Reactive Oxygen Species and Long Non-Coding RNAs, an Unexpected Crossroad in Cancer Cells
Abstract
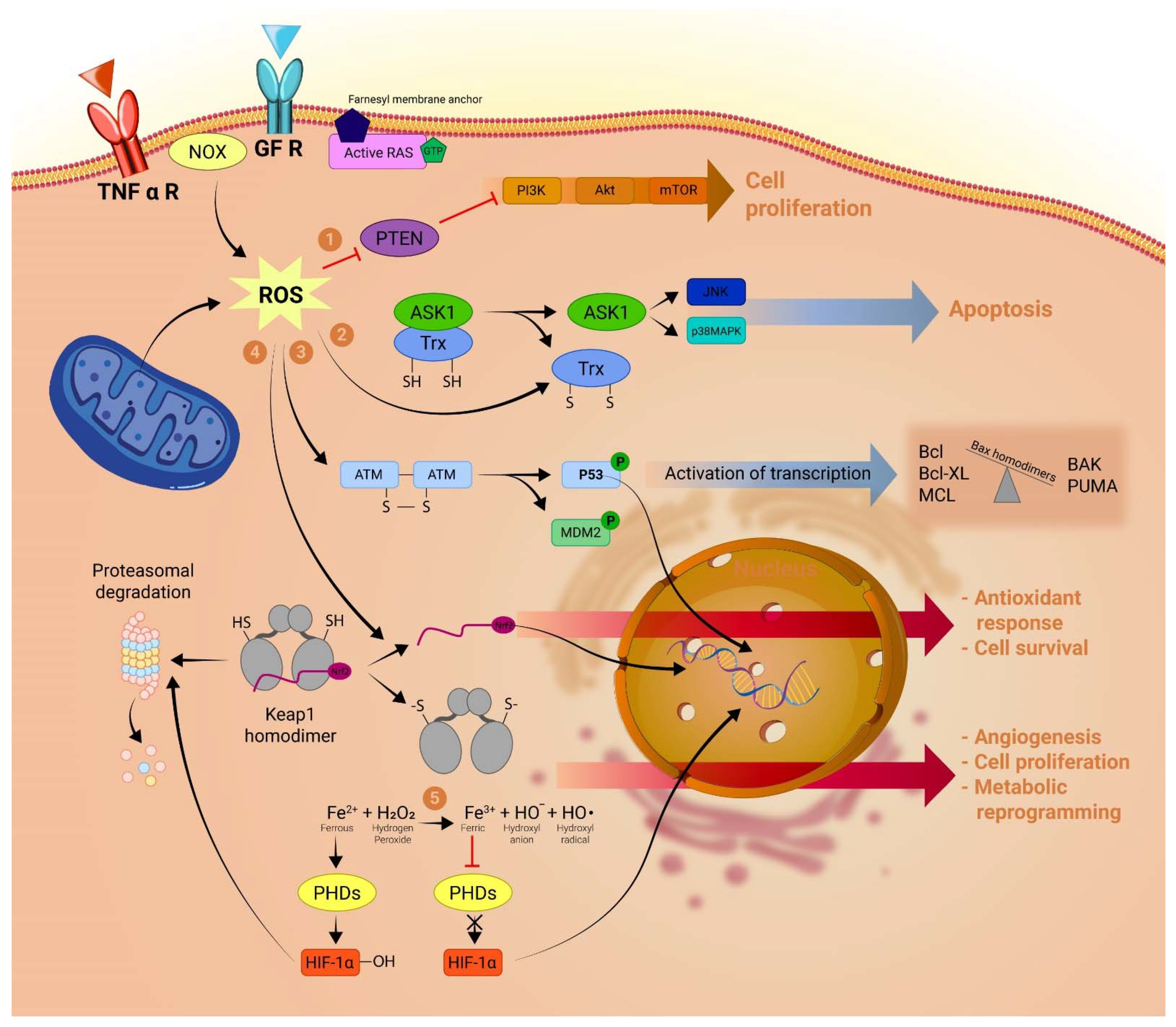
:1. Introduction
2. LncRNAs and the Generation of Mitochondrial ROS in Cancer

3. LncRNAs and Nrf2. A Key Point in Cancer Cell’s Oxidative Stress Modulation and Chemoresistance
3.1. LncRNAs as Regulators of Nrf2
3.2. LncRNAs Regulated by Nrf2
4. A LncRNA Perspective on the Warburg Effect. Tipping the Redox Balance
5. ROS and LncRNAs Determine Cancer Cell Fate. Possible Therapeutic Strategies
5.1. ROS, lncRNAs and Cell Proliferation
5.2. ROS, lncRNAs and Cell Death
5.3. The Road Forward. Possible Therapeutic Strategies
6. Conclusions and Further Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cancer over Time. Available online: https://gco.iarc.fr/overtime/en/dataviz/cohorts?populations=84000&sexes=2&cancers=0&multiple_populations=0&key=total&age_end=16&years=2017&cohort=cohort&cohort_type=time (accessed on 11 April 2022).
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer Statistics, 2010. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Hayyan, M.; Hashim, M.A.; Alnashef, I.M. Superoxide Ion: Generation and Chemical Implications. Chem. Rev. 2016, 116, 3029–3085. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Imlay, J.A.; Chin, S.M.; Linn, S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 1988, 240, 640–642. [Google Scholar] [CrossRef]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Nogueira, V.; Park, Y.; Chen, C.C.; Xu, P.Z.; Chen, M.L.; Tonic, I.; Unterman, T.; Hay, N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 2008, 14, 458. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Chen, Y.; St. Clair, D.K. ROS and p53: Versatile partnership. Free Radic. Biol. Med. 2008, 44, 1529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Murugesan, P.; Huang, K.; Cai, H. NADPH oxidases and oxidase crosstalk in cardiovascular diseases: Novel therapeutic targets. Nat. Rev. Cardiol. 2020, 17, 170–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, T.; Wu, X.; Nice, E.C.; Huang, C.; Zhang, Y. Oxidative stress and diabetes: Antioxidative strategies. Front. Med. 2020, 14, 583–600. [Google Scholar] [CrossRef]
- Hamarsheh, S.; Osswald, L.; Saller, B.S.; Unger, S.; De Feo, D.; Vinnakota, J.M.; Konantz, M.; Uhl, F.M.; Becker, H.; Lübbert, M.; et al. Oncogenic Kras G12D causes myeloproliferation via NLRP3 inflammasome activation. Nat. Commun. 2020, 11, 1659. [Google Scholar] [CrossRef]
- Stanicka, J.; Russell, E.G.; Woolley, J.F.; Cotter, T.G. NADPH oxidase-generated hydrogen peroxide induces DNA damage in mutant FLT3-expressing leukemia cells. J. Biol. Chem. 2015, 290, 9348–9361. [Google Scholar] [CrossRef]
- Szatrowski, T.P.; Nathan, C.F. Production of Large Amounts of Hydrogen Peroxide by Human Tumor Cells. Cancer Res. 1991, 51, 794–798. [Google Scholar]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.A.; Sethi, G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef]
- Lin, X.; Jia, Y.; Dong, X.; Shen, J.; Jin, Y.; Li, Y.; Wang, F.; Anenberg, E.; Zhou, J.; Zhu, J.; et al. Diplatin, a Novel and Low-Toxicity Anti-Lung Cancer Platinum Complex, Activation of Cell Death in Tumors via a ROS/JNK/p53-Dependent Pathway, and a Low Rate of Acquired Treatment Resistance. Front. Pharmacol. 2019, 10, 982. [Google Scholar] [CrossRef]
- Guo, Z.; Kozlov, S.; Lavin, M.F.; Person, M.D.; Paull, T.T. ATM activation by oxidative stress. Science 2010, 330, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yang, M.; Deng, J.; Li, P.; Su, W.; Jiang, R. Upregulation and activation of p53 by erastin-induced reactive oxygen species contribute to cytotoxic and cytostatic effects in A549 lung cancer cells. Oncol. Rep. 2018, 40, 2363–2370. [Google Scholar] [CrossRef]
- Chandra, J.; Samali, A.; Orrenius, S. Triggering and modulation of apoptosis by oxidative stress. Free Radic. Biol. Med. 2000, 29, 323–333. [Google Scholar] [CrossRef]
- Denicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106. [Google Scholar] [CrossRef] [PubMed]
- Sander, C.S.; Hamm, F.; Elsner, P.; Thiele, J.J. Oxidative stress in malignant melanoma and non-melanoma skin cancer. Br. J. Dermatol. 2003, 148, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Bisevac, J.P.; Djukic, M.; Stanojevic, I.; Stevanovic, I.; Mijuskovic, Z.; Djuric, A.; Gobeljic, B.; Banovic, T.; Vojvodic, D. Association Between Oxidative Stress and Melanoma Progression. J. Med. Biochem. 2018, 37, 12. [Google Scholar] [CrossRef]
- Davison, C.A.; Durbin, S.M.; Thau, M.R.; Zellmer, V.R.; Chapman, S.E.; Diener, J.; Wathen, C.; Leevy, W.M.; Schafer, Z.T. Antioxidant enzymes mediate survival of breast cancer cells deprived of extracellular matrix. Cancer Res. 2013, 73, 3704–3715. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Yousef, M.H.; El-Fawal, H.A.N.; Abdelnaser, A. Hepigenetics: A Review of Epigenetic Modulators and Potential Therapies in Hepatocellular Carcinoma. Biomed Res. Int. 2020, 2020, 9593254. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Liao, Y.; Gong, D.; Zhao, X.; Ji, W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018, 502, 194–201. [Google Scholar] [CrossRef]
- Yang, T.; He, X.; Chen, A.; Tan, K.; Du, X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene 2018, 670, 114–122. [Google Scholar] [CrossRef]
- Ginn, L.; Shi, L.; La Montagna, M.; Garofalo, M. LncRNAs in non-small-cell lung cancer. Non-Coding RNA 2020, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zitello, E.; Guo, R.; Deng, Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin. Transl. Med. 2021, 11, e367. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Song, G.; Zhang, X.; Gao, S.; Liu, H. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019, 450, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Esfandi, F.; Taheri, M.; Omrani, M.D.; Shadmehr, M.B.; Arsang-Jang, S.; Shams, R.; Ghafouri-Fard, S. Expression of long non-coding RNAs (lncRNAs) has been dysregulated in non-small cell lung cancer tissues. BMC Cancer 2019, 19, 222. [Google Scholar] [CrossRef]
- Takayama, K.I.; Fujimura, T.; Suzuki, Y.; Inoue, S. Identification of long non-coding RNAs in advanced prostate cancer associated with androgen receptor splicing factors. Commun. Biol. 2020, 3, 393. [Google Scholar] [CrossRef]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef]
- Jianfeng, W.; Yutao, W.; Jianbin, B. Long non-coding RNAs correlate with genomic stability in prostate cancer: A clinical outcome and survival analysis. Genomics 2021, 113, 3141–3151. [Google Scholar] [CrossRef]
- Song, Y.; Wang, R.; Li, L.W.; Liu, X.; Wang, Y.F.; Wang, Q.X.; Zhang, Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2019, 54, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Qi-Dong, X.; Yang, X.; Lu, J.L.; Liu, C.Q.; Sun, J.X.; Li, C.; Wang, S.G. Development and Validation of a Nine-Redox-Related Long Noncoding RNA Signature in Renal Clear Cell Carcinoma. Oxid. Med. Cell. Longev. 2020, 2020, 6634247. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Wang, Y.; Qi, H.; Liu, Y.; Duan, C.; Liu, X.; Xia, T.; Chen, D.; Piao, H.L.; Liu, H.X. The double-edged roles of ROS in cancer prevention and therapy. Theranostics 2021, 11, 4839–4857. [Google Scholar] [CrossRef]
- Ahmadov, U.; Picard, D.; Bartl, J.; Silginer, M.; Trajkovic-Arsic, M.; Qin, N.; Blümel, L.; Wolter, M.; Lim, J.K.M.; Pauck, D.; et al. The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 2021, 12, 885. [Google Scholar] [CrossRef]
- Zheng, P.; Xiong, Q.; Wu, Y.; Chen, Y.; Chen, Z.; Fleming, J.; Gao, D.; Bi, L.; Ge, F. Quantitative Proteomics Analysis Reveals Novel Insights into Mechanisms of Action of Long Noncoding RNA Hox Transcript Antisense Intergenic RNA (HOTAIR) in HeLa Cells. Mol. Cell. Proteomics 2015, 14, 1447. [Google Scholar] [CrossRef]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014, 7, 1–10. [Google Scholar] [CrossRef]
- Orre, C.; Dieu, X.; Guillon, J.; Gueguen, N.; Ahmadpour, S.T.; Dumas, J.F.; Khiati, S.; Reynier, P.; Lenaers, G.; Coqueret, O.; et al. The long non-coding RNA SAMMSON is a regulator of chemosensitivity and metabolic orientation in MCF-7 doxorubicin-resistant breast cancer cells. Biology 2021, 10, 1156. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiong, R.; Li, C.; Xu, M.; Guo, M. LncRNA TUG1 promotes cisplatin resistance in esophageal squamous cell carcinoma cells by regulating Nrf2. Acta Biochim. Biophys. Sin. 2019, 51, 826–833. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, G.; Cheng, H. Transcription factor Nrf2 induces the up-regulation of lncRNA TUG1 to promote progression and adriamycin resistance in urothelial carcinoma of the bladder. Cancer Manag. Res. 2019, 11, 6079. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizaveh, S.; Ashrafizadeh, M.; Zarrabi, A.; Husmandi, K.; Zabolian, A.; Shahinozzaman, M.; Aref, A.R.; Hamblin, M.R.; Nabavi, N.; Crea, F.; et al. Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett. 2021, 508, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, K.; Huang, X.; Zhao, C.; Mei, Y.; Li, X.; Jiao, L.; Yang, H. lncRNA SLC7A11-AS1 Promotes Chemoresistance by Blocking SCFβ-TRCP-Mediated Degradation of NRF2 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 2020, 19, 974–985. [Google Scholar] [CrossRef]
- Luo, P.; Wu, S.; Ji, K.; Yuan, X.; Li, H.; Chen, J.; Tian, Y.; Qiu, Y.; Zhong, X. LncRNA MIR4435-2HG mediates cisplatin resistance in HCT116 cells by regulating Nrf2 and HO-1. PLoS ONE 2020, 15, e0223035. [Google Scholar] [CrossRef]
- Wu, L.; Cai, W.; Lei, X.; Shi, K.; Lin, X.; Shi, L. NRAL mediates cisplatin resistance in hepatocellular carcinoma via miR-340-5p/Nrf2 axis. J. Cell Commun. Signal. 2019, 13, 99. [Google Scholar] [CrossRef]
- Wu, L.; Pan, C.; Wei, X.; Shi, Y.; Zheng, J.; Lin, X.; Shi, L. lncRNA KRAL reverses 5-fluorouracil resistance in hepatocellular carcinoma cells by acting as a ceRNA against miR-141. Cell Commun. Signal. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Gutschner, T.; Hämmerle, M.; Eißmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Groß, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef]
- Wang, Z.; Katsaros, D.; Biglia, N.; Shen, Y.; Fu, Y.; Loo, L.W.M.; Jia, W.; Obata, Y.; Yu, H. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res. Treat. 2018, 171, 261. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Piao, H.L.; Kim, B.J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Roche, V.; Chew, X.H.; Fadieieva, A.; Tay, Y. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer 2018, 143, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Eastlack, S.C.; Dong, S.; Mo, Y.Y.; Alahari, S.K. Expression of long noncoding RNA MALAT1 correlates with increased levels of Nischarin and inhibits oncogenic cell functions in breast cancer. PLoS ONE 2018, 13, e0198945. [Google Scholar] [CrossRef]
- Cruz, C.R.V.; Ferrer, J.L.M.; Garcia, R.L. Concomitant and decoupled effects of cigarette smoke and SCAL1 upregulation on oncogenic phenotypes and ROS detoxification in lung adenocarcinoma cells. Sci. Rep. 2021, 11, 18345. [Google Scholar] [CrossRef]
- Thai, P.; Statt, S.; Chen, C.H.; Liang, E.; Campbell, C.; Wu, R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am. J. Respir. Cell Mol. Biol. 2013, 49, 204–211. [Google Scholar] [CrossRef]
- Zhang, C.-L.; Zhu, K.-P.; Shen, G.-Q.; Zhu, Z.-S. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumor Biol. 2016, 37, 2737–2748. [Google Scholar] [CrossRef]
- Gao, M.; Zhao, B.; Chen, M.; Liu, Y.; Xu, M.; Wang, Z.; Liu, S.; Zhang, C. Nrf-2-driven long noncoding RNA ODRUL contributes to modulating silver nanoparticle-induced effects on erythroid cells. Biomaterials 2017, 130, 14–27. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, D.; Zheng, T.; Yang, G.; Wang, J.; Meng, F.; Liu, Y.; Zhang, G.; Zhang, L.; Han, J.; et al. lncRNA-SOX2OT promotes hepatocellular carcinoma invasion and metastasis through miR-122-5p-mediated activation of PKM2. Oncogenesis 2020, 9, 54. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Yan, S.; Wang, H.; Shao, X.; Xiao, M.; Yang, B.; Qin, G.; Kong, R.; Chen, R.; et al. Interactome analysis reveals that lncRNA HULC promotes aerobic glycolysis through LDHA and PKM2. Nat. Commun. 2020, 11, 3162. [Google Scholar] [CrossRef]
- Xin, X.; Wu, M.; Meng, Q.; Wang, C.; Lu, Y.; Yang, Y.; Li, X.; Zheng, Q.; Pu, H.; Gui, X.; et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 2018, 17, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon, L.M.; Gautier, M.; Allan, R.; Ilié, M.; Nottet, N.; Pons, N.; Paquet, A.; Lebrigand, K.; Truchi, M.; Fassy, J.; et al. The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA contributes to an aggressive phenotype in lung adenocarcinoma through regulation of oxidative stress. Oncogene 2019, 38, 7146–7165. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, G.; Cheng, Z.; Dai, L.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T.; et al. Knockdown of LncRNA-XIST Suppresses Proliferation and TGF-β1-Induced EMT in NSCLC Through the Notch-1 Pathway by Regulation of miR-137. Genet. Test. Mol. Biomark. 2018, 22, 333–342. [Google Scholar] [CrossRef]
- Zhou, K.; Yang, J.; Li, X.; Chen, W. Long non-coding RNA XIST promotes cell proliferation and migration through targeting miR-133a in bladder cancer. Exp. Ther. Med. 2019, 18, 3475. [Google Scholar] [CrossRef]
- Liu, J.; Yao, L.; Zhang, M.; Jiang, J.; Yang, M.; Wang, Y. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging 2019, 11, 7830–7846. [Google Scholar] [CrossRef]
- Wen, J.F.; Jiang, Y.Q.; Li, C.; Dai, X.K.; Wu, T.; Yin, W.Z. LncRNA-XIST promotes the oxidative stress-induced migration, invasion, and epithelial-to-mesenchymal transition of osteosarcoma cancer cells through miR-153-SNAI1 axis. Cell Biol. Int. 2020, 44, 1991–2001. [Google Scholar] [CrossRef]
- Chen, S.; Liang, H.; Yang, H.; Zhou, K.; Xu, L.; Liu, J.; Lai, B.; Song, L.; Luo, H.; Peng, J.; et al. LincRNa-p21: Function and mechanism in cancer. Med. Oncol. 2017, 34, 98. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, M.; Cui, X.; O’Connell, D.; Yang, Y. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022, 29, 1850–1863. [Google Scholar] [CrossRef]
- Li, X.; Sun, D.; Zhao, T.; Zhang, Z. Long non-coding RNA ROR confers arsenic trioxide resistance to HepG2 cells by inhibiting p53 expression. Eur. J. Pharmacol. 2020, 872, 172982. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Y.; Zhang, X.; Cai, H.; Zhang, C.; Qu, H.; Liu, L.; Zhang, M.; Fu, J.; Zhang, J.; et al. Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell Death Dis. 2021, 12, 90. [Google Scholar] [CrossRef]
- Zhang, N.; Zeng, X.; Sun, C.; Guo, H.; Wang, T.; Wei, L.; Zhang, Y.; Zhao, J.; Ma, X. LncRNA LINC00963 Promotes Tumorigenesis and Radioresistance in Breast Cancer by Sponging miR-324-3p and Inducing ACK1 Expression. Mol. Ther. Nucleic Acids 2019, 18, 871–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Mourtada-Maarabouni, M.; Williams, G.T. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Jin, F.; Xia, R.; Kong, R.; Li, J.; Xu, T.; Liu, Y.; Zhang, E.; Liu, X.; De, W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014, 14, 319. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Yi, Z.; Jiang, L.; Li, Y.; Han, Q.; Yang, Y.; Zhang, Q.; Yang, Z.; Kuang, Y.; et al. LncRNA GAS5 regulates redox balance and dysregulates the cell cycle and apoptosis in malignant melanoma cells. J. Cancer Res. Clin. Oncol. 2019, 145, 637. [Google Scholar] [CrossRef]
- Xu, W.; Yan, Z.; Hu, F.; Wei, W.; Yang, C.; Sun, Z. Long non-coding RNA GAS5 accelerates oxidative stress in melanoma cells by rescuing EZH2-mediated CDKN1C downregulation. Cancer Cell Int. 2020, 20, 116. [Google Scholar] [CrossRef]
- Xie, C.; Wu, W.; Tang, A.; Luo, N.; Tan, Y. IncRNA GAS5/miR-452-5p reduces oxidative stress and pyroptosis of high-glucose-stimulated renal tubular cells. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 2609–2617. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, X.; Yang, M.; Shangguan, J.; Yin, Y. GAS5 knockdown suppresses inflammation and oxidative stress induced by oxidized low-density lipoprotein in macrophages by sponging miR-135a. Mol. Cell. Biochem. 2021, 476, 949–957. [Google Scholar] [CrossRef]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta Bioenerg. 2010, 1797, 1171–1177. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. Regulation of autophagy by ROS: Physiology and pathology. Trends Biochem. Sci. 2011, 36, 30–38. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Stanicka, J.; Cotter, T.G. Subcellular localization of the FLT3-ITD oncogene plays a significant role in the production of NOX- and p22phox-derived reactive oxygen species in acute myeloid leukemia. Leuk. Res. 2017, 52, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Badana, A.K.; G, M.M.; G, S.; Malla, R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 1177271918755391. [Google Scholar] [CrossRef] [PubMed]
- Guzy, R.D.; Schumacker, P.T. Oxygen sensing by mitochondria at complex III: The paradox of increased reactive oxygen species during hypoxia. Exp. Physiol. 2006, 91, 807–819. [Google Scholar] [CrossRef]
- Waypa, G.B.; Schumacker, P.T. Oxygen sensing in hypoxic pulmonary vasoconstriction: Using new tools to answer an age-old question. Exp. Physiol. 2008, 93, 133–138. [Google Scholar] [CrossRef]
- Hoffman, D.L.; Salter, J.D.; Brookes, P.S. Response of mitochondrial reactive oxygen species generation to steady-state oxygen tension: Implications for hypoxic cell signaling. Am. J. Physiol. Hear. Circ. Physiol. 2007, 292, H101–H108. [Google Scholar] [CrossRef]
- JF, T.; A, A.; AL, L. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985, 237, 408–414. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, P.; Wang, L.; Piao, H.L.; Ma, L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim. Biophys. Sin. 2014, 46, 1–5. [Google Scholar] [CrossRef]
- Wan, Y.; Chang, H.Y. HOTAIR: Flight of noncoding RNAs in cancer metastasis. Cell Cycle 2010, 9, 3391–3392. [Google Scholar] [CrossRef]
- D’Eletto, M.; Rossin, F.; Occhigrossi, L.; Farrace, M.G.; Faccenda, D.; Desai, R.; Marchi, S.; Refolo, G.; Falasca, L.; Antonioli, M.; et al. Transglutaminase Type 2 Regulates ER-Mitochondria Contact Sites by Interacting with GRP75. Cell Rep. 2018, 25, 3573–3581.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suski, J.M.; Lebiedzinska, M.; Bonora, M.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Relation between mitochondrial membrane potential and ROS formation. Methods Mol. Biol. 2012, 810, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, S.S.; Skulachev, V.P.; Starkov, A.A. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997, 416, 15–18. [Google Scholar] [CrossRef]
- Sgarbi, G.; Barbato, S.; Costanzini, A.; Solaini, G.; Baracca, A. The role of the ATPase inhibitor factor 1 (IF1) in cancer cells adaptation to hypoxia and anoxia. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Faccenda, D.; Nakamura, J.; Gorini, G.; Dhoot, G.K.; Piacentini, M.; Yoshida, M.; Campanella, M. Control of Mitochondrial Remodeling by the ATPase Inhibitory Factor 1 Unveils a Pro-survival Relay via OPA1. Cell Rep. 2017, 18, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Santacatterina, F.; Sánchez-Cenizo, L.; Formentini, L.; Mobasher, M.A.; Casas, E.; Rueda, C.B.; Martínez-Reyes, I.; de Arenas, C.N.; García-Bermúdez, J.; Zapata, J.M.; et al. Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumor-promoter metabolic state. Oncotarget 2015, 7, 490–508. [Google Scholar] [CrossRef]
- Boveris, A.; Cadenas, E.; Stoppani, A.O. Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem. J. 1976, 156, 435–444. [Google Scholar] [CrossRef]
- Hammad, A.; Namani, A.; Elshaer, M.; Wang, X.J.; Tang, X. “NRF2 addiction” in lung cancer cells and its impact on cancer therapy. Cancer Lett. 2019, 467, 40–49. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, H.; Chen, F.; Fu, J.; Xu, Y.; Hou, Y.; Kou, H.H.; Zhai, C.; Nelson, M.B.; Zhang, Q.; et al. An overview of chemical inhibitors of the Nrf2-ARE signaling pathway and their potential applications in cancer therapy. Free Radic. Biol. Med. 2016, 99, 544–556. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Li, J.; Dashwood, R.H. Emerging crosstalk between long non-coding RNAs and Nrf2 signaling. Cancer Lett. 2020, 490, 154–164. [Google Scholar] [CrossRef]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef] [Green Version]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Zeng, R.; Zhang, R.; Song, X.; Ni, L.; Lai, Z.; Liu, C.; Ye, W. The long non-coding RNA MALAT1 activates Nrf2 signaling to protect human umbilical vein endothelial cells from hydrogen peroxide. Biochem. Biophys. Res. Commun. 2018, 495, 2532–2538. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297. [Google Scholar] [CrossRef]
- Christofk, H.R.; Vander Heiden, M.G.; Wu, N.; Asara, J.M.; Cantley, L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008, 452, 181–186. [Google Scholar] [CrossRef]
- Hitosugi, T.; Kang, S.; Vander Heiden, M.G.; Chung, T.W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z.; et al. Tyrosine phosphorylation inhibits PKM2 to promote the warburg effect and tumor growth. Sci. Signal. 2009, 2, ra73. [Google Scholar] [CrossRef]
- Anastasiou, D.; Poulogiannis, G.; Asara, J.M.; Boxer, M.B.; Jiang, J.K.; Shen, M.; Bellinger, G.; Sasaki, A.T.; Locasale, J.W.; Auld, D.S.; et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011, 334, 1278–1283. [Google Scholar] [CrossRef]
- Kirsch, M.; Groot, H. NAD(P)H, a directly operating antioxidant? FASEB J. 2001, 15, 1569–1574. [Google Scholar] [CrossRef]
- Halliwell, B. Antioxidant defence mechanisms: From the beginning to the end (of the beginning). Free Radic. Res. 1999, 31, 261–272. [Google Scholar] [CrossRef]
- Zhang, H.; Liao, Z.; Liu, F.; Su, C.; Zhu, H.; Li, Y.; Tao, R.; Liang, H.; Zhang, B.; Zhang, X. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging 2019, 11, 9111. [Google Scholar] [CrossRef]
- Dombrauckas, J.D.; Santarsiero, B.D.; Mesecar, A.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 2005, 44, 9417–9429. [Google Scholar] [CrossRef]
- Ikeda, S.; Kitadate, A.; Abe, F.; Takahashi, N.; Tagawa, H. Hypoxia-inducible KDM3A addiction in multiple myeloma. Blood Adv. 2018, 2, 323–334. [Google Scholar] [CrossRef]
- Callapina, M.; Zhou, J.; Schmid, T.; Köhl, R.; Brüne, B. NO restores HIF-1α hydroxylation during hypoxia: Role of reactive oxygen species. Free Radic. Biol. Med. 2005, 39, 925–936. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr. Opin. Cell Biol. 2009, 21, 894. [Google Scholar] [CrossRef]
- Pan, Y.; Mansfield, K.D.; Bertozzi, C.C.; Rudenko, V.; Chan, D.A.; Giaccia, A.J.; Simon, M.C. Multiple Factors Affecting Cellular Redox Status and Energy Metabolism Modulate Hypoxia-Inducible Factor Prolyl Hydroxylase Activity In Vivo and In Vitro. Mol. Cell. Biol. 2007, 27, 912–925. [Google Scholar] [CrossRef]
- Jadaliha, M.; Zong, X.; Malakar, P.; Ray, T.; Singh, D.K.; Freier, S.M.; Jensen, T.; Prasanth, S.G.; Karni, R.; Ray, P.S.; et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016, 7, 40418–40436. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, L.; Jiang, C.; Bai, J.; Chi, Y.; Zhang, H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumor Biol. 2015, 36, 9179–9188. [Google Scholar] [CrossRef]
- Li, X.; Deng, S.J.; Zhu, S.; Jin, Y.; Cui, S.P.; Chen, J.Y.; Xiang, C.; Li, Q.Y.; He, C.; Zhao, S.F.; et al. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget 2016, 7, 6000–6014. [Google Scholar] [CrossRef]
- Xue, M.; Li, X.; Li, Z.; Chen, W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumor Biol. 2014, 35, 6901–6912. [Google Scholar] [CrossRef]
- Deng, S.J.; Chen, H.Y.; Ye, Z.; Deng, S.C.; Zhu, S.; Zeng, Z.; He, C.; Liu, M.L.; Huang, K.; Zhong, J.X.; et al. Hypoxia-induced LncRNA-bx111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene 2018, 37, 5811–5828. [Google Scholar] [CrossRef]
- Cesi, G.; Walbrecq, G.; Zimmer, A.; Kreis, S.; Haan, C. ROS production induced by BRAF inhibitor treatment rewires metabolic processes affecting cell growth of melanoma cells. Mol. Cancer 2017, 16, 102. [Google Scholar] [CrossRef] [Green Version]
- Liu-Smith, F.; Dellinger, R.; Meyskens, F.L. Updates of reactive oxygen species in melanoma etiology and progression. Arch. Biochem. Biophys. 2014, 563, 51–55. [Google Scholar] [CrossRef]
- Yamaura, M.; Mitsushita, J.; Furuta, S.; Kiniwa, Y.; Ashida, A.; Goto, Y.; Shang, W.H.; Kubodera, M.; Kato, M.; Takata, M.; et al. NADPH oxidase 4 contributes to transformation phenotype of melanoma cells by regulating G2-M cell cycle progression. Cancer Res. 2009, 69, 2647–2654. [Google Scholar] [CrossRef]
- Wittgen, H.G.M.; Van Kempen, L.C.L.T. Reactive oxygen species in melanoma and its therapeutic implications. Melanoma Res. 2007, 17, 400–409. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Wu, Y.-Z.; Su, Y.-H.; Kuo, C.-Y. Stressing the Regulatory Role of Long Non-Coding RNA in the Cellular Stress Response during Cancer Progression and Therapy. Biomedicines 2022, 10, 1212. [Google Scholar] [CrossRef]
- Liu, C.; Li, G.; Yang, N.; Su, Z.; Zhang, S.; Deng, T.; Ren, S.; Lu, S.; Tian, Y.; Liu, Y.; et al. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer Cell Int. 2017, 17, 1–7. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461. [Google Scholar] [CrossRef]
- Sahu, K.; Langeh, U.; Singh, C.; Singh, A. Crosstalk between anticancer drugs and mitochondrial functions. Curr. Res. Pharmacol. Drug Discov. 2021, 2, 100047. [Google Scholar] [CrossRef]
- Dasari, S.; Bernard Tchounwou, P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364. [Google Scholar] [CrossRef]
- Mirzaei, S.; Hushmandi, K.; Zabolian, A.; Saleki, H.; Torabi, S.M.R.; Ranjbar, A.; Seyedsaleh, S.; Sharifzadeh, S.O.; Khan, H.; Ashrafizadeh, M.; et al. Elucidating Role of Reactive Oxygen Species (ROS) in Cisplatin Chemotherapy: A Focus on Molecular Pathways and Possible Therapeutic Strategies. Molecules 2021, 26, 2382. [Google Scholar] [CrossRef]
- Choi, Y.M.; Kim, H.K.; Shim, W.; Anwar, M.A.; Kwon, J.W.; Kwon, H.K.; Kim, H.J.; Jeong, H.; Kim, H.M.; Hwang, D.; et al. Mechanism of Cisplatin-Induced Cytotoxicity Is Correlated to Impaired Metabolism Due to Mitochondrial ROS Generation. PLoS ONE 2015, 10, e0135083. [Google Scholar] [CrossRef]
- Shandilya, M.; Sharma, S.; Das, P.P.; Charak, S. Molecular-Level Understanding of the Anticancer Action Mechanism of Anthracyclines. In Advances in Precision Medicine Oncology; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Beretta, G.L.; Zunino, F. Molecular mechanisms of anthracycline activity. Top. Curr. Chem. 2008, 283, 1–19. [Google Scholar] [CrossRef]
- Siddik, Z.H. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef]
- Gewirtz, D.A. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999, 57, 727–741. [Google Scholar] [CrossRef]
- Ozben, T. Oxidative stress and apoptosis: Impact on cancer therapy. J. Pharm. Sci. 2007, 96, 2181–2196. [Google Scholar] [CrossRef]
- Kim, W.; Lee, S.; Seo, D.; Kim, D.; Kim, K.; Kim, E.; Kang, J.; Seong, K.M.; Youn, H.; Youn, B. Cellular stress responses in radiotherapy. Cells 2019, 8, 1105. [Google Scholar] [CrossRef]
- Look, M.P.; Musch, E. Lipid Peroxides in the Polychemotherapy of Cancer Patients. Chemotherapy 1994, 40, 8–15. [Google Scholar] [CrossRef]
- McDonald, E.S.; Windebank, A.J. Cisplatin-Induced Apoptosis of DRG Neurons Involves Bax Redistribution and Cytochrome cRelease But Not fas Receptor Signaling. Neurobiol. Dis. 2002, 9, 220–233. [Google Scholar] [CrossRef]
- Cashman, C.R.; Höke, A. Mechanisms of distal axonal degeneration in peripheral neuropathies. Neurosci. Lett. 2015, 596, 33–50. [Google Scholar] [CrossRef] [Green Version]
- Areti, A.; Yerra, V.G.; Naidu, V.G.M.; Kumar, A. Oxidative stress and nerve damage: Role in chemotherapy induced peripheral neuropathy. Redox Biol. 2014, 2, 289–295. [Google Scholar] [CrossRef]
- Aydinoz, S.; Uzun, G.; Cermik, H.; Atasoyu, E.M.; Yildiz, S.; Karagoz, B.; Evrenkaya, R. Effects of different doses of hyperbaric oxygen on cisplatin-induced nephrotoxicity. Ren. Fail. 2007, 29, 257–263. [Google Scholar] [CrossRef]
- Baliga, R.; Zhang, Z.; Baliga, M.; Ueda, N.; Shah, S.V. Role of cytochrome P-450 as a source of catalytic iron in cisplatin-induced nephrotoxicity. Kidney Int. 1998, 54, 1562–1569. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Gutschner, T.; Richtig, G.; Haemmerle, M.; Pichler, M. From biomarkers to therapeutic targets-the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev. 2018, 37, 83–105. [Google Scholar] [CrossRef]
- Thakore, P.I.; D’Ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 12, 1143–1149. [Google Scholar] [CrossRef]
- Gutschner, T.; Baas, M.; Diederichs, S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011, 21, 1944–1954. [Google Scholar] [CrossRef]
- Goyal, A.; Myacheva, K.; Groß, M.; Klingenberg, M.; Duran Arqué, B.; Diederichs, S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017, 45, e12. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, Y.F.; Zhang, J.; Wang, Q.X.; Han, L.; Mei, M.; Kang, C.S. Targeted design and identification of AC1NOD4Q to block activity of HOTAIR by abrogating the scaffold interaction with EZH2. Clin. Epigenetics 2019, 11, 29. [Google Scholar] [CrossRef]
- Blanco, M.R.; Guttman, M. Re-evaluating the foundations of lncRNA-Polycomb function. EMBO J. 2017, 36, 964–966. [Google Scholar] [CrossRef] [Green Version]
- Portoso, M.; Ragazzini, R.; Brenčič, Ž.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017, 36, 981. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689. [Google Scholar] [CrossRef]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Kim, S.S.; Harford, J.B.; Moghe, M.; Rait, A.; Pirollo, K.F.; Chang, E.H. Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res. 2018, 46, 1424–1440. [Google Scholar] [CrossRef]
- Dhawan, M.S.; Aggarwal, R.R.; Boyd, E.; Comerford, K.; Zhang, J.; Méndez, B.; Valenzuela, P.; Grabowsky, J.; Thomas, S.; Munster, P.N. Phase 1 study of ANDES-1537: A novel antisense oligonucleotide against non-coding mitochondrial DNA in advanced solid tumors. J. Clin. Oncol. 2018, 36, 2557. [Google Scholar] [CrossRef]
- Zuo, J.; Zhang, Z.; Li, M.; Yang, Y.; Zheng, B.; Wang, P.; Huang, C.; Zhou, S. The crosstalk between reactive oxygen species and noncoding RNAs: From cancer code to drug role. Mol. Cancer 2022, 21, 30. [Google Scholar] [CrossRef]


| LncRNA | Expression in Cancer Cells | Role in Oxidative Stress Regulation | ROS Level | Impact on Cancer Cells | Refs. |
|---|---|---|---|---|---|
| lncRNA HOTAIR | ↑ (increased) in lung cancer | confers structural stability to complex III | ↓(decreased) | ↑ chemoresistance and survival | [49,50,51] |
| ↑ in glioblastoma ↑ in cervical cancer | ↑ invasion and metastasis | ||||
| lncRNA SAMMSON | ↑ in breast cancer | inhibits complex I protein transcription and translation | ↓ | ↑ chemoresistance and survival | [52] |
| lncRNA TUG1 | ↑ in oesophageal SCC | ↑ antioxidant response by potentiating Nrf2 expression expression is also stimulated by Nrf2 | ↓ | ↑ chemoresistance and survival | [53,54,55] |
| ↑ in urothelial carcinoma of the bladder | ↑ cell proliferation ↓ apoptosis | ||||
| lncRNA SLC7A11-AS1 | ↑ in pancreatic adenocarcinoma | prevents proteosomal degradation of Nrf2 | ↓ | ↑ chemoresistance | [56] |
| lncRNA MIR4435-2HG | ↑ in colon cancer | increases Nrf2 expression | ↓ | ↑ chemoresistance ↑ cell proliferation | [57] |
| lncRNA NRAL | ↑ in HCC | acts as an ceRNA by binding miRNA-miR340–5p and thereby freeing Nrf2 antioxidant effects | ↓ | ↑ chemoresistance | [58] |
| lncRNA KRAL | ↓ in HCC | acts as an ceRNA by binding miRNA miR-141 thereby freeing KEAP1 and preventing nuclear translocation of Nrf2 | ↑ | ↑ chemoresistance | [59] |
| lncRNA MALAT1 | ↑ in NSCLC | HIF 1 α dependent transcription | ↓ | ↑ cell survival and proliferation | [60,61,62,63,64,65] |
| ↓ KEAP1 activity | possible tumor suppressive role | ||||
| lncRNA SCAL1 | ↑ after exposure to cigarette smoke | effector of Nrf2 antioxidant response | ↓ | ↑cryoprotection against cigarette smoke–induced toxicity | [66,67] |
| ↑ in NSCLC | ↓ apoptosis in NSCLC | ||||
| lncRNA ODRUL | ↑ in osteosarcoma | mediates Nrf2 pro-apoptotic effects | ↓ | ↑ doxorubicin resistance | [68,69] |
| lncRNA SOX-2-OT | ↑ in HCC | inducing the PKM2 isoform of the enzyme PK | ↓ | metabolic reprogramming | [70] |
| ↑ invasion and metastasis | |||||
| lncRNA HULC | ↑ in HCC | inducing the PKM2 isoform of the enzyme PK | ↓ | metabolic reprogramming | [71,72] |
| lncRNA H19 | ↑ in HCC | decreases SOD activity via MAPK/ERK pathway | ↓ | ↑ cell viability | [33] |
| ↓ cell apoptosis | |||||
| ↑ chemoresistance | |||||
| lncRNA NLUCAT | ↑ in lung adenocarcinoma | effector of Nrf2 antioxidant response | ↓ | ↑ cell proliferation | [73] |
| ↓ cisplatin susceptibility | |||||
| lncRNA XIST | ↑ in NSCLC ↑ in osteosarcoma | acts as a sponge for miR-335, inducing an increase in SOD2 expression acts as a sponge for miR-153, increasing Snail expression | ↓ | prevents pyroptosis ↑ cell proliferation ↑ EMT | [74,75,76,77] |
| lincRNA-p21 | ↓ in NSCLC | induces apoptosis as a result of ROS-mediated p53 activation | ↑ | ↓chemoresistance ↓cell survival | [78] |
| ↓ in HCC | |||||
| ↓ in breast cancer | |||||
| lncRNA NEAT1 | ↑ in HCC | induces apoptosis as a result of ROS-mediated p53 activation | ↑ apoptosis | [79] | |
| lncRNA ROR | ↑ in HCC | Inhibits p53 activity | ↓ cell apoptosis | [80] | |
| lncRNA NORAD | ↑ in gastric cancer | increases the expression of autophagy related genes ATG-5 and ATG-12 | ↓ | ↑cell survival | [81] |
| lincRNA 00963 | ↑ in breast cancer | sponges miR324-3p | ↓ | ↑cell survival ↑ chemoresistance | [82] |
| lncRNA GAS-5 | ↓ in breast cancer | inhibits NOX4 protein expression Inhibits G6PD protein expression | ↑/↓ | ↓ tumor suppressor effect | [83,84,85,86,87,88,89] |
| ↓ in prostate cancer | |||||
| ↓ in gastric cancer | ↑cell survival and proliferation | ||||
| ↓ in melanoma |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kacso, T.P.; Zahu, R.; Tirpe, A.; Paslari, E.V.; Nuțu, A.; Berindan-Neagoe, I. Reactive Oxygen Species and Long Non-Coding RNAs, an Unexpected Crossroad in Cancer Cells. Int. J. Mol. Sci. 2022, 23, 10133. https://doi.org/10.3390/ijms231710133
Kacso TP, Zahu R, Tirpe A, Paslari EV, Nuțu A, Berindan-Neagoe I. Reactive Oxygen Species and Long Non-Coding RNAs, an Unexpected Crossroad in Cancer Cells. International Journal of Molecular Sciences. 2022; 23(17):10133. https://doi.org/10.3390/ijms231710133
Chicago/Turabian StyleKacso, Teodor Paul, Renata Zahu, Alexandru Tirpe, Elina Valeria Paslari, Andreea Nuțu, and Ioana Berindan-Neagoe. 2022. "Reactive Oxygen Species and Long Non-Coding RNAs, an Unexpected Crossroad in Cancer Cells" International Journal of Molecular Sciences 23, no. 17: 10133. https://doi.org/10.3390/ijms231710133
APA StyleKacso, T. P., Zahu, R., Tirpe, A., Paslari, E. V., Nuțu, A., & Berindan-Neagoe, I. (2022). Reactive Oxygen Species and Long Non-Coding RNAs, an Unexpected Crossroad in Cancer Cells. International Journal of Molecular Sciences, 23(17), 10133. https://doi.org/10.3390/ijms231710133









