Peroxisome-Mediated Reactive Oxygen Species Signals Modulate Programmed Cell Death in Plants
Abstract
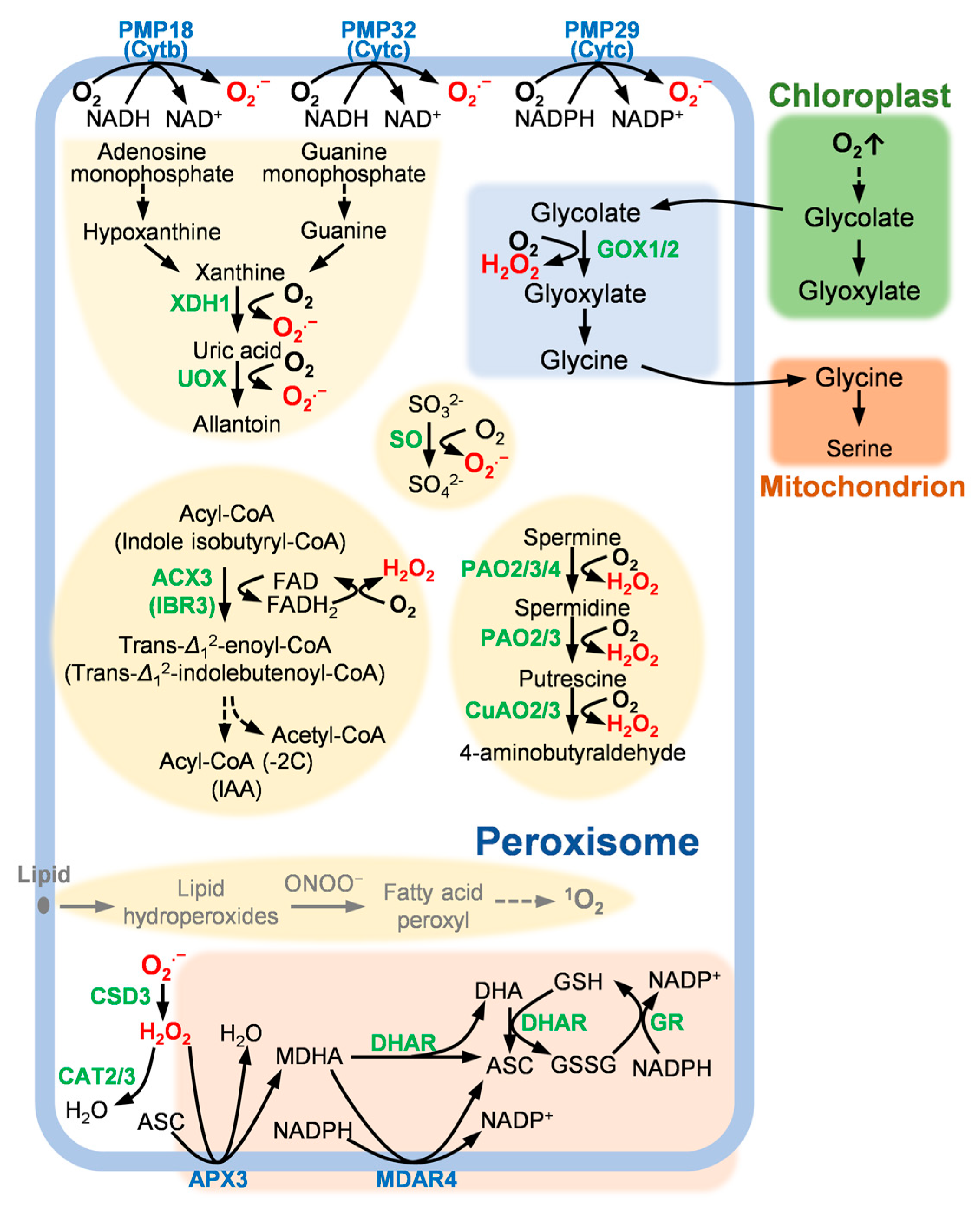
:1. Introduction
2. Peroxisomal ROS Generation Mechanism and PCD
2.1. The Purine Base Degradation Pathway and PCD
2.2. Photorespiratory Cycle and PCD
2.3. The Fatty Acid β-Oxidation Pathway and PCD
2.4. The Polyamine Oxidation Pathway and PCD
2.5. Other ROS Generation Mechanism
3. Peroxisomal ROS Scavenging Mechanism and PCD
3.1. SOD and PCD
3.2. CAT and PCD
3.3. The ASC–GSH Cycle and PCD
4. Challenges and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant Peroxisomes: A Factory of Reactive Species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef]
- Mor, A.; Koh, E.; Weiner, L.; Rosenwasser, S.; Sibony-Benyamini, H.; Fluhr, R. Singlet Oxygen Signatures Are Detected Independent of Light or Chloroplasts in Response to Multiple Stresses. Plant Physiol. 2014, 165, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L.; Mu, J.; Zuo, J. LESION SIMULATING DISEASE1 Interacts with Catalases to Regulate Hypersensitive Cell Death in Arabidopsis. Plant Physiol. 2013, 163, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wang, J.; Leng, H.-N.; Wang, X.; Liu, Y.; Lu, H.; Lu, M.-Z.; Zhang, J. Transcriptional Regulation and Signaling of Developmental Programmed Cell Death in Plants. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Saifullah; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Janků, M.; Luhová, L.; Petřivalský, M. On the Origin and Fate of Reactive Oxygen Species in Plant Cell Compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Gross, F.; Durner, J.; Gaupels, F. Nitric oxide, antioxidants and prooxidants in plant defence responses. Front. Plant Sci. 2013, 4, 419. [Google Scholar] [CrossRef]
- Sandalio, L.M.; Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Río, L.A. Role of Peroxisomes as a Source of Reactive Oxygen Species (ROS) Signaling Molecules. Subcell. Biochem. 2013, 69, 231–255. [Google Scholar] [CrossRef]
- Costa, T.J.; Barros, P.R.; Arce, C.; Santos, J.D.; da Silva-Neto, J.; Egea, G.; Dantas, A.P.; Tostes, R.C.; Jimenez-Altayo, F. The homeostatic role of hydrogen peroxide, superoxide anion and nitric oxide in the vasculature. Free Radic. Biol. Med. 2021, 162, 615–635. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M.; Sandalio, L.M.; Valderrama, R.; Barroso, J.B.; del Río, L.A. Peroxisomal xanthine oxidoreductase: Characterization of the enzyme from pea (Pisum sativum L.) leaves. J. Plant Physiol. 2008, 165, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Sandalio, L.M.; Fernandez, V.M.; Ruperez, F.L.; del Rio, L.A. Superoxide Free Radicals are Produced in Glyoxysomes. Plant Physiol. 1988, 87, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Hauck, O.K.; Scharnberg, J.; Escobar, N.M.; Wanner, G.; Giavalisco, P.; Witte, C.-P. Uric Acid Accumulation in an Arabidopsis Urate Oxidase Mutant Impairs Seedling Establishment by Blocking Peroxisome Maintenance. Plant Cell 2014, 26, 3090–3100. [Google Scholar] [CrossRef] [PubMed]
- Soltabayeva, A.; Bekturova, A.; Kurmanbayeva, A.; Oshanova, D.; Nurbekova, Z.; Srivastava, S.; Standing, D.; Sagi, M. Ureides are accumulated similarly in response to UV-C irradiation and wounding in Arabidopsis leaves but are remobilized differently during recovery. J. Exp. Bot. 2022, 73, 1016–1032. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Srivastava, S.; Kurmanbayeva, A.; Bekturova, A.; Fluhr, R.; Sagi, M. Early Senescence in Older Leaves of Low Nitrate-Grown Atxdh1 Uncovers a Role for Purine Catabolism in N Supply. Plant Physiol. 2018, 178, 1027–1044. [Google Scholar] [CrossRef]
- Byrne, R.S.; Hänsch, R.; Mendel, R.R.; Hille, R. Oxidative Half-reaction of Arabidopsis thaliana Sulfite Oxidase. J. Biol. Chem. 2009, 284, 35479–35484. [Google Scholar] [CrossRef]
- Lopez-Huertas, E.; Corpas, F.J.; Sandalio, L.M.; Del Rio, L.A. Characterization of membrane polypeptides from pea leaf peroxisomes involved in superoxide radical generation. Biochem. J. 1999, 337, 531–536. [Google Scholar] [CrossRef]
- Noctor, G.; Veljovic-Jovanovic, S.; Driscoll, S.; Novitskaya, L.; Foyer, C.H. Drought and oxidative load in the leaves of C3 plants: A predominant role for photorespiration? Ann. Bot. 2002, 89, 841–850. [Google Scholar] [CrossRef]
- Launay, A.; Jolivet, S.; Clement, G.; Zarattini, M.; Dellero, Y.; Le Hir, R.; Jossier, M.; Hodges, M.; Expert, D.; Fagard, M. DspA/E-Triggered Non-Host Resistance against E. amylovora Depends on the Arabidopsis GLYCOLATE OXIDASE 2 Gene. Int. J Mol. Sci. 2022, 23, 4224. [Google Scholar] [CrossRef]
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J.; et al. Lack of GLYCOLATE OXIDASE1, but Not GLYCOLATE OXIDASE2, Attenuates the Photorespiratory Phenotype of CATALASE2-Deficient Arabidopsis. Plant Physiol. 2016, 171, 1704–1719. [Google Scholar] [CrossRef]
- Dellero, Y.; Jossier, M.; Glab, N.; Oury, C.; Tcherkez, G.; Hodges, M. Decreased glycolate oxidase activity leads to altered carbon allocation and leaf senescence after a transfer from high CO2 to ambient air in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 3149–3163. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Graham, I.A.; Holdsworth, M.; Smith, S.M.; Theodoulou, F.L. Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci. 2006, 11, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Fan, J.; Xu, C. Peroxisomal fatty acid beta-oxidation negatively impacts plant survival under salt stress. Plant Signal. Behav. 2019, 14, 1561121. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.; Chung, H.S.; Kobayashi, Y.; Howe, G.A. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 2006, 281, 33511–33520. [Google Scholar] [CrossRef]
- Zolman, B.K.; Nyberg, M.; Bartel, B. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 2007, 64, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Kaurilind, E.; Xu, E.; Brosche, M. A genetic framework for H2O2 induced cell death in Arabidopsis thaliana. BMC Genom. 2015, 16, 837. [Google Scholar] [CrossRef]
- Khan, B.R.; Adham, A.R.; Zolman, B.K. Peroxisomal Acyl-CoA oxidase 4 activity differs between Arabidopsis accessions. Plant Mol. Biol. 2011, 78, 45–58. [Google Scholar] [CrossRef]
- Yuan, H.M.; Liu, W.C.; Lu, Y.T. CATALASE2 Coordinates SA-Mediated Repression of Both Auxin Accumulation and JA Biosynthesis in Plant Defenses. Cell Host Microbe 2017, 21, 143–155. [Google Scholar] [CrossRef]
- Fincato, P.; Moschou, P.N.; Spedaletti, V.; Tavazza, R.; Angelini, R.; Federico, R.; Roubelakis-Angelakis, K.A.; Tavladoraki, P. Functional diversity inside the Arabidopsis polyamine oxidase gene family. J. Exp. Bot. 2011, 62, 1155–1168. [Google Scholar] [CrossRef]
- Planas-Portell, J.; Gallart, M.; Tiburcio, A.F.; Altabella, T. Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Ono, Y.; Kim, D.W.; Watanabe, K.; Sasaki, A.; Niitsu, M.; Berberich, T.; Kusano, T.; Takahashi, Y. Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 2012, 42, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Shao, G.; Jiao, G.; Sheng, Z.; Xie, L.; Hu, S.; Tang, S.; Wei, X.; Hu, P. Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol. Plant 2021, 14, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Tisi, A.; Federico, R.; Moreno, S.; Lucretti, S.; Moschou, P.N.; Roubelakis-Angelakis, K.A.; Angelini, R.; Cona, A. Perturbation of Polyamine Catabolism Can Strongly Affect Root Development and Xylem Differentiation. Plant Physiol. 2011, 157, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chen, Y.; Li, M.; Xu, X.; Wu, G. Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J. Integr. Plant Biol. 2011, 53, 375–387. [Google Scholar] [CrossRef]
- Miyamoto, S.; Ronsein, G.E.; Prado, F.M.; Uemi, M.; Correa, T.C.; Toma, I.N.; Bertolucci, A.; Oliveira, M.C.; Motta, F.D.; Medeiros, M.H.; et al. Biological hydroperoxides and singlet molecular oxygen generation. IUBMB Life 2007, 59, 322–331. [Google Scholar] [CrossRef] [PubMed]
- op den Camp, R.G.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Gobel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Prakash Sanyal, R.; Samant, A.; Prashar, V.; Sharan Misra, H.; Saini, A. Biochemical and functional characterization of OsCSD3, a novel CuZn superoxide dismutase from rice. Biochem. J. 2018, 475, 3105–3121. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Kuo, W.Y.; Weiss, C.; Jinn, T.L. Copper chaperone-dependent and -independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 2012, 158, 737–746. [Google Scholar] [CrossRef]
- Xing, Y.; Cao, Q.; Zhang, Q.; Qin, L.; Jia, W.; Zhang, J. MKK5 regulates high light-induced gene expression of Cu/Zn superoxide dismutase 1 and 2 in Arabidopsis. Plant Cell Physiol. 2013, 54, 1217–1227. [Google Scholar] [CrossRef]
- Holzmeister, C.; Gaupels, F.; Geerlof, A.; Sarioglu, H.; Sattler, M.; Durner, J.; Lindermayr, C. Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J. Exp. Bot. 2015, 66, 989–999. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; del Río, L.A.; Palma, J.M. Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 2019, 61, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Chaouch, S.; Noctor, G. Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 2010, 188, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Vanderauwera, S.; Suzuki, N.; Miller, G.; van de Cotte, B.; Morsa, S.; Ravanat, J.L.; Hegie, A.; Triantaphylides, C.; Shulaev, V.; Van Montagu, M.C.; et al. Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1711–1716. [Google Scholar] [CrossRef] [PubMed]
- Tyutereva, E.V.; Dobryakova, K.S.; Schiermeyer, A.; Shishova, M.F.; Pawlowski, K.; Demidchik, V.; Reumann, S.; Voitsekhovskaja, O.V. The levels of peroxisomal catalase protein and activity modulate the onset of cell death in tobacco BY-2 cells via reactive oxygen species levels and autophagy. Funct. Plant Biol. 2018, 45, 247–258. [Google Scholar] [CrossRef]
- Mateo, A.; Muhlenbock, P.; Rusterucci, C.; Chang, C.C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 2004, 136, 2818–2830. [Google Scholar] [CrossRef]
- Han, Y.; Chaouch, S.; Mhamdi, A.; Queval, G.; Zechmann, B.; Noctor, G. Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Sign. 2013, 18, 2106–2121. [Google Scholar] [CrossRef]
- Hu, L.; Zhou, K.; Ren, G.; Yang, S.; Liu, Y.; Zhang, Z.; Li, Y.; Gong, X.; Ma, F. Myo-inositol mediates reactive oxygen species-induced programmed cell death via salicylic acid-dependent and ethylene-dependent pathways in apple. Hortic. Res. 2020, 7, 138. [Google Scholar] [CrossRef]
- Kang, S.; Shin, K.D.; Kim, J.H.; Chung, T. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018, 37, 653–664. [Google Scholar] [CrossRef]
- Wang, Y.; Nishimura, M.T.; Zhao, T.; Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011, 68, 74–87. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Song, R.-F.; Li, T.-T.; Liu, W.-C. Jasmonic Acid Impairs Arabidopsis Seedling Salt Stress Tolerance Through MYC2-Mediated Repression of CAT2 Expression. Front. Plant Sci. 2021, 12, 730228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, Y.; Xie, Z.; Li, X.; He, Z.H.; Peng, X.X. Association-Dissociation of Glycolate Oxidase with Catalase in Rice: A Potential Switch to Modulate Intracellular H2O2 Levels. Mol. Plant 2016, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Narendra, S. The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 2006, 57, 3033–3042. [Google Scholar] [CrossRef]
- Eastmond, P.J. MONODEHYROASCORBATE REDUCTASE4 Is Required for Seed Storage Oil Hydrolysis and Postgerminative Growth in Arabidopsis. Plant Cell 2007, 19, 1376–1387. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The Heart of the Redox Hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Panchuk, I.I.; Zentgraf, U.; Volkov, R.A. Expression of the APX gene family during leaf senescence of Arabidopsis thaliana. Planta 2005, 222, 926–932. [Google Scholar] [CrossRef]
- Rosa, S.B.; Caverzan, A.; Teixeira, F.K.; Lazzarotto, F.; Silveira, J.A.G.; Ferreira-Silva, S.L.; Abreu-Neto, J.; Margis, R.; Margis-Pinheiro, M. Cytosolic APX knockdown indicates an ambiguous redox responses in rice. Phytochemistry 2010, 71, 548–558. [Google Scholar] [CrossRef]
- Bulley, S.M.; Cooney, J.M.; Laing, W. Elevating Ascorbate in Arabidopsis Stimulates the Production of Abscisic Acid, Phaseic Acid, and to a Lesser Extent Auxin (IAA) and Jasmonates, Resulting in Increased Expression of DHAR1 and Multiple Transcription Factors Associated with Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 6743. [Google Scholar] [CrossRef]
- Theodoulou, F.L.; Eastmond, P.J. Seed storage oil catabolism: A story of give and take. Curr. Opin. Plant Biol. 2012, 15, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yan, C.; Roston, R.; Shanklin, J.; Xu, C. Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward beta-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 2014, 26, 4119–4134. [Google Scholar] [CrossRef] [PubMed]
- Wink, D.A.; Hanbauer, I.; Grisham, M.B.; Laval, F.; Nims, R.W.; Laval, J.; Cook, J.; Pacelli, R.; Liebmann, J.; Krishna, M.; et al. Chemical biology of nitric oxide: Regulation and protective and toxic mechanisms. Curr. Top. Cell Regul. 1996, 34, 159–187. [Google Scholar] [CrossRef]
- Lozano-Juste, J.; Colom-Moreno, R.; Leon, J. In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3501–3517. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Mata-Pérez, C.; Valderrama, R.; Padilla, M.N.; López-Jaramillo, J.; Luque, F.; Corpas, F.J.; Barroso, J.B. Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation. J. Exp. Bot. 2015, 66, 5983–5996. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, M.; González-Gordo, S.; Cañas, A.; Campos, M.J.; Paradela, A.; Corpas, F.J.; Palma, J.M. Sweet Pepper (Capsicum annuum L.) Fruits Contain an Atypical Peroxisomal Catalase That Is Modulated by Reactive Oxygen and Nitrogen Species. Antioxidants 2019, 8, 374. [Google Scholar] [CrossRef]
- Qin, X.; Wu, C.; Niu, D.; Qin, L.; Wang, X.; Wang, Q.; Li, Y. Peroxisome inspired hybrid enzyme nanogels for chemodynamic and photodynamic therapy. Nat. Commun. 2021, 12, 5243. [Google Scholar] [CrossRef]
- Shen, B.R.; Wang, L.M.; Lin, X.L.; Yao, Z.; Xu, H.W.; Zhu, C.H.; Teng, H.Y.; Cui, L.L.; Liu, E.E.; Zhang, J.J.; et al. Engineering a New Chloroplastic Photorespiratory Bypass to Increase Photosynthetic Efficiency and Productivity in Rice. Mol. Plant 2019, 12, 199–214. [Google Scholar] [CrossRef]
- Kim, M.C.; Kim, T.H.; Park, J.H.; Moon, B.Y.; Lee, C.H.; Cho, S.H. Expression of rice acyl-CoA oxidase isoenzymes in response to wounding. J. Plant Physiol. 2007, 164, 665–668. [Google Scholar] [CrossRef]
- Chou, T.S.; Chao, Y.Y.; Kao, C.H. Involvement of hydrogen peroxide in heat shock- and cadmium-induced expression of ascorbate peroxidase and glutathione reductase in leaves of rice seedlings. J. Plant Physiol. 2012, 169, 478–486. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, I.S.; Bae, M.J.; Choe, Y.H.; Kim, Y.H.; Park, H.M.; Kang, H.G.; Yoon, H.S. Homologous expression of cytosolic dehydroascorbate reductase increases grain yield and biomass under paddy field conditions in transgenic rice (Oryza sativa L. japonica). Planta 2013, 237, 1613–1625. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Kim, D.W.; Niitsu, M.; Berberich, T.; Kusano, T. Oryza sativa polyamine oxidase 1 back-converts tetraamines, spermine and thermospermine, to spermidine. Plant Cell Rep. 2014, 33, 143–151. [Google Scholar] [CrossRef] [PubMed]

| Gene Name (Arabidopsis thaliana) | Gene ID | ||
|---|---|---|---|
| Dicotyledon | Monocotyledon (Oryza sativa) | ||
| Arabidopsis thaliana | Populus trichocarpa | ||
| ACX3 | AT1G06290 [28] | Potri.019G092600 | LOC_Os06g24704 [69] |
| APX3 | AT4G35000 [58] | Potri.009G134100 | LOC_Os08g43560 [70] |
| CAT2 | AT4G35090 [43] | Potri.002G009800 | LOC_Os03g03910 [68] |
| CAT3 | AT1G20620 [3] | Potri.005G251600 | LOC_Os02g02400 |
| CSD3 | AT5G18100 [38] | Potri.019G035800 | LOC_Os07g46990 |
| CuAO2 | AT1G31710 [30] | Potri.008G151900 | LOC_Os07g38440 |
| CuAO3 | AT2G42490 [30] | Potri.015G082900 | LOC_Os04g40040 |
| DHAR1 | AT1G19570 [60] | Potri.008G049300 | LOC_Os05g02530 [71] |
| GOX1 | AT3G14420 [20] | Potri.011G112700 | LOC_Os07g05820 [54] |
| GOX2 | AT3G14415 [19] | Potri.011G112700 | LOC_Os07g05820 [54] |
| IBR3 | AT3G06810 [25] | Potri.T030600 | LOC_Os07g47820 |
| MDAR4/SDP2 | AT3G27820 [56] | Potri.001G346200 | LOC_Os02g47800 |
| PAO2 | AT2G43020 [29] | Potri.005G207300 | LOC_Os04g53190 [72] |
| PAO3 | AT3G59050 [29] | Potri.002G055300 | LOC_Os04g53190 [72] |
| PAO4 | AT1G65840 [29] | Potri.004G075800 | LOC_Os04g57560 [32] |
| UOX | AT2G26230 [13] | Potri.010G242600 | LOC_Os01g64520 |
| XDH1 | AT4G34890 [14] | Potri.009G054600 | LOC_Os03g31550 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Liu, Y.; Wang, X.; Jiang, C.; Zhao, Y.; Lu, M.; Zhang, J. Peroxisome-Mediated Reactive Oxygen Species Signals Modulate Programmed Cell Death in Plants. Int. J. Mol. Sci. 2022, 23, 10087. https://doi.org/10.3390/ijms231710087
Huang L, Liu Y, Wang X, Jiang C, Zhao Y, Lu M, Zhang J. Peroxisome-Mediated Reactive Oxygen Species Signals Modulate Programmed Cell Death in Plants. International Journal of Molecular Sciences. 2022; 23(17):10087. https://doi.org/10.3390/ijms231710087
Chicago/Turabian StyleHuang, Lichao, Yijing Liu, Xiaqin Wang, Cheng Jiang, Yanqiu Zhao, Mengzhu Lu, and Jin Zhang. 2022. "Peroxisome-Mediated Reactive Oxygen Species Signals Modulate Programmed Cell Death in Plants" International Journal of Molecular Sciences 23, no. 17: 10087. https://doi.org/10.3390/ijms231710087






