayal, S.; Naji, M.; Ezelle, H.J.; Zeng, C.; Zhou, A.; Hassel, B.A. RNase L interacts with Filamin A to regulate actin dynamics and barrier function for viral entry. mBio 2014, 5, e02012. [Google Scholar] [CrossRef]
- Sorgeloos, F.; Jha, B.K.; Silverman, R.H.; Michiels, T. Evasion of antiviral innate immunity by Theiler’s virus L* protein through direct inhibition of RNase L. PLoS Pathog. 2013, 9, e1003474. [Google Scholar] [CrossRef]
- Shin, D.L.; Chludzinski, E.; Wu, N.H.; Peng, J.Y.; Ciurkiewicz, M.; Sawatsky, B.; Pfaller, C.K.; Baechlein, C.; von Messling, V.; Haas, L.; et al. Overcoming the Barrier of the Respiratory Epithelium during Canine Distemper Virus Infection. mBio 2022, 13, e0304321. [Google Scholar] [CrossRef]
- Carvalho, O.V.; Saraiva, G.L.; Ferreira, C.G.; Felix, D.M.; Fietto, J.L.; Bressan, G.C.; Almeida, M.R.; Silva Júnior, A. In-vitro antiviral efficacy of ribavirin and interferon-alpha against canine distemper virus. Can. J. Vet. Res. 2014, 78, 283–289. [Google Scholar]
- Röthlisberger, A.; Wiener, D.; Schweizer, M.; Peterhans, E.; Zurbriggen, A.; Plattet, P. Two domains of the V protein of virulent canine distemper virus selectively inhibit STAT1 and STAT2 nuclear import. J. Virol. 2010, 84, 6328–6343. [Google Scholar] [CrossRef] [PubMed]
- Palosaari, H.; Parisien, J.P.; Rodriguez, J.J.; Ulane, C.M.; Horvath, C.M. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003, 77, 7635–7644. [Google Scholar] [CrossRef] [PubMed]
- Caignard, G.; Guerbois, M.; Labernardière, J.L.; Jacob, Y.; Jones, L.M.; Wild, F.; Tangy, F.; Vidalain, P.O. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 2007, 368, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Svitek, N.; Gerhauser, I.; Goncalves, C.; Grabski, E.; Döring, M.; Kalinke, U.; Anderson, D.E.; Cattaneo, R.; von Messling, V. Morbillivirus control of the interferon response: Relevance of STAT2 and mda5 but not STAT1 for canine distemper virus virulence in ferrets. J. Virol. 2014, 88, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Goodbourn, S.; Randall, R.E. The regulation of type I interferon production by paramyxoviruses. J. Interferon Cytokine Res. 2009, 29, 539–547. [Google Scholar] [CrossRef]
- Yang, L.; Xu, L.; Li, Y.; Li, J.; Bi, Y.; Liu, W. Molecular and functional characterization of canine interferon-epsilon. J. Interf. Cytokine Res. 2013, 33, 760–768. [Google Scholar] [CrossRef]
- Shingai, M.; Ebihara, T.; Begum, N.A.; Kato, A.; Honma, T.; Matsumoto, K.; Saito, H.; Ogura, H.; Matsumoto, M.; Seya, T. Differential type I IFN-inducing abilities of wild-type versus vaccine strains of measles virus. J. Immunol. 2007, 179, 6123–6133. [Google Scholar] [CrossRef]
- Vandevelde, M.; Zurbriggen, A.; Higgins, R.J.; Palmer, D. Spread and distribution of viral antigen in nervous canine distemper. Acta Neuropathol. 1985, 67, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Vandevelde, M.; Kristensen, F.; Kristensen, B.; Steck, A.J.; Kihm, U. Immunological and pathological findings in demyelinating encephalitis associated with canine distemper virus infection. Acta Neuropathol. 1982, 56, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Raw, M.E.; Pearson, G.R.; Brown, P.J.; Baumgärtner, W. Canine distemper infection associated with acute nervous signs in dogs. Vet. Rec. 1992, 130, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Summers, B.A.; Greisen, H.A.; Appel, M.J. Early events in canine distemper demyelinating encephalomyelitis. Acta Neuropathol. 1979, 46, 1–10. [Google Scholar] [CrossRef]
- Summers, B.A.; Appel, M.J. Demyelination in canine distemper encephalomyelitis: An ultrastructural analysis. J. Neurocytol. 1987, 16, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Higgins, R.J.; Krakowka, S.G.; Metzler, A.E.; Koestner, A. Primary demyelination in experimental canine distemper virus induced encephalomyelitis in gnotobiotic dogs. Sequential immunologic and morphologic findings. Acta Neuropathol. 1982, 58, 1–8. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000research 2016, 5, 1438. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Puff, C.; Krudewig, C.; Imbschweiler, I.; Baumgärtner, W.; Alldinger, S. Influence of persistent canine distemper virus infection on expression of RECK, matrix-metalloproteinases and their inhibitors in a canine macrophage/monocytic tumour cell line (DH82). Vet. J. 2009, 182, 100–107. [Google Scholar] [CrossRef]
- Schwartz, M.; Puff, C.; Stein, V.M.; Baumgärtner, W.; Tipold, A. Pathogenetic factors for excessive IgA production: Th2-dominated immune response in canine steroid-responsive meningitis-arteritis. Vet. J. 2011, 187, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Gröne, A.; Frisk, A.L.; Baumgärtner, W. Cytokine mRNA expression in whole blood samples from dogs with natural canine distemper virus infection. Vet. Immunol. Immunopathol. 1998, 65, 11–27. [Google Scholar] [PubMed]
- Von Smolinski, D.; Leverkoehne, I.; von Samson-Himmelstjerna, G.; Gruber, A.D. Impact of formalin-fixation and paraffin-embedding on the ratio between mRNA copy numbers of differently expressed genes. Histochem. Cell Biol. 2005, 124, 177–188. [Google Scholar] [CrossRef]
- Spitzbarth, I.; Bock, P.; Haist, V.; Stein, V.M.; Tipold, A.; Wewetzer, K.; Baumgärtner, W.; Beineke, A. Prominent microglial activation in the early proinflammatory immune response in naturally occurring canine spinal cord injury. J. Neuropathol. Exp. Neurol. 2011, 70, 703–714. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]

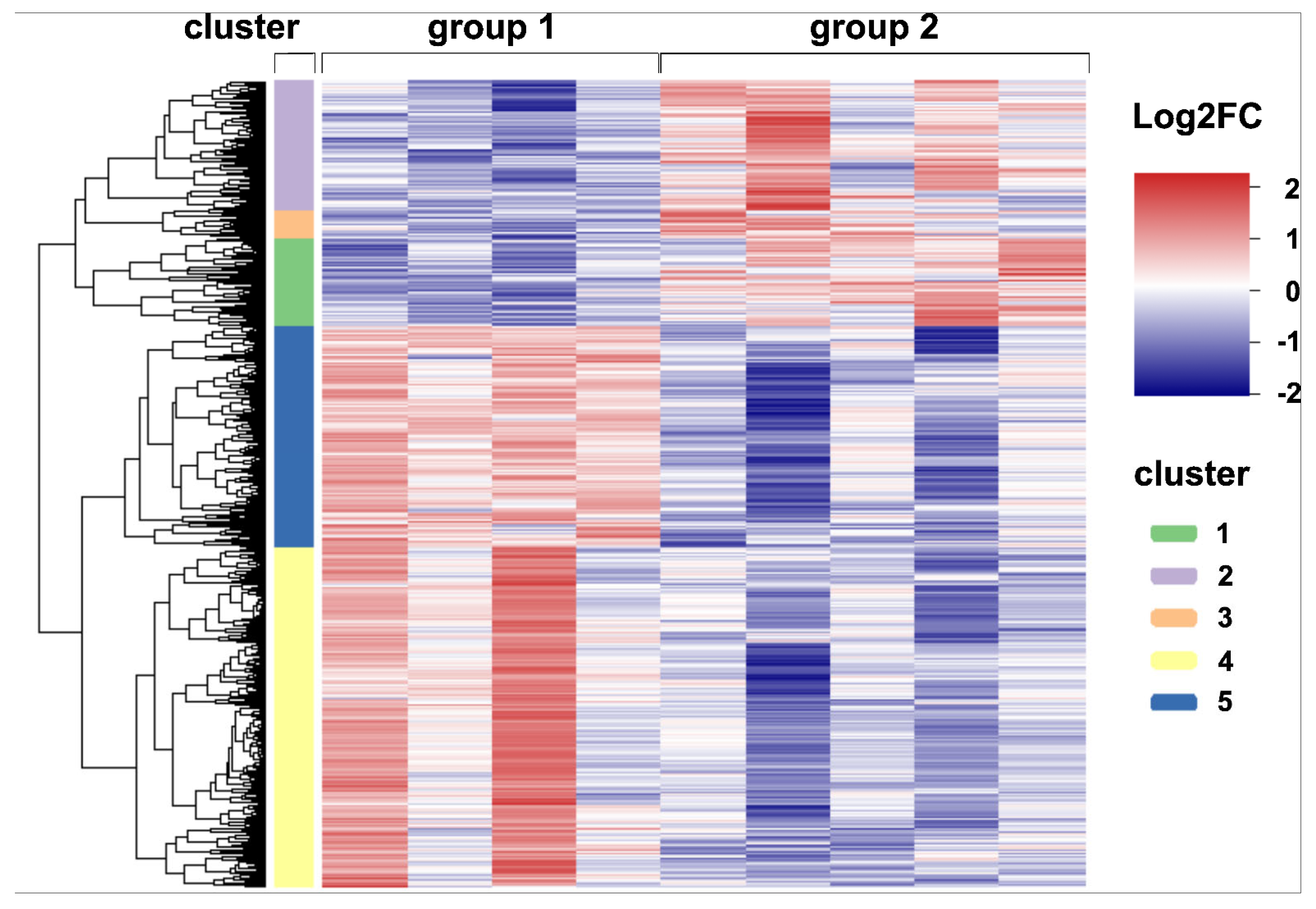
| Gene Set | GO Terms (Biological Function) | Adjusted p-Value |
|---|---|---|
| DEGs in cluster 1 (n = 353) | defense response to virus | 0.000 |
| negative regulation of viral process | 0.001 | |
| negative regulation of viral genome replication | 0.000 | |
| positive regulation of innate immune response | 0.002 | |
| leukocyte activation involved in immune response | 0.028 | |
| response to type I interferon | 0.013 | |
| DEGs in cluster 2 (n = 515) | mononuclear cell proliferation | 0.000 |
| regulation of leukocyte activation | 0.000 | |
| T-helper 1 type immune response | 0.001 | |
| positive regulation of cytokine production | 0.000 | |
| negative regulation of cytokine production | 0.031 | |
| cellular response to cytokine stimulus | 0.000 | |
| cytokine-mediated signaling pathway | 0.000 | |
| tumor necrosis factor superfamily cytokine production | 0.010 | |
| positive regulation of apoptotic signaling pathway | 0.035 | |
| regulation of apoptotic process | 0.000 | |
| receptor signaling pathway via STAT | 0.000 | |
| DEGs in cluster 3 (n = 118) | no enriched GO terms | |
| DEGs in cluster 4 (n = 1373) | cilium assembly | 0.000 |
| cilium movement | 0.000 | |
| DEGs in cluster 5 (n = 893) | epithelial cell proliferation | 0.040 |
| Epitope | Specificity | Source | Cat No | Species | Clone | Dilution |
|---|---|---|---|---|---|---|
| CDV-NP | CDV-infected cells | A. Zurbriggen, University of Bern, Bern, Switzerland | - | mouse | D110 | 1:1000 * |
| CDV-NP | CDV-infected cells | Santa Cruz Biotechnology, Dallas, TX, USA | sc-57660 | mouse | DV2-12 | 1:100 ** |
| MHC-II | antigen-presenting cells | Dako, Glostrup, Denmark | M0746 | mouse | TAL.1B5 | 1:80 * |
| Iba-1 | histiocytic cells | Invitrogen™, Thermo Fisher Scientific, Langenselbold, Germany | PA5-27436 | rabbit | polyclonal | 1:500 *; 1:200 ** |
| CD204 | histiocytic cells | Abnova Corporation, Taipei, Taiwan | MAB1710 | mouse | SRA-E5 | 1:500 * |
| CD3 | T lymphocytes | Dako, Glostrup, Denmark | A0452 | rabbit | polyclonal | 1:500 * |
| CD20 | B lymphocytes | Lab Vision, Thermo Fisher Scientific, Langenselbold, Germany | RB9013P | rabbit | polyclonal | 1:300 * |
| CK | epithelial cells | Dako, Glostrup, Denmark | Z0622 | rabbit | polyclonal | 1:50 ** |
| TNF-α | TNF-α | Novus Biologicals, Centennial, CO, USA | NBP2-34303 | mouse | TNF706 | 1:50 *; 1:20 ** |
| CC-3 | apoptosis | Cell Signaling Technology, Danvers, MA, USA | 9664 | rabbit | 5A1E | 1:200 * |
| Mx | antiviral immune response | O. Haller and G. Kochs, University Medical Center Freiburg, Freiburg, Germany | - | mouse | M143 | 1:800 * |
| PKR | antiviral immune response | Abcam, Cambridge, MA, USA | ab32036 | rabbit | E120 | 1:600 * |
| ISG15 | antiviral immune response | Boster Biological Technology, Pleasanton, CA, USA | PB9951 | rabbit | polyclonal | 1:750 * |
| Gene | Primer Direction | Primer Sequence (5′–3′) | Position | Amplicon Length (bp) | Acc No. | Reference |
|---|---|---|---|---|---|---|
| GAPDH | forward | AAG GTC GGA GTC AAC GGA TT | 7–26 | 365 | AB038240 | Puff et al., 2008 [176] |
| reverse | GCA GAA GGA GCA GAG ATG ATG | 371–351 | ||||
| CDV | forward | ACA GGA TTG CTG AGG ACC TAT | 769–789 | 287 | AF378705 | Markus et al., 2002 [41] |
| reverse | CAA GAT AAC CAT GTA CGG TGC | 1055–1035 | ||||
| IL-1-β | forward | TCC AAT GTG AAG TGC TGC TG | 14–33 | 262 | Z70047 | Primer-BLAST [181] |
| reverse | GCA TGG CTG CAT CAC TCA TA | 275–256 | ||||
| IL-2 | forward | ACC TCA ACT CCT GCC ACA AT | 14–33 | 289 | D30710 | Schwartz et al., 2011 [177] |
| reverse | GCA CTT CCT CCA GGT TTT TG | 302–283 | ||||
| IL-6 | forward | TCT CCA CAA GCG CCT TCT CC | 68–87 | 318 | U12234 | Gröne et al., 1998 [178] |
| reverse | TTC TTG TCA AGC AGG TCT CC | 385–366 | ||||
| IL-8 | forward | ACT TCC AAG CTG GCT GTT GC | 39–58 | 172 | D28772 | Gröne et al., 1998 [178] |
| reverse | GGC CAC TGT CAA TCA CTC TC | 210–191 | ||||
| TNF-α | forward | CCA AGT GAC AAG CCA GTA GC | 32–51 | 274 | Z70046 | Gröne et al., 1998 [178] |
| reverse | TCT TGA TGG CAG AGA GTA GG | 305–287 |
| Gene | Plasmid Sequence (5’–3’) | Position | Gene Size (bp) | Acc No |
|---|---|---|---|---|
| IL-4 | ACT GAT TCC AAC TCT GGT CTG CTT ACT AGC ACT CAC CAG CAC CTT TGT CCA CGG ACA TAA CTT CAA TAT TAC TAT TAA AGA GAT CAT CAA AAT GTT GAA CAT CCT CAC AGC GAG AAA CGA CTC GTG CAT GGA GCT GAC TGT CAA GGA CGT CTT CAC TGC TCC AAA GAA CAC AAG CGA TAA GGA AAT CTT CTG CAG AGC TGC TAC TGT ACT GCG GCA GAT CTA TAC ACA CAA CTG CTC CAA CAG ATA TCT CAG AGG ACT CTA CAG GAA CCT CAG CAG CAT GGC AAA CAA GA | 81–370 | 290 | AF239917 |
| IL-10 | ATG CCC CGG GCT GAG AAC CAC GAC CCA GAC ATC AAG AAC CAC GTG AAC TCC CTG GGA GAG AAG CTC AAG ACC CTC AGG CTG AGA CTG AGG CTG CGA CGC TGT CAC CGA TTT CTT CCC TGT GAG AAT AAG AGC AAG GCG GTG GAG CAG GTG AAG AGC GCA TTT AG | 287–450 | 164 | U33843 |
| IL-12 | ATG CAT CCT CAG CAG TTG GT C ATC TCC TGG TTT TCC CTC GTT TTG CTG GCG TCT TCC CTC ATG ACC ATA TGG GAA CTG GAG AAA GAT GTT TAT GTT GTA GAG TTG GAC TGG CAC CCT GAT GCC CCC GGA GAA ATG GTG GTC CTC ACC TGC CAT ACC CCT GAA GAA GAT GAC ATC ACT TGG ACC TCA GCG CAG AGC AGT GAA GTC CTA GGT TCT GGT AAA ACT CTG ACC ATC CAA GTC AAA GAA TTT GGA GAT GCT GGC CAG TAT ACC TGC CAT AAA GGA | 1–279 | 279 | U49100 |
| TGF-β | GGA GCT GTA CCA GAA ATA TAG CAA TGA TTC CTG GCG CTA CCT CAG CAA CCG GCT GCT GGC GCC CAG CGA CAC GCC AGA ATG GCT GTC CTT TGA TGT CAC TGG AGT CGT GAG GCA GTG GCT GAG CCA TGG AGG GGA AGT CGA GGG CTT TCG CCT CAG TGC CCA CTG TTC CTG TGA CAG CAA AGA TAA CAC A | 561–750 | 190 | L34956 |
| IFN-γ | CCA GAT GTA TCG GAC GGT GGG TCT CTT TTC GTA GAT ATT TTG AAG AAA TGG AGA GAG GAG AGT GAC AAA ACA ATC ATT CAG AGC CAA ATT GTC TCT TTC TAC TTG AAA CTG TTT GAC AAC TTT AAA GAT AAC CAG ATC ATT CAA AGG AGC ATG GAT ACC ATC AAG GAA GAC ATG CTT GGC AAG TTC TTA AAT AGC AGC ACC AGT AAG AGG GAG GAC TTC CTT AAG CTG ATT CAA ATT CCT GTG AAC GAT CTG CAG GTC CAG CGC AAG GCG ATA A | 76–349 | 274 | S41201 |
| Gene | Primer Direction | Primer Sequence (5′–3′) | Position | Amplicon Length (bp) | Acc No | Reference |
|---|---|---|---|---|---|---|
| GAPDH | forward | GTC ATC AAC GGG AAG TCC ATC TC | 196–218 | 84 | AB038240 | von Smolinski et al., 2005 [179] |
| reverse | AAC ATA CTC AGC ACC AGC ATC AC | 279–257 | ||||
| CDV | forward | GCT CTT GGG TTG CAT GAG TT | 954–973 | 83 | AF378705 | Puff et al., 2008 [176] |
| reverse | GCT GTT TCA CCC ATC TGT TG | 1036–1017 | ||||
| IL-1-β | forward | TGT CAG TCA TTG TAG CTT TG | 123–142 | 113 | Z70047 | Primer-BLAST [181] |
| reverse | GCA GAT GAT AGG TTC TTC TT | 235–216 | ||||
| IL-2 | forward | CCA ACT CTC CAG GAT GCT CAC | 196–216 | 81 | D30710 | Schwartz et al., 2011 [177] |
| reverse | TCT GCT AGA CAT TGA AGG TGT GTA | 276–252 | ||||
| IL-4 | forward | CTC CAA AGA ACA CAA GCG ATA AGG | 239–262 | 84 | AF239917 | Schwartz et al., 2011 [177] |
| reverse | TGT TGG AGC AGT TGT GTG TAT AGA | 322–299 | ||||
| IL-6 | forward | TGA TGC CAC TTC AAA TAG TCT ACC A | 156–180 | 89 | U12234 | Spitzbarth et al., 2011 [180] |
| reverse | TCA GTG CAG AGA TTT TGC CGA GGA | 244–221 | ||||
| IL-8 | forward | TTC GAT GCC AGT GTA TAA AA | 130–149 | 74 | D28772 | Primer-BLAST [181] |
| reverse | GTC AAT CAC TCT CAG TTC TT | 203–184 | ||||
| IL-10 | forward | ACC ACG ACC CAG ACA TCA AGA A | 303–324 | 120 | U33843 | Primer-BLAST [181] |
| reverse | CCT TGC TCT TAT TCT CAC AGG GAA G | 422–398 | ||||
| IL-12 | forward | CTC GTT TTG CTG GCG TCT TC | 37–56 | 153 | U49100 | Primer-BLAST [181] |
| reverse | CGC TGA GGT CCA AGT GAT GT | 189–170 | ||||
| TNF-α | forward | GGA GCT GAC AGA CAA CCA GCT GA | 133–155 | 91 | Z70046 | Spitzbarth et al., 2011 [180] |
| reverse | GGA AGG GCA CCC TTG GCC CT | 223–204 | ||||
| TGF-β | forward | TGG CGC TAC CTC AGC AAC CG | 592–611 | 115 | L34956 | Spitzbarth et al., 2011 [180] |
| reverse | AGC CCT CGA CTT CCC CTC CA | 706–687 | ||||
| IFN-γ | forward | AGC ATG GAT ACC ATC AAG GAA GA | 223–245 | 104 | S41201 | Schwartz et al., 2011 [177] |
| reverse | AGA TCG TTC ACA GGA ATT TGA ATC A | 326–302 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chludzinski, E.; Klemens, J.; Ciurkiewicz, M.; Geffers, R.; Pöpperl, P.; Stoff, M.; Shin, D.-L.; Herrler, G.; Beineke, A. Phenotypic and Transcriptional Changes of Pulmonary Immune Responses in Dogs Following Canine Distemper Virus Infection. Int. J. Mol. Sci. 2022, 23, 10019. https://doi.org/10.3390/ijms231710019
Chludzinski E, Klemens J, Ciurkiewicz M, Geffers R, Pöpperl P, Stoff M, Shin D-L, Herrler G, Beineke A. Phenotypic and Transcriptional Changes of Pulmonary Immune Responses in Dogs Following Canine Distemper Virus Infection. International Journal of Molecular Sciences. 2022; 23(17):10019. https://doi.org/10.3390/ijms231710019
Chicago/Turabian StyleChludzinski, Elisa, Johanna Klemens, Małgorzata Ciurkiewicz, Robert Geffers, Pauline Pöpperl, Melanie Stoff, Dai-Lun Shin, Georg Herrler, and Andreas Beineke. 2022. "Phenotypic and Transcriptional Changes of Pulmonary Immune Responses in Dogs Following Canine Distemper Virus Infection" International Journal of Molecular Sciences 23, no. 17: 10019. https://doi.org/10.3390/ijms231710019
APA StyleChludzinski, E., Klemens, J., Ciurkiewicz, M., Geffers, R., Pöpperl, P., Stoff, M., Shin, D.-L., Herrler, G., & Beineke, A. (2022). Phenotypic and Transcriptional Changes of Pulmonary Immune Responses in Dogs Following Canine Distemper Virus Infection. International Journal of Molecular Sciences, 23(17), 10019. https://doi.org/10.3390/ijms231710019





