Genome-Wide Identification and Expression Patterns of the SWEET Gene Family in Bletilla striata and its Responses to Low Temperature and Oxidative Stress
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of BsSWEET Genes
2.2. Multiple Sequence Alignment and Phylogenetic Relationship of BsSWEET Genes
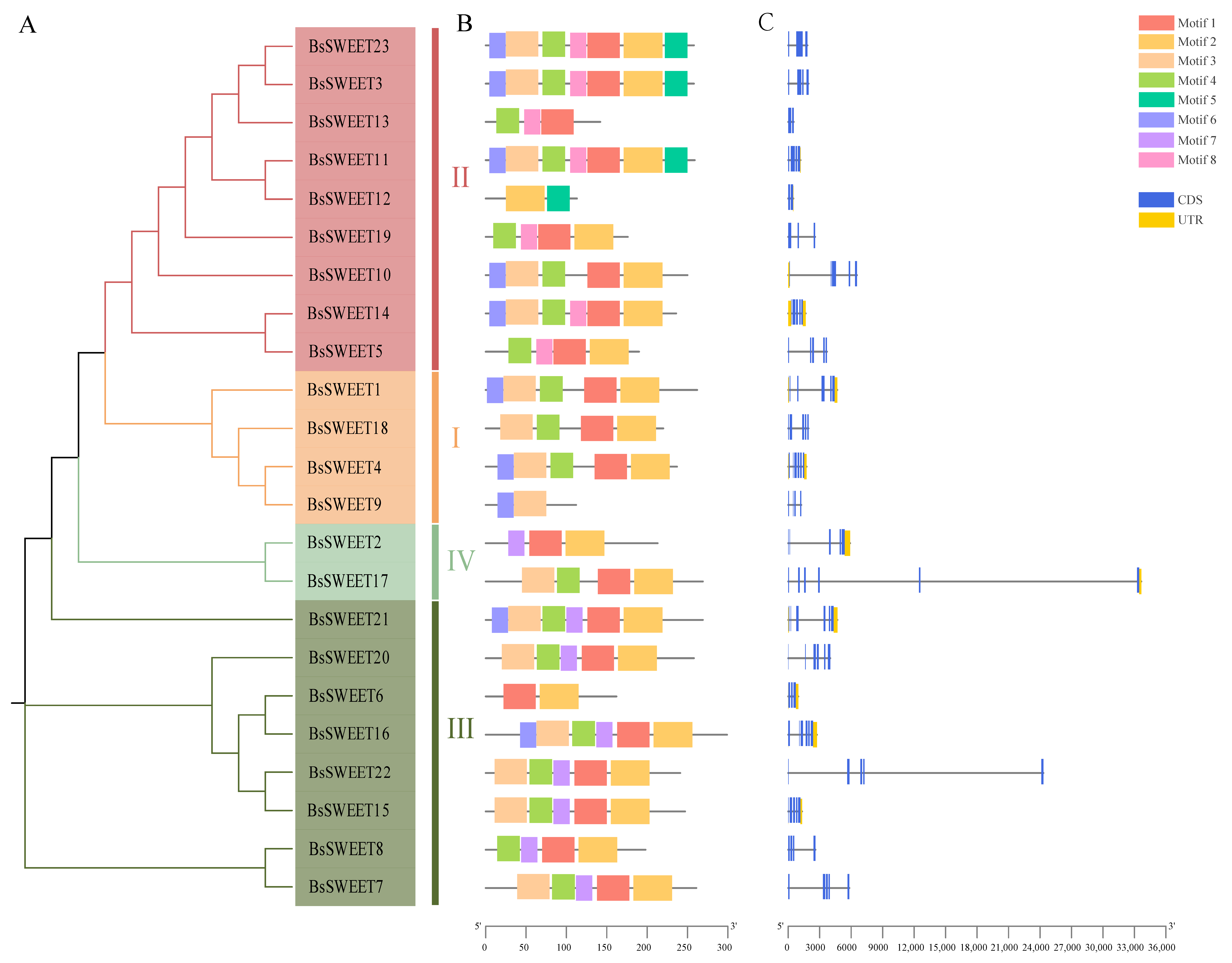
2.3. Gene Structures and Conserved Motifs of BsSWEET Genes
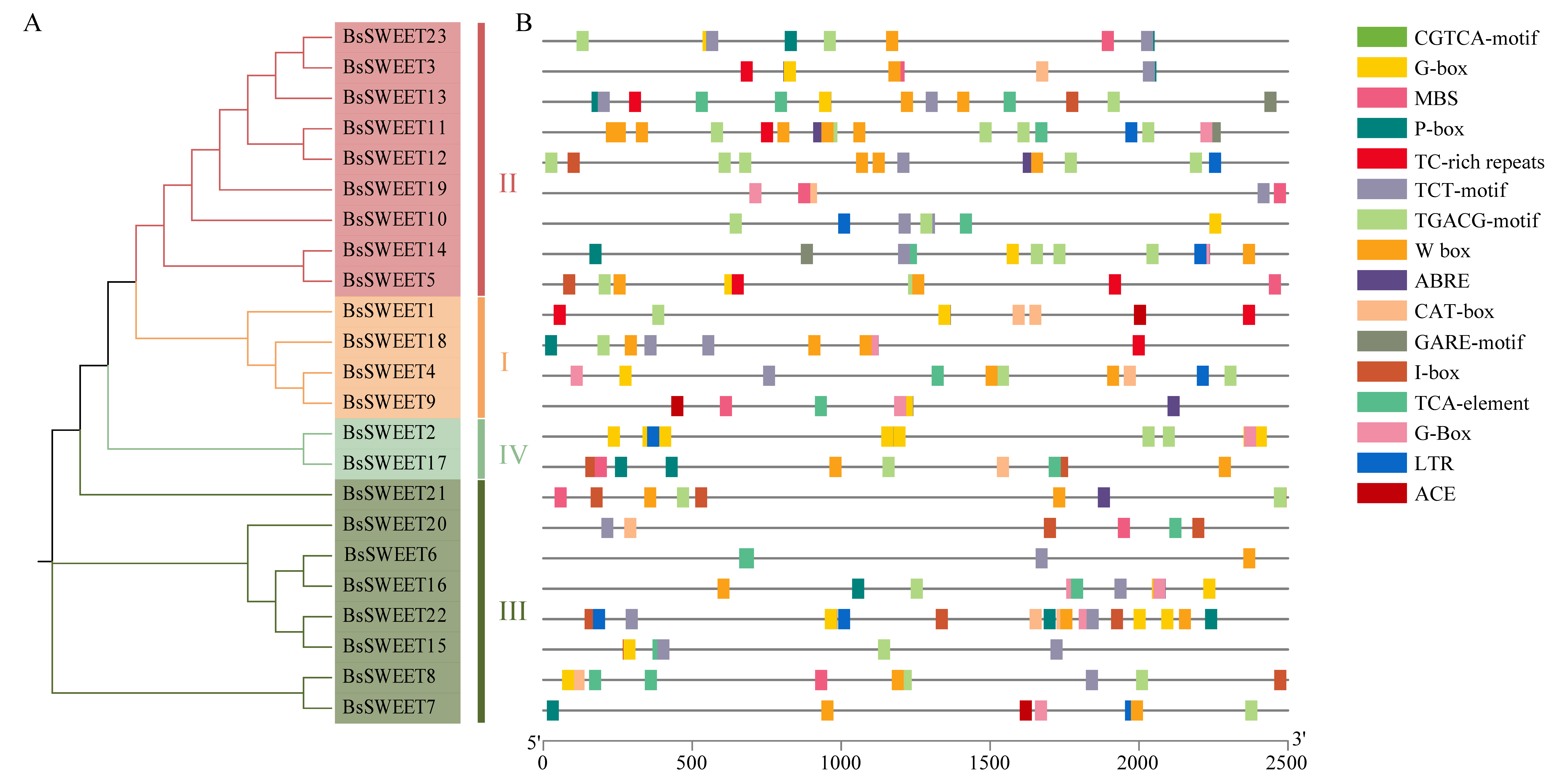
2.4. Cis-Element Analysis in Promoters of BsSWEET Genes
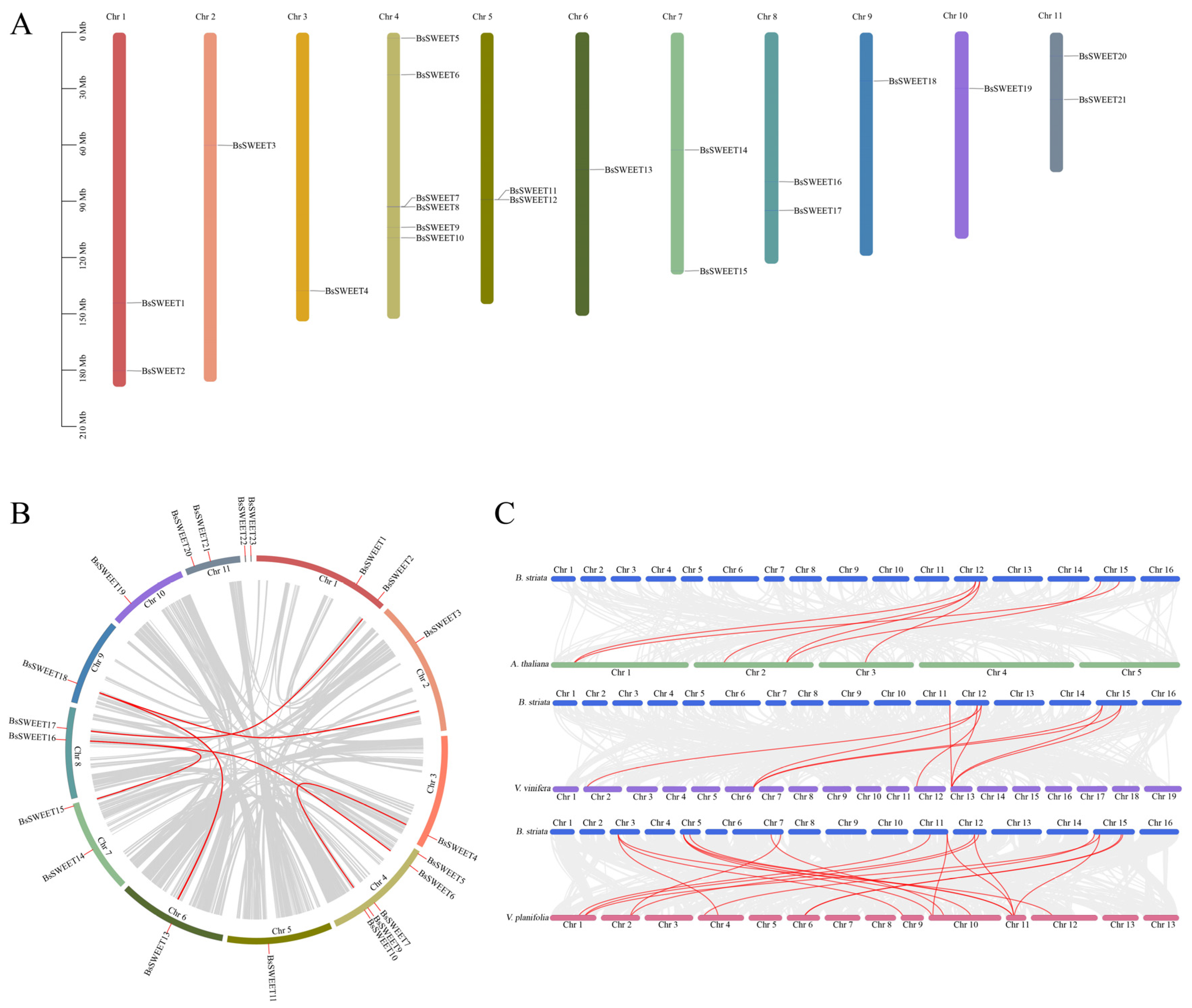
2.5. Synteny and ω Analysis of BsSWEET Genes
2.6. Tissue-Specific Expression Patterns of BsSWEETs
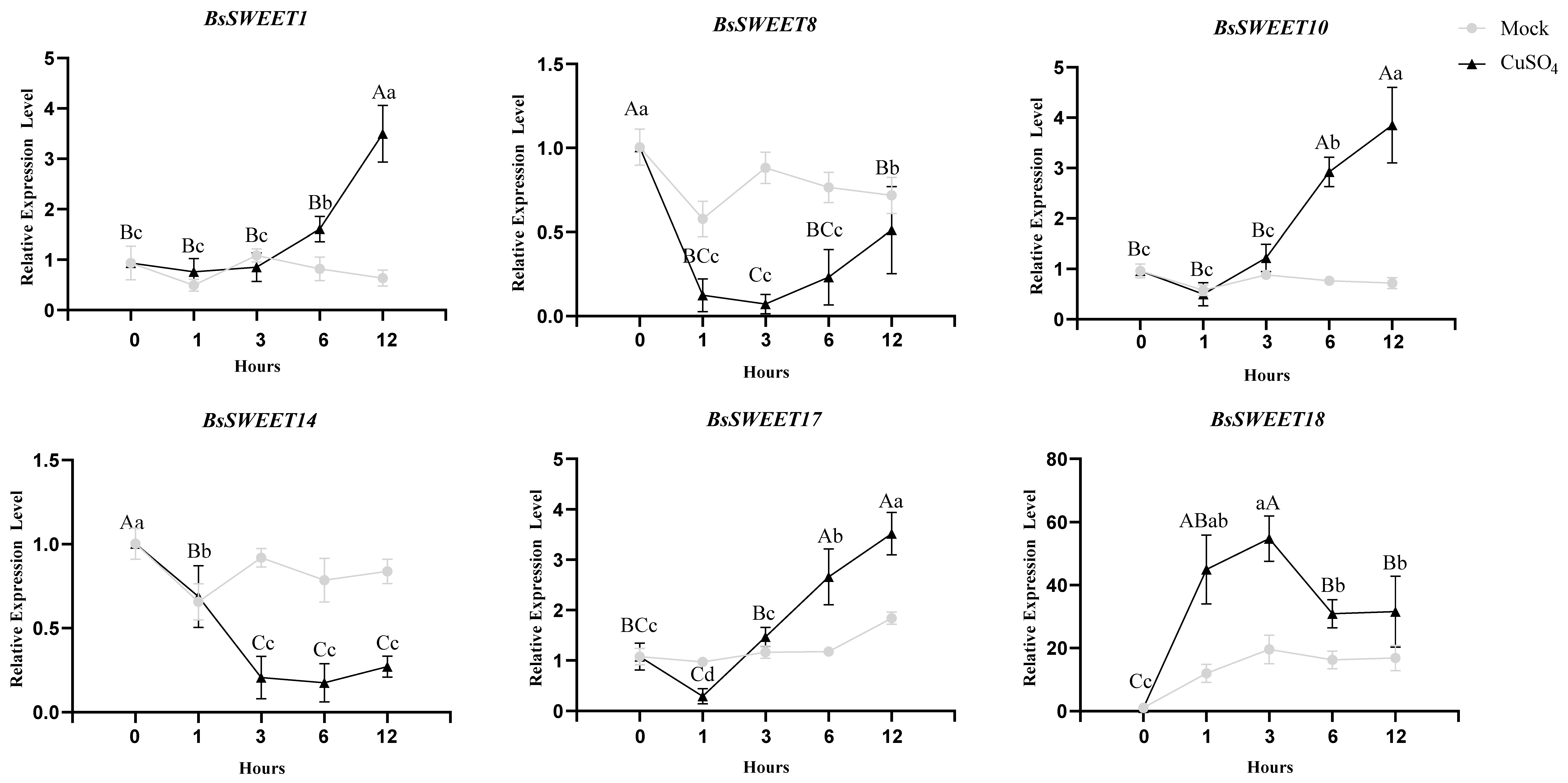
2.7. Expression Patterns of BsSWEETs in Responses to Low Temperature and Oxidative Stress
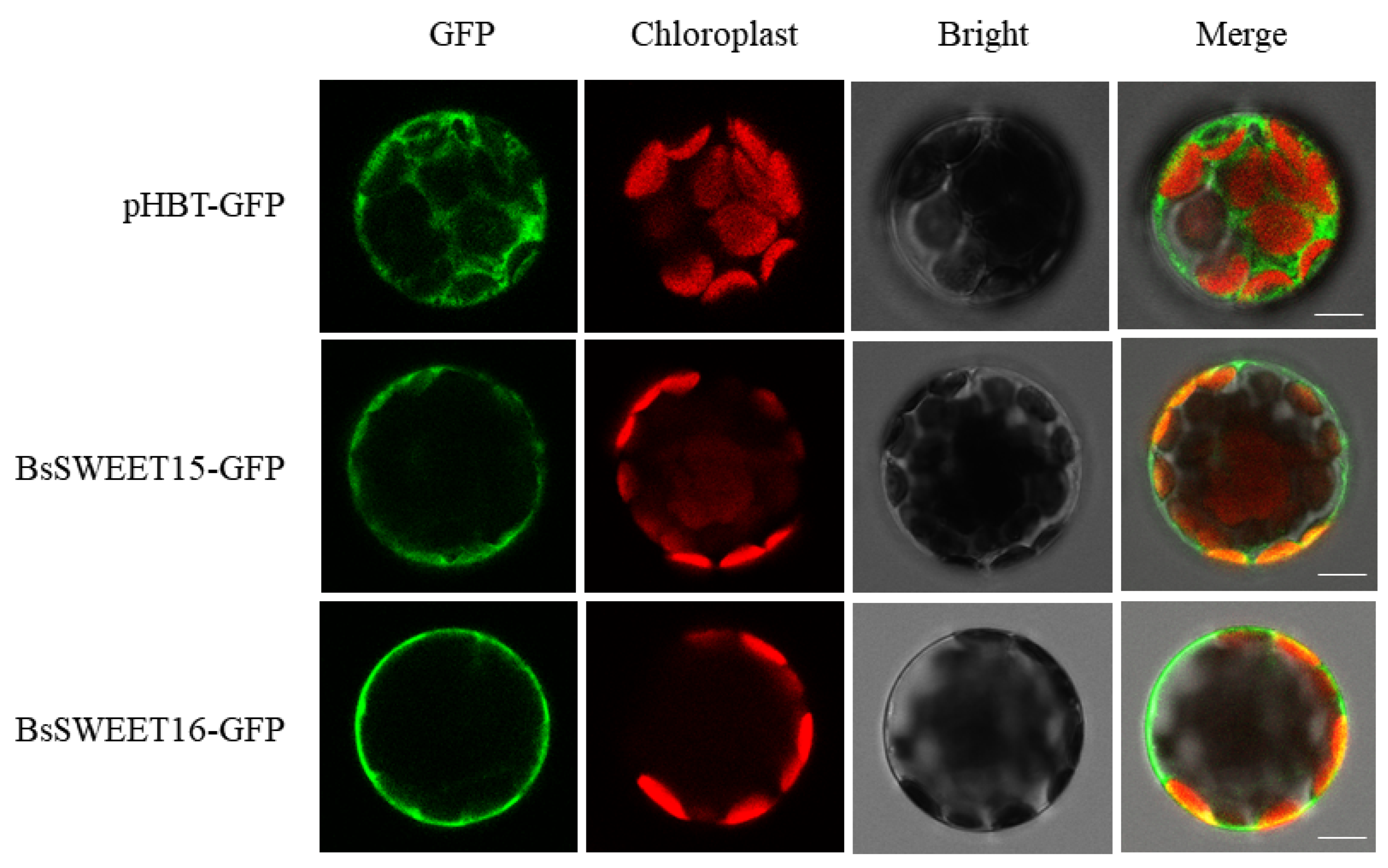
2.8. The Subcellular Localization of BsSWEET15 and BsSWEET16
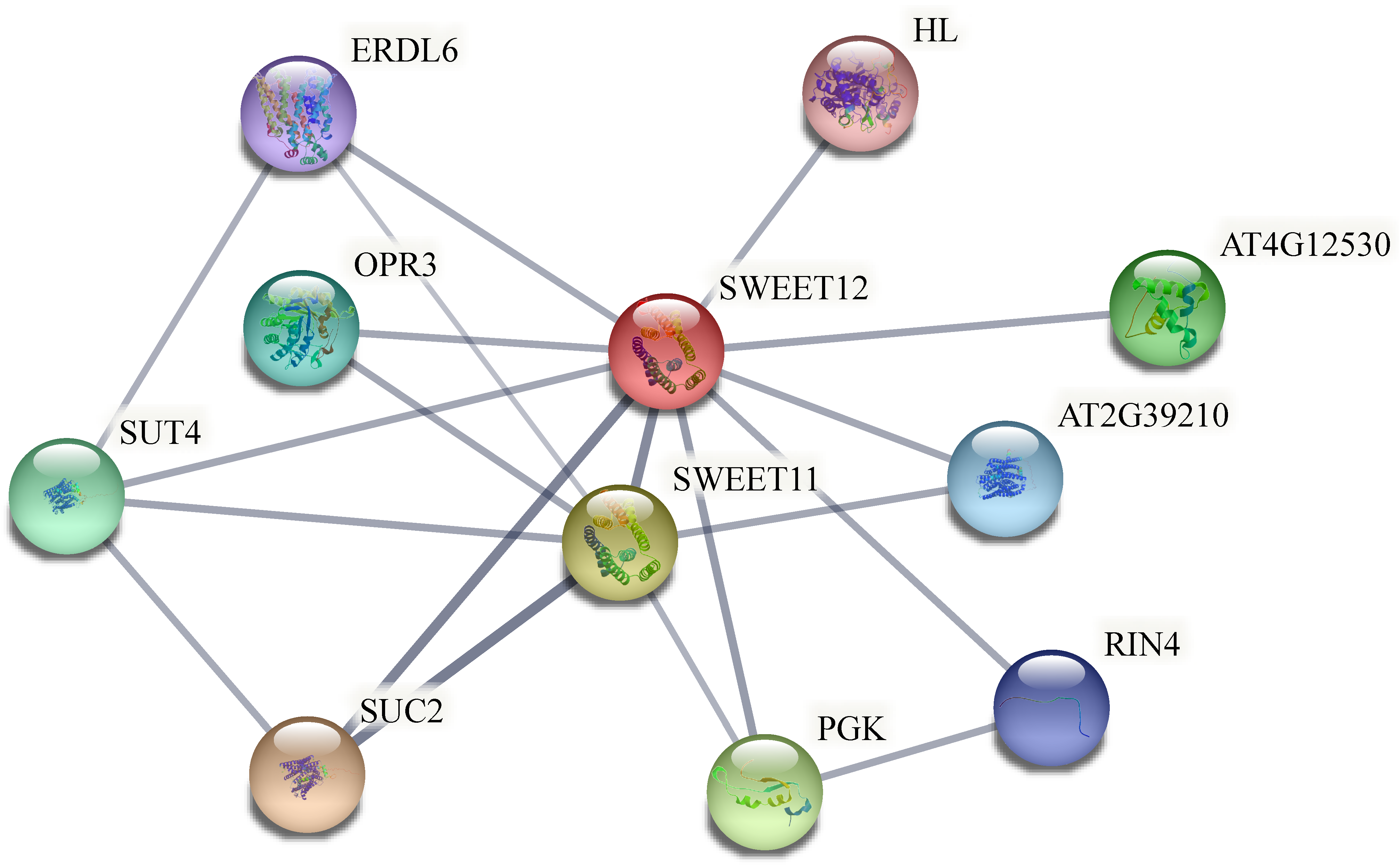
2.9. Protein Interaction Analysis of BsSWEETs
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification, Protein Features, and Chromosomal Locations of BsSWEET Genes
4.2. Phylogenetic Analysis of SWEET Genes
4.3. Analysis of Conserved Motifs, Gene Structures, and Cis-Acting Elements of the BsSWEET Genes
4.4. Analysis of Gene Duplication and Synteny in B. striata
4.5. Plant Materials and Treatments
4.6. RNA Extraction and q-PCR Analysis
4.7. Expression Patterns of BsSWEET Genes
4.8. The Subcellular Localization of BsSWEET15 and BsSWEET16
4.9. Interactive Network Analysis of BsSWEET Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lalonde, S.; Wipf, D.; Frommer, W.B. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 2004, 55, 341–372. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef] [PubMed]
- Ruhland, W.; Wolf, J. Metabolism of Carbohydrates and Organic Acids in Plants (Exclusive of Bacteria and Fungi). Annu. Rev. Biochem. 1936, 5, 485–512. [Google Scholar] [CrossRef]
- Ayre, B.G. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 2011, 43, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S. Sugar-Induced Signal Transduction in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 49–81. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Mccourt, P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 2001, 4, 387–391. [Google Scholar] [CrossRef]
- Neuhaus, H.E. Transport of primary metabolites across the plant vacuolar membrane. FEBS Lett. 2007, 58112, 2223–2226. [Google Scholar] [CrossRef]
- Kuhn, C.; Grof, C.P. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 2010, 133, 288–298. [Google Scholar] [CrossRef]
- Roitsch, T. Source-sink regulation by sugar and stress. Curr. Opin. Plant Biol. 1999, 2, 198–206. [Google Scholar] [CrossRef]
- Riesmeier, J.W.; Willmitzer, L.; Frommer, W.B. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992, 11, 4705–4713. [Google Scholar] [CrossRef]
- Lemoine, R. Sucrose transporters in plants: Update on function and structure. Biochim. Biophys. Acta 2000, 1465, 246–262. [Google Scholar] [CrossRef]
- Ruiz-Medrano, R.; Xoconostle-Cázares, B.; Lucas, W.J. The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol. 2001, 4, 202–209. [Google Scholar] [CrossRef]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Bush, D.R. Carbohydrate export from the leaf: A highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol. 2011, 1551, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.F.; Leach, K.A.; Braun, D.M. SWEET as sugar: New sucrose effluxers in plants. Mol. Plant 2012, 54, 766–768. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol. Plant 2013, 63, 665–674. [Google Scholar] [CrossRef]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET Family of Sugar Transporters: Structure, Evolution and Biological Functions. Biomolecules 2022, 122, 205. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, 3685–3694. [Google Scholar] [CrossRef]
- Braun, D.M. Phloem Loading and Unloading of Sucrose: What a Long, Strange Trip from Source to Sink. Annu. Rev. Plant Biol. 2022, 73, 553–584. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. Ruptured Pollen Grain1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008, 1472, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.X.; Huang, X.Y.; Yang, J.; Guan, Y.F.; Yang, Z.N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013, 262, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed]
- Antony, G.; Zhou, J.; Huang, S.; Li, T.; Liu, B.; White, F.; Yang, B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N. Plant Cell 2010, 2211, 3864–3876. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.; Sonnewald, U.; Bachem, C. Source-Sink Regulation Is Mediated by Interaction of an FT Homolog with a SWEET Protein in Potato. Curr. Biol. 2019, 29, 1178–1186. [Google Scholar] [CrossRef]
- Bown, D. The Royal Horticultural Society Encyclopedia of Herbs & Their Uses; Dorling Kindersley Limited: New York, NY, USA, 1995. [Google Scholar]
- Wiart, C. Medicinal Plants of China, Korea, and Japan: Bioresources for Tomorrow’s Drugs and Cosmetics; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Tan, K.W. The systematic status of the genus Bletilla (Orchidaceae). Brittonia 1969, 21, 202–214. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, M.; Jiang, L.; Xie, Q.; Yuan, H.; Yang, Y.; Zafar, S.; Liu, Y.; Jian, Y.; Li, B.; et al. The medicinal uses of the genus Bletilla in traditional Chinese medicine: A phytochemical and pharmacological review. J. Ethnopharmacol. 2021, 280, 114263. [Google Scholar] [CrossRef]
- Dietz, K.J.; Baier, M.; Krämer, U. Free Radicals and Reactive Oxygen Species as Mediators of Heavy Metal Toxicity in Plants; Springer: Berlin/Heidelberg, Germany, 1999. [Google Scholar]
- Liu, S.; Lu, C.; Jiang, G.; Zhou, R.; Chang, Y.; Wang, S.; Wang, D.; Niu, J.; Wang, Z. Comprehensive functional analysis of the PYL-PP2C-SnRK2s family in Bletilla striata reveals that BsPP2C22 and BsPP2C38 interact with BsPYLs and BsSnRK2s in response to multiple abiotic stresses. Front. Plant Sci. 2022, 13, 963069. [Google Scholar] [CrossRef]
- Cui, B.; Cheng, S.; Yuan, X.; Zhou, Y.; Hao, P.; Junlin, L.I.; Jie, M.A. Effects of Low Temperature Stress on the Photosynthetic Characteristics and Chlorophyll Fluorescence Parameters of Bletilla striata. Chin. J. Trop. Crops 2019, 40, 891–897. [Google Scholar] [CrossRef]
- Li, R.; Wang, W. Research survey and countermeasure on resources utilization in stem tuber of Bletilla striata. Chin. Tradit. Herb. Drugs 2006, 37, 1751–1755. [Google Scholar]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 2015, 677, 461–471. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dedaldechamp, F.; Perez-Garcia, M.D.; Ogé, L.; Hamama, L.; Atanassova, R. The Sugar-Signaling Hub: Overview of Regulators and Interaction with the Hormonal and Metabolic Network. Int. J. Mol. Sci. 2018, 199, 2506. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xu, X.; Xu, D.; Zhang, H.; Zhang, C.; Li, G. WRKY18 and WRKY53 Coordinate with HISTONE ACETYLTRANSFERASE1 to Regulate Rapid Responses to Sugar. Plant Physiol. 2019, 1804, 2212–2226. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.S.; Karrer, E.E.; Thomas, B.R.; Chen, L.; Rodriguez, R.L. Three cis-elements required for rice α-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 1998, 363, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, T.; Lu, J.; Zhou, Y.; Liu, S.; Jiao, P.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Characterization and Functional Analysis of ZmSWEET15a in Maize. DNA Cell Biol. 2022, 416, 564–574. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 5732, 261–272. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Jiang, S.; Balan, B.; Assis, R.; Sagawa, C.; Wan, X.; Han, S.; Wang, L.; Zhang, L.; Zaini, P.A.; Walawage, S.L.; et al. Genome-Wide Profiling and Phylogenetic Analysis of the SWEET Sugar Transporter Gene Family in Walnut and Their Lack of Responsiveness to Xanthomonas arboricola pv. juglandis Infection. Int. J. Mol. Sci. 2020, 21, 1251. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Lan, G.; Si, F.; Zeng, Z.; Wang, C.; Yadav, V.; Wei, C.; Zhang, X. Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon. Int. J. Mol. Sci. 2021, 22, 8407. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 94, e94210. [Google Scholar] [CrossRef]
- Klemens, P.A.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 1633, 1338–1352. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, G.Q.; Zhang, D.; Liu, X.D.; Xu, X.Y.; Sun, W.H.; Yu, X.; Zhu, X.; Wang, Z.W.; Zhao, X.; et al. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 2021, 81, 183. [Google Scholar] [CrossRef]
- Geng, Y.; Wu, M.; Zhang, C. Sugar Transporter ZjSWEET2.2 Mediates Sugar Loading in Leaves of Ziziphus jujuba Mill. Front. Plant Sci. 2020, 11, 1081. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, C.; Wang, M.; Li, T.; Liu, X.; Jiang, J. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic. Res. 2021, 81, 186. [Google Scholar] [CrossRef]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Ren, Z.; Wang, Z.; Sun, K.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Song, C.; Zhou, X.; et al. Evolution and Stress Responses of Gossypium hirsutum SWEET Genes. Int. J. Mol. Sci. 2018, 19, 769. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status. Nucleic Acids Res. 2015, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 309, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 374, 420–423. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 3053, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R. Clustal W and Clustal X version 2. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 46, 1633–1649. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2. Bioinformatics 2007, 2321, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 337, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 138, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 407, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 199, 1639–1645. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 2000, 155, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 27, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Ye, J.; Chang, Y.; Mi, Z.; Liu, S.; Wang, D.; Wang, Z.; Niu, J. Genome-Wide Identification and Expression Patterns of the SWEET Gene Family in Bletilla striata and its Responses to Low Temperature and Oxidative Stress. Int. J. Mol. Sci. 2022, 23, 10057. https://doi.org/10.3390/ijms231710057
Lu C, Ye J, Chang Y, Mi Z, Liu S, Wang D, Wang Z, Niu J. Genome-Wide Identification and Expression Patterns of the SWEET Gene Family in Bletilla striata and its Responses to Low Temperature and Oxidative Stress. International Journal of Molecular Sciences. 2022; 23(17):10057. https://doi.org/10.3390/ijms231710057
Chicago/Turabian StyleLu, Chan, Jun Ye, Yuanqing Chang, Zeyuan Mi, Shuai Liu, Donghao Wang, Zhezhi Wang, and Junfeng Niu. 2022. "Genome-Wide Identification and Expression Patterns of the SWEET Gene Family in Bletilla striata and its Responses to Low Temperature and Oxidative Stress" International Journal of Molecular Sciences 23, no. 17: 10057. https://doi.org/10.3390/ijms231710057
APA StyleLu, C., Ye, J., Chang, Y., Mi, Z., Liu, S., Wang, D., Wang, Z., & Niu, J. (2022). Genome-Wide Identification and Expression Patterns of the SWEET Gene Family in Bletilla striata and its Responses to Low Temperature and Oxidative Stress. International Journal of Molecular Sciences, 23(17), 10057. https://doi.org/10.3390/ijms231710057





