Genetic Variation among Pharmacogenes in the Sardinian Population
Abstract
:1. Introduction
2. Results
2.1. Haplotype and Phenotype Calling
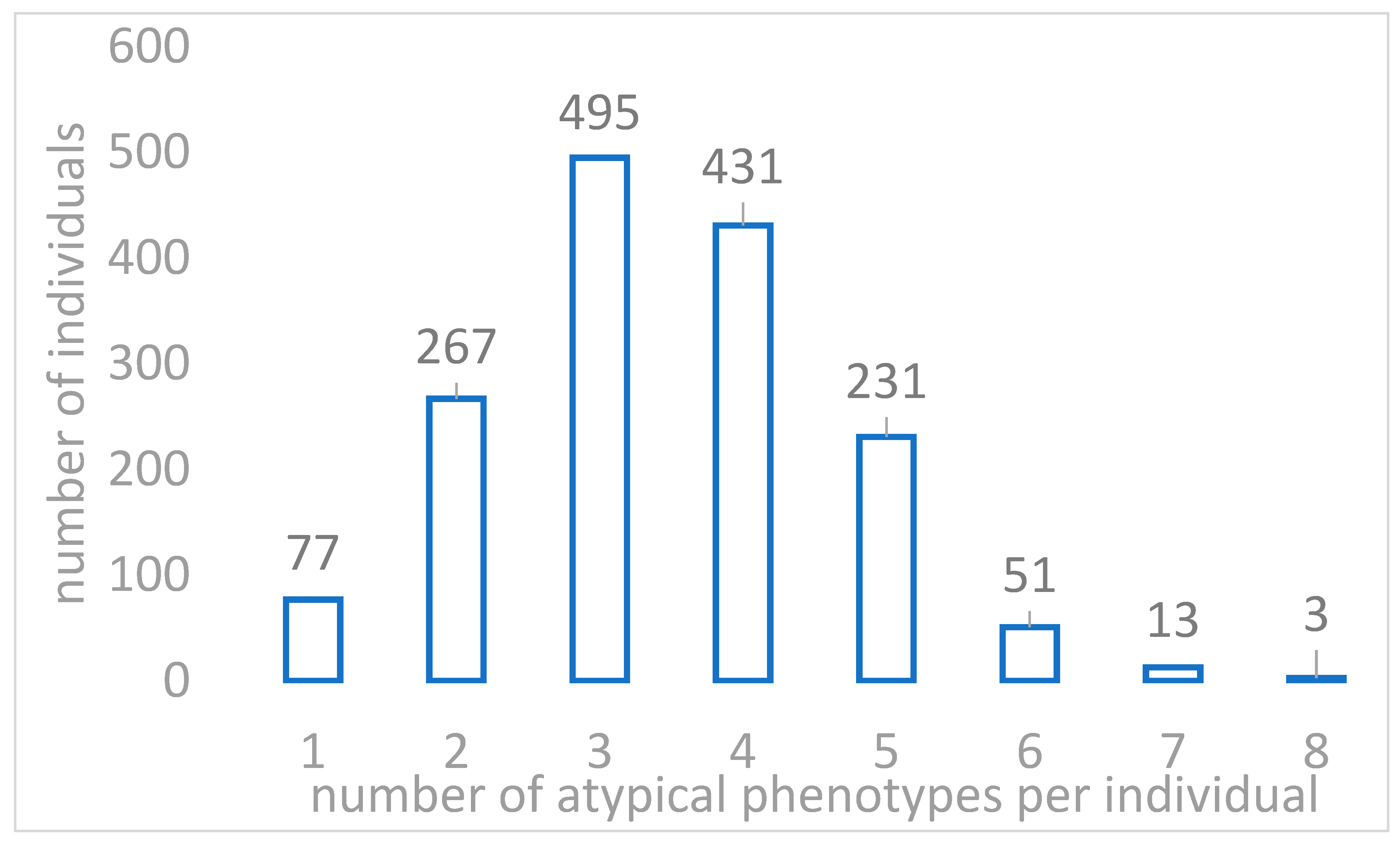
2.2. Actionable Pharmacogenomic Variants in Sardinian Genomes
3. Discussion
4. Material and Methods
4.1. Study Population and Data Sets
4.1.1. Dataset
4.1.2. Genotyping and Imputation
4.2. Variant Calling and Relatedness
4.3. Pharmacogenetics Resources
4.3.1. Haplotype and Phenotype Calling
4.3.2. Allele Frequency Analysis of PGx Actionable Variants from PharmGKB
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bouvy, J.C.; De Bruin, M.L.; Koopmanschap, M.A. Epidemiology of Adverse Drug Reactions in Europe: A Review of Recent Observational Studies. Drug Saf. 2015, 38, 437–453. [Google Scholar] [CrossRef]
- Pirmohamed, M.; James, S.; Meakin, S.; Green, C.; Scott, A.K.; Walley, T.J.; Farrar, K.; Park, B.K.; Breckenridge, A.M. Adverse drug reactions as cause of admission to hospital: Prospective analysis of 18 820 patients. BMJ 2004, 329, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.V.; Da Silva, G.O.; Gibbon, S. Pharmacogenomics, human genetic diversity and the incorporation and rejection of color/race in Brazil. BioSocieties 2014, 10, 48–69. [Google Scholar] [CrossRef] [PubMed]
- Auwerx, C.; Sadler, M.C.; Reymond, A.; Kutalik, Z. From pharmacogenetics to pharmaco-omics: Milestones and future directions. Hum. Genet. Genom. Adv. 2022, 3, 100100. [Google Scholar] [CrossRef] [PubMed]
- Gaedigk, A.; Riffel, A.K.; Leeder, J.S. CYP2D6 Haplotype Determination Using Long Range Allele-Specific Amplification: Resolution of a Complex Genotype and a Discordant Genotype Involving the CYP2D6*59 Allele. J. Mol. Diagn. 2015, 17, 740–748. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; McDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; Klein, T.E. Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef]
- Sim, S.C.; Altman, R.B.; Ingelman-Sundberg, M. Databases in the area of pharmacogenetics. Hum. Mutat. 2011, 32, 526–531. [Google Scholar] [CrossRef]
- Badagnani, I.; Sorani, M.; Edwards, R.H.; Brown, C.; Castro, R.A.; Huang, C.C.; Stryke, U.; Kawamoto, M.; Johns, S.J.; Carlson, E.J.; et al. PharmGKB Submission Update: VI. PMT Submissions of Genetic Variations in Neurotransmitter Transporters (SLC6, SLC17, and SLC18) to the PharmGKB Network. Pharmacol. Rev. 2006, 58, 5–6. [Google Scholar] [CrossRef]
- Sahana, S.; Bhoyar, R.C.; Sivadas, A.; Jain, A.; Imran, M.; Rophina, M.; Senthivel, V.; Diwakar, M.K.; Sharma, D.; Mishra, A.; et al. Pharmacogenomic landscape of Indian population using whole genomes. Clin. Transl. Sci. 2022, 15, 866–877. [Google Scholar] [CrossRef]
- Sivadas, A.; Scaria, V. Pharmacogenomic survey of Qatari populations using whole-genome and exome sequences. Pharmacogenom. J. 2018, 18, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Oslin, D.W.; Lynch, K.G.; Shih, M.-C.; Ingram, E.P.; Wray, L.O.; Chapman, S.R.; Kranzler, H.R.; Gelernter, J.; Pyne, J.M.; Stone, A.; et al. Effect of Pharmacogenomic Testing for Drug-Gene Interactions on Medication Selection and Remission of Symptoms in Major Depressive Disorder. JAMA 2022, 328, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Cecchin, E.; Roncato, R.; Guchelaar, H.J.; Toffoli, G.; for the Ubiquitous Pharmacogenomics Consortium. Ubiquitous Pharmacogenomics (U-PGx): The Time for Implementation is Now. An Horizon2020 Program to Drive Pharmacogenomics into Clinical Practice. Curr. Pharm. Biotechnol. 2017, 18, 204–209. [Google Scholar] [CrossRef]
- van der Wouden, C.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.; Dávila-Fajardo, C.; Deneer, V.; Dolžan, V.; Ingelman-Sundberg, M.; Jönsson, S.; Karlsson, M.; et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2016, 101, 341–358. [Google Scholar] [CrossRef]
- Marcus, J.H.; Posth, C.; Ringbauer, H.; Lai, L.; Skeates, R.; Sidore, C.; Beckett, J.; Furtwängler, A.; Olivieri, A.; Chiang, C.W.K.; et al. Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nat. Commun. 2020, 11, 939. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.W.K.; Marcus, J.H.; Sidore, C.; Biddanda, A.; Al-Asadi, H.; Zoledziewska, M.; Pitzalis, M.; Busonero, F.; Maschio, A.; Pistis, G.; et al. Genomic history of the Sardinian population. Nat. Genet. 2018, 50, 1426–1434. [Google Scholar] [CrossRef]
- Zoledziewska, M.; Sidore, C.; Chiang, C.W.K.; Sanna, S.; Mulas, A.; Steri, A.M.; Busonero, F.; Marcus, J.; Marongiu, M.; Maschio, A.; et al. Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. 2015, 47, 1352–1356. [Google Scholar] [CrossRef]
- Danjou, F.; Zoledziewska, M.; Sidore, C.; Steri, M.; Busonero, F.; Maschio, A.; Mulas, A.; Perseu, L.; Barella, S.; Porcu, E.; et al. Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat. Genet. 2015, 47, 1264–1271. [Google Scholar] [CrossRef]
- Sidore, C.; Busonero, F.; Maschio, A.; Porcu, E.; Naitza, S.; Zoledziewska, M.; Mulas, A.; Pistis, G.; Steri, A.M.; Danjou, F.; et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 2015, 47, 1272–1281. [Google Scholar] [CrossRef]
- Steri, M.; Orrù, V.; Idda, M.L.; Pitzalis, M.; Pala, M.; Zara, I.; Sidore, C.; Faà, V.; Floris, M.; Deiana, M.; et al. Overexpression of the Cytokine BAFF and Autoimmunity Risk. N. Engl. J. Med. 2017, 376, 1615–1626. [Google Scholar] [CrossRef]
- McInnes, G.; Lavertu, A.; Sangkuhl, K.; Klein, T.E.; Whirl-Carrillo, M.; Altman, R.B. Pharmacogenetics at Scale: An Analysis of the UK Biobank. Clin. Pharmacol. Ther. 2020, 109, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Sangkuhl, K.; Whirl-Carrillo, M.; Whaley, R.M.; Woon, M.; Lavertu, A.; Altman, R.; Carter, L.; Verma, A.; Ritchie, M.D.; Klein, T.E. Pharmacogenomics Clinical Annotation Tool (Pharm CAT). Clin. Pharmacol. Ther. 2019, 107, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Numanagić, I.; Malikić, S.; Ford, M.; Qin, X.; Toji, L.; Radovich, M.; Skaar, T.C.; Pratt, V.M.; Berger, B.; Scherer, S.; et al. Allelic decomposition and exact genotyping of highly polymorphic and structurally variant genes. Nat. Commun. 2018, 9, 828. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Caudle, K.; Gong, L.; Whirl-Carrillo, M.; Stein, C.; Scott, S.; Lee, M.; Gage, B.; Kimmel, S.; Perera, M.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef]
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef]
- Zito, S.; Fortinguerra, F.; Pierantozzi, A.; Ambrosino, F.; Traversa, G.; Trotta, F.; Da Cas, R.; Cangini, A. Medicines use in Italy. 2020 OsMed National Report. Recenti Prog. Med. 2021, 112, 659–667. [Google Scholar] [CrossRef]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef]
- Lee, C.R.; Luzum, J.A.; Sangkuhl, K.; Gammal, R.S.; Sabatine, M.S.; Stein, C.M.; Kisor, D.F.; Limdi, N.A.; Lee, Y.M.; Scott, S.A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2C19 Genotype and Clopidogrel Therapy: 2022 Update. Clin. Pharmacol. Ther. 2022. [CrossRef]
- Quartuccio, L.; Fabris, M.; Pontarini, E.; Salvin, S.; Zabotti, A.; Benucci, M.; Manfredi, M.; Biasi, D.; Ravagnani, V.; Atzeni, F.; et al. The 158VV Fcgamma receptor 3A genotype is associated with response to rituximab in rheumatoid arthritis: Results of an Italian multicentre study. Ann. Rheum. Dis. 2013, 73, 716–721. [Google Scholar] [CrossRef]
- Rishavy, M.A.; Hallgren, K.W.; Wilson, L.; Singh, S.; Runge, K.W.; Berkner, K.L. Warfarin alters vitamin K metabolism: A surprising mechanism of VKORC1 uncoupling necessitates an additional reductase. Blood 2018, 131, 2826–2835. [Google Scholar] [CrossRef] [Green Version]
- Limdi, N.A.; Wadelius, M.; Cavallari, L.; Eriksson, N.; Crawford, D.; Lee, M.T.M.; Chen, C.-H.; Motsinger-Reif, A.; Sagreiya, H.; Liu, N.; et al. Warfarin pharmacogenetics: A single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 2010, 115, 3827–3834. [Google Scholar] [CrossRef] [PubMed]
- Hershfield, M.S.; Callaghan, J.T.; Tassaneeyakul, W.; Mushiroda, T.; Thorn, C.F.; Klein, T.E.; Lee, M.T.M. Clinical Pharmacogenetics Implementation Consortium Guidelines for Human Leukocyte Antigen-B Genotype and Allopurinol Dosing. Clin. Pharmacol. Ther. 2012, 93, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Génin, E.; Schumacher, M.; Roujeau, J.-C.; Naldi, L.; Liss, Y.; Kazma, R.; Sekula, P.; Hovnanian, A.; Mockenhaupt, M. Genome-wide association study of Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in Europe. Orphanet J. Rare Dis. 2011, 6, 52. [Google Scholar] [CrossRef]
- Cheng, L.; Sun, B.; Xiong, Y.; Hu, L.; Gao, L.; Lv, Q.; Zhou, M.; Li, J.; Chen, X.; Zhang, W.; et al. The minor alleles HCP5 rs3099844 A and PSORS1C1 rs3131003 G are associated with allopurinol-induced severe cutaneous adverse reactions in Han Chinese: A multicentre retrospective case-control clinical study. Br. J. Dermatol. 2017, 178, e191–e193. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- García-Nieto, P.E.; Morrison, A.J.; Fraser, H.B. The somatic mutation landscape of the human body. Genome Biol. 2019, 20, 1–20. [Google Scholar] [CrossRef]
- Brazhnik, K.; Sun, S.; Alani, O.; Kinkhabwala, M.; Wolkoff, A.W.; Maslov, A.Y.; Dong, X.; Vijg, J. Single-cell analysis reveals different age-related somatic mutation profiles between stem and differentiated cells in human liver. Sci. Adv. 2020, 6, eaax2659. [Google Scholar] [CrossRef]
| Number of Individuals | Number of Drugs for Which an Atypical Response Is Expected |
|---|---|
| 1 | 39 |
| 3 | 38 |
| 6 | 37 |
| 12 | 36 |
| 16 | 35 |
| 14 | 34 |
| 5 | 33 |
| 7 | 32 |
| 5 | 31 |
| 19 | 30 |
| 38 | 29 |
| 57 | 28 |
| 47 | 27 |
| 49 | 26 |
| 40 | 25 |
| 45 | 24 |
| 43 | 23 |
| 51 | 22 |
| 36 | 21 |
| 34 | 20 |
| 42 | 19 |
| 92 | 18 |
| 132 | 17 |
| 125 | 16 |
| 127 | 15 |
| 89 | 14 |
| 58 | 13 |
| 58 | 12 |
| 22 | 11 |
| 22 | 10 |
| 12 | 9 |
| 11 | 8 |
| 2 | 7 |
| 9 | 6 |
| 27 | 5 |
| 49 | 4 |
| 68 | 3 |
| 55 | 2 |
| 40 | 1 |
| TOT = 1568 (99.43%) |
| Phenotypes | Pharmacogenes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFTR | CYP2B6 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A5 | CYP4F2 | DPYD | IFNL3 | NUDT15 | SLCO1B1 | TPMT | UGT1A1 | VKORC1 | |
| Decreased Function | 324 | |||||||||||||
| Decreased warfarin dose | 414 | |||||||||||||
| Favorable response genotype | 721 | |||||||||||||
| Increased dose phenotype | 268 | |||||||||||||
| Indeterminate | 50 | 207 | 11 | 133 | 2 | |||||||||
| Intermediate Function | 101 | |||||||||||||
| Intermediate Metabolizer | 552 | 450 | 606 | 304 | 117 | 2 | ||||||||
| Ivacaftor irrelevant | 1555 | |||||||||||||
| Ivacaftor non-responsive | 14 | |||||||||||||
| Ivacaftor responsive | 8 | |||||||||||||
| Normal dose phenotype | 534 | |||||||||||||
| Normal Function | 533 | 1472 | ||||||||||||
| Normal Metabolizer | 862 | 715 | 925 | 622 | 16 | 1460 | 1575 | 815 | ||||||
| Normal warfarin dose | 385 | |||||||||||||
| Not available | 8 | 2 | 748 | 14 | 764 | 201 | 762 | |||||||
| Poor Function | 51 | 2 | ||||||||||||
| Poor Metabolizer | 85 | 32 | 44 | 1243 | ||||||||||
| Possible increased function | 335 | |||||||||||||
| Possibly decreased warfarin dose | 778 | |||||||||||||
| Rapid Metabolizer | 20 | 343 | ||||||||||||
| Ultrarapid Metabolizer | 37 | |||||||||||||
| Unfavorable response genotype | 856 | |||||||||||||
| Total non-typical phenotypes | 14 | 552 | 519 | 650 | 0 | 1547 | 268 | 117 | 856 | 0 | 375 | 103 | 0 | 1192 |
| Gene | Related Drugs | Non-Typical Response Phenotypes | N SARD | Freq SARD | Delta Freq. | p-Value |
|---|---|---|---|---|---|---|
| CFTR | Ivacaftor | ivacaftor irrelevant | 1555 | 0.98604946 | 0.02922937 | 7.77 × 10−8 |
| ivacaftor non-responsive | 14 | 0.00887762 | −0.0240865 | 5.38 × 10−7 | ||
| ivacaftor responsive | 8 | 0.00507292 | −0.00509874 | 0.10 | ||
| Not available | 0 | 0 | 0 | 1 | ||
| CYP2B6 | efavirenz | Rapid Metabolizer | 20 | 0.01268231 | 0.01235134 | 3.82 × 10−65 |
| Indeterminate | 50 | 0.03170577 | −0.01226846 | 0.04 | ||
| Intermediate Metabolizer | 552 | 0.35003171 | 0.00655614 | 0.79 | ||
| Poor Metabolizer | 85 | 0.05389981 | −0.00298206 | 0.83 | ||
| Not available | 8 | 0.00507292 | −0.00231863 | 0.54 | ||
| Normal Metabolizer | 862 | 0.54660748 | −0.00122801 | 1 | ||
| Ultrarapid Metabolizer | 0 | 0 | −0.00011032 | 1 | ||
| CYP2C19 | amitriptyline, citalopram, sertraline, clopidogrel, escitalopram, imipramine, clomipramine, doxepin, trimipramine, voriconazole | Normal Metabolizer | 715 | 0.45339252 | 0.05749207 | 1.63 × 10−5 |
| Rapid Metabolizer | 343 | 0.21750159 | −0.05355882 | 9.34 × 10−6 | ||
| Intermediate Metabolizer | 450 | 0.28535193 | 0.02347028 | 0.07 | ||
| Ultrarapid Metabolizer | 37 | 0.02346227 | −0.02161518 | 0.00 | ||
| Poor Metabolizer | 32 | 0.02029169 | −0.00393495 | 0.54 | ||
| Not available | 0 | 0 | −0.00145625 | 0.39 | ||
| Indeterminate | 0 | 0 | −0.00019858 | 1 | ||
| Likely Intermediate Metabolizer | 0 | 0 | −0.00019858 | 1 | ||
| Likely Poor Metabolizer | 0 | 0 | 0 | - | ||
| CYP2C9 | ibuprofen, piroxicam, celecoxib, meloxicam, phenytoin, flurbiprofen, tenoxicam, lornoxicam, warfarin | Normal Metabolizer | 925 | 0.58655675 | −0.05549347 | 2.20 × 10−5 |
| Intermediate Metabolizer | 606 | 0.38427394 | 0.04975649 | 0.00 | ||
| Poor Metabolizer | 44 | 0.02790108 | 0.00590293 | 0.23 | ||
| Indeterminate | 0 | 0 | −0.00028684 | 1 | ||
| Not available | 2 | 0.00126823 | 0.00012088 | 1 | ||
| CYP3A5 | tacrolimus | Poor Metabolizer | 1243 | 0.78820545 | −0.08090889 | 1.09 × 10−19 |
| Intermediate Metabolizer | 304 | 0.19277108 | 0.06775453 | 1.17 × 10−14 | ||
| Normal Metabolizer | 16 | 0.01014585 | 0.0042988 | 0.078017 | ||
| Indeterminate | 0 | 0 | 0 | 1 | ||
| CYP4F2 | warfarin | Not available | 764 | 0.48446417 | 0.18778706 | 4.30 × 10−56 |
| Normal dose phenotype | 534 | 0.33861763 | −0.15050465 | 5.15 × 10−31 | ||
| Increased dose phenotype | 268 | 0.16994293 | −0.04138931 | 2.07 × 10−4 | ||
| Indeterminate | 11 | 0.00697527 | 0.00410691 | 0.02 | ||
| DPYD | fluorouracil, capecitabine | Intermediate Metabolizer | 117 | 0.0741915 | 0.00471089 | 0.68 |
| Normal Metabolizer | 1460 | 0.9258085 | −0.0040269 | 0.76 | ||
| Poor Metabolizer | 0 | 0 | −0.00046335 | 1 | ||
| Not available | 0 | 0 | −0.00022064 | 1 | ||
| IFNL3 | ribavirin, peginterferon alfa-2a, peginterferon alfa-2b | Unfavorable response genotype | 856 | 0.54280279 | 0.03075566 | 0.03 |
| Favorable response genotype | 721 | 0.45719721 | −0.03075566 | 0.03 | ||
| NUDT15 | azathioprine, mercaptopurine, thioguanine | Normal Metabolizer | 1575 | 0.99873177 | 0.01104367 | 2.84 × 10−04 |
| Intermediate Metabolizer | 0 | 0 | −0.00849477 | 9.58 × 10−4 | ||
| Indeterminate | 2 | 0.00126823 | −0.00164426 | 0.52 | ||
| Not available | 0 | 0 | −0.00083844 | 0.68 | ||
| Poor Metabolizer | 0 | 0 | 0 | 1 | ||
| Possible Intermediate Metabolizer | 0 | 0 | 0 | 1 | ||
| SLCO1B1 | fluvastatin, lovastatin, pitavastatin, pravastatin, rosuvastatin, simvastatin | Normal Function | 533 | 0.33798351 | −0.22189911 | 1.25 × 10−66 |
| Possible Decreased Function | 335 | 0.21242866 | 0 | 0 | ||
| Possible Increased Function | 0 | 0 | −0.08680111 | 2.87 × 10−33 | ||
| Not available | 201 | 0.1274572 | 0.07320099 | 7.12 × 10−34 | ||
| Indeterminate | 133 | 0.08433735 | 0.02026251 | 3.54 × 10−3 | ||
| Poor Function | 51 | 0.03233989 | 0.01027555 | 0.02 | ||
| Decreased Function | 324 | 0.20545339 | −0.00737923 | 0.68 | ||
| Possible Poor Function | 0 | 0 | 0 | NA | ||
| TPMT | azathioprine, mercaptopurine, thioguanine | Normal Function | 1472 | 0.93341788 | 0.0372306 | 7.17 × 10−6 |
| Intermediate Function | 101 | 0.06404566 | −0.03380969 | 2.74 × 10−5 | ||
| Indeterminate | 2 | 0.00126823 | −0.00173252 | 0.50 | ||
| Poor Function | 2 | 0.00126823 | −0.00144568 | 0.58 | ||
| Possible Intermediate Metabolizer | 0 | 0 | −0.00017651 | 1 | ||
| Not available | 0 | 0 | 0 | 1 | ||
| UGT1A1 | atazanavir, irinotecan | Normal Metabolizer | 815 | 0.51680406 | 0.05715974 | 2.50 × 10−5 |
| Intermediate Metabolizer | 0 | 0 | −0.04417281 | 1.10 × 10−16 | ||
| Not available | 762 | 0.48319594 | −0.00903741 | 0.67 | ||
| Poor Metabolizer | 0 | 0 | −0.00366268 | 0.05 | ||
| Indeterminate | 0 | 0 | −0.00028684 | 1 | ||
| VKORC1 | warfarin | Normal warfarin dose | 385 | 0.24413443 | −0.15075106 | 1.39 × 10−32 |
| Decreased warfarin dose | 414 | 0.26252378 | 0.12201807 | 1.24 × 10−40 | ||
| Possibly decreased warfarin dose | 778 | 0.49334179 | 0.02873299 | 0.05 |
| CHR | BP | RSID | Gene | A2 | SARD_A2_FRQ | ALFA_A2_FRQ | Delta |
| 1 | 161514542 | rs396991 | FCGR3A | C | 0.528 | 0.317 | 0.211 |
| 16 | 31104509 | rs8050894 | VKORC1 | G | 0.523 | 0.374 | 0.148 |
| 19 | 15990431 | rs2108622 | CYP4F2 | T | 0.415 | 0.294 | 0.120 |
| 16 | 31107689 | rs9923231 | VKORC1 | T | 0.510 | 0.392 | 0.117 |
| 16 | 31104878 | rs9934438 | VKORC1 | A | 0.510 | 0.395 | 0.114 |
| 6 | 39325078 | rs20455 | KIF6 | G | 0.463 | 0.364 | 0.099 |
| 1 | 11856378 | rs1801133 | MTHFR | A | 0.424 | 0.349 | 0.075 |
| 16 | 31105554 | rs2884737 | VKORC1 | C | 0.353 | 0.278 | 0.074 |
| 1 | 97981395 | rs1801159 | DPYD | C | 0.252 | 0.199 | 0.053 |
| CHR | BP | RSID | Gen | A2 | SARD_A2_FRQ | ALFA_EUR_FRQ | Delta |
| 1 | 98348885 | rs1801265 | DPYD | G | 0.159 | 0.215 | −0.056 |
| 21 | 46957794 | rs1051266 | SLC19A1 | C | 0.511 | 0.568 | −0.057 |
| 6 | 31543031 | rs1800629 | TNF | A | 0.050 | 0.159 | −0.109 |
| 16 | 31103796 | rs2359612 | VKORC1 | G | 0.490 | 0.604 | −0.114 |
| Clinical Annotation ID | Variant/Haplotypes | is_top | Gene | Level of Evidence | Phenotype Category | Drug(s) | Phenotype(s) | Pediatric Population |
|---|---|---|---|---|---|---|---|---|
| 1184661194 | rs2108622 | rs2108622 | CYP4F2 | 2A | Dosage | acenocoumarol | Atrial Fibrillation | 0 |
| 981204044 | rs9923231 | rs9923231 | VKORC1 | 1A | Dosage | acenocoumarol | 0 | |
| 1183704228 | rs9934438 | rs9934438 | VKORC1 | 2A | Dosage | acenocoumarol | 0 | |
| 1451237940 | rs9923231 | rs9923231 | VKORC1 | 1A | Dosage | phenprocoumon | 1 | |
| 1451244040 | rs9934438 | rs9934438 | VKORC1 | 2A | Dosage | phenprocoumon | 1 | |
| 655385400 | rs2108622 | rs2108622 | CYP4F2 | 1A | Dosage | warfarin | 1 | |
| 982035703 | rs2884737 | rs2884737 | VKORC1 | 2A | Dosage | warfarin | 0 | |
| 655385392 | rs9934438 | rs9934438 | VKORC1 | 1B | Dosage | warfarin | 1 | |
| 655385028 | rs8050894 | rs8050894 | VKORC1 | 1B | Dosage | warfarin | 0 | |
| 655385012 | rs9923231 | rs9923231 | VKORC1 | 1A | Dosage | warfarin | 1 | |
| 655385024 | rs2359612 | rs2359612 | VKORC1 | 1B | Dosage | warfarin | 0 | |
| 655384799 | rs1800629 | rs1800629 | TNF | 2B | Efficacy | etanercept | Arthritis, Psoriatic; Arthritis, Rheumatoid; Crohn Disease; Inflammation; Psoriasis; Spondylitis, Ankylosing | 0 |
| 1451245360 | rs1051266 | rs1051266 | SLC19A1 | 2A | Efficacy | methotrexate | Arthritis, Rheumatoid | 0 |
| 655384621 | rs20455 | rs20455 | KIF6 | 2B | Efficacy | pravastatin | Coronary Disease; Myocardial Infarction | 0 |
| 1444608384 | rs396991 | rs396991 | FCGR3A | 2B | Efficacy | rituximab | Arthritis, Rheumatoid; Neuromyelitis Optica | 0 |
| 1447672998 | rs9923231 | rs9923231 | VKORC1 | 2A | Efficacy | warfarin | time to therapeutic inr | 1 |
| 1447673015 | rs9923231 | rs9923231 | VKORC1 | 2A | Efficacy | warfarin | time in therapeutic range | 1 |
| 1451286320 | rs1801159 | rs1801159 | DPYD | 1A | Toxicity | capecitabine | Neoplasms | 0 |
| 1451287240 | rs1801265 | rs1801265 | DPYD | 1A | Toxicity | capecitabine | Neoplasms | 0 |
| 981201981 | rs1801265 | rs1801265 | DPYD | 1A | Toxicity | fluorouracil | Neoplasms | 1 |
| 981201962 | rs1801159 | rs1801159 | DPYD | 1A | Toxicity | fluorouracil | Neoplasms | 0 |
| 827848365 | rs1801133 | rs1801133 | MTHFR | 2A | Toxicity | methotrexate | Drug Toxicity;hematotoxicity; Leukopenia; Lymphoma; mucositis; Neoplasms; Neutropenia; Osteosarcoma; Precursor Cell Lymphoblastic Leukemia-Lymphoma; primary central nervous system lymphoma; Thrombocytopenia; Toxic liver disease | 1 |
| 655385307 | rs1801133 | rs1801133 | MTHFR | 2A | Toxicity | methotrexate | Arthritis, Juvenile Rheumatoid; Arthritis, Psoriatic; Arthritis, Rheumatoid; Drug Toxicity | 1 |
| 1451243676 | rs9923231 | rs9923231 | VKORC1 | 2A | Toxicity | phenprocoumon | Hemorrhage;over-anticoagulation; time above therapeutic range | 0 |
| 1449269910 | rs9923231 | rs9923231 | VKORC1 | 2A | Toxicity | warfarin | Hemorrhage | 1 |
| 1447673005 | rs9923231 | rs9923231 | VKORC1 | 1B | Toxicity | warfarin | over-anticoagulation | 1 |
| CHR | POS | RSID | Gene | A2 | SARD_A2_FRQ | ALFA_A2_FRQ | Delta |
|---|---|---|---|---|---|---|---|
| 1 | 1:207753621 | rs2274567 | CR1 | G | 0.608 | 0.195 | 0.413 |
| 6 | 6:31093482 | rs3131003 | PSORS1C1 | A | 0.739 | 0.432 | 0.308 |
| 6 | 6:31093587 | rs3815087 | PSORS1C1 | A | 0.507 | 0.217 | 0.290 |
| 13 | 12:13953118 | rs2058878 | GRIN2B | A | 0.602 | 0.338 | 0.265 |
| 16 | 12:85243681 | rs6539870 | IL1B | G | 0.486 | 0.221 | 0.264 |
| 4 | 3:45732515 | rs2742421 | SACM1L | G | 0.687 | 0.427 | 0.260 |
| 6 | 6:31603770 | rs11229 | PRRC2A | G | 0.435 | 0.176 | 0.259 |
| 15 | 15:30193316 | rs813676 | TJP1 | C | 0.761 | 0.504 | 0.257 |
| 6 | 6:31107361 | rs2233945 | PSORS1C1 | A | 0.417 | 0.163 | 0.254 |
| 6 | 6:31604591 | rs10885 | PRRC2A | T | 0.436 | 0.182 | 0.254 |
| 6 | 6:31018546 | rs2523864 | HCG22 | T | 0.679 | 0.433 | 0.245 |
| 19 | 19:15959200 | rs2189784 | CYP4F2 | A | 0.667 | 0.423 | 0.245 |
| 6 | 6:31022113 | rs3873352 | HCG22 | G | 0.336 | 0.091 | 0.245 |
| 6 | 6:43737794 | rs13207351 | VEGFA | G | 0.611 | 0.367 | 0.244 |
| 3 | 3:151090996 | rs9859552 | P2RY12 | T | 0.338 | 0.094 | 0.244 |
| 6 | 6:31543101 | rs361525 | TNF | A | 0.292 | 0.054 | 0.238 |
| 6 | 6:31542308 | rs1799964 | TNF | C | 0.447 | 0.214 | 0.233 |
| 7 | 6:78173281 | rs130058 | HTR1B | A | 0.414 | 0.183 | 0.231 |
| 5 | 5:158750769 | rs3213094 | IL12B | T | 0.427 | 0.206 | 0.221 |
| 7 | 7:86331756 | rs2189814 | GRM3 | C | 0.375 | 0.159 | 0.216 |
| 11 | 10:32202069 | rs2799018 | ARHGAP12 | T | 0.611 | 0.396 | 0.215 |
| 1 | 1:161514542 | rs396991 | FCGR3A | C | 0.528 | 0.317 | 0.211 |
| 11 | 10:92619161 | rs7905446 | HTR7, RPP30 | G | 0.480 | 0.276 | 0.205 |
| 6 | 6:33047612 | rs3097671 | HLA-DPB1 | C | 0.366 | 0.163 | 0.203 |
| 10 | 1:29161999 | rs2236855 | OPRD1 | A | 0.386 | 0.185 | 0.201 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Idda, M.L.; Zoledziewska, M.; Urru, S.A.M.; McInnes, G.; Bilotta, A.; Nuvoli, V.; Lodde, V.; Orrù, S.; Schlessinger, D.; Cucca, F.; et al. Genetic Variation among Pharmacogenes in the Sardinian Population. Int. J. Mol. Sci. 2022, 23, 10058. https://doi.org/10.3390/ijms231710058
Idda ML, Zoledziewska M, Urru SAM, McInnes G, Bilotta A, Nuvoli V, Lodde V, Orrù S, Schlessinger D, Cucca F, et al. Genetic Variation among Pharmacogenes in the Sardinian Population. International Journal of Molecular Sciences. 2022; 23(17):10058. https://doi.org/10.3390/ijms231710058
Chicago/Turabian StyleIdda, Maria Laura, Magdalena Zoledziewska, Silvana Anna Maria Urru, Gregory McInnes, Alice Bilotta, Viola Nuvoli, Valeria Lodde, Sandro Orrù, David Schlessinger, Francesco Cucca, and et al. 2022. "Genetic Variation among Pharmacogenes in the Sardinian Population" International Journal of Molecular Sciences 23, no. 17: 10058. https://doi.org/10.3390/ijms231710058
APA StyleIdda, M. L., Zoledziewska, M., Urru, S. A. M., McInnes, G., Bilotta, A., Nuvoli, V., Lodde, V., Orrù, S., Schlessinger, D., Cucca, F., & Floris, M. (2022). Genetic Variation among Pharmacogenes in the Sardinian Population. International Journal of Molecular Sciences, 23(17), 10058. https://doi.org/10.3390/ijms231710058









