Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model
Abstract
:1. Introduction
2. Results
2.1. Complete Prophages Are Common in P. aeruginosa Genomes
2.2. Pseudomonas Phage Lysins Are Functionally and Genetically Diverse
2.3. Expression and Antibacterial Activity of Phage Lysins: Putative Lysins Are Lysins de Facto
2.4. Pa7 and Pa119 Have Peptidoglycan Hydrolase Activity
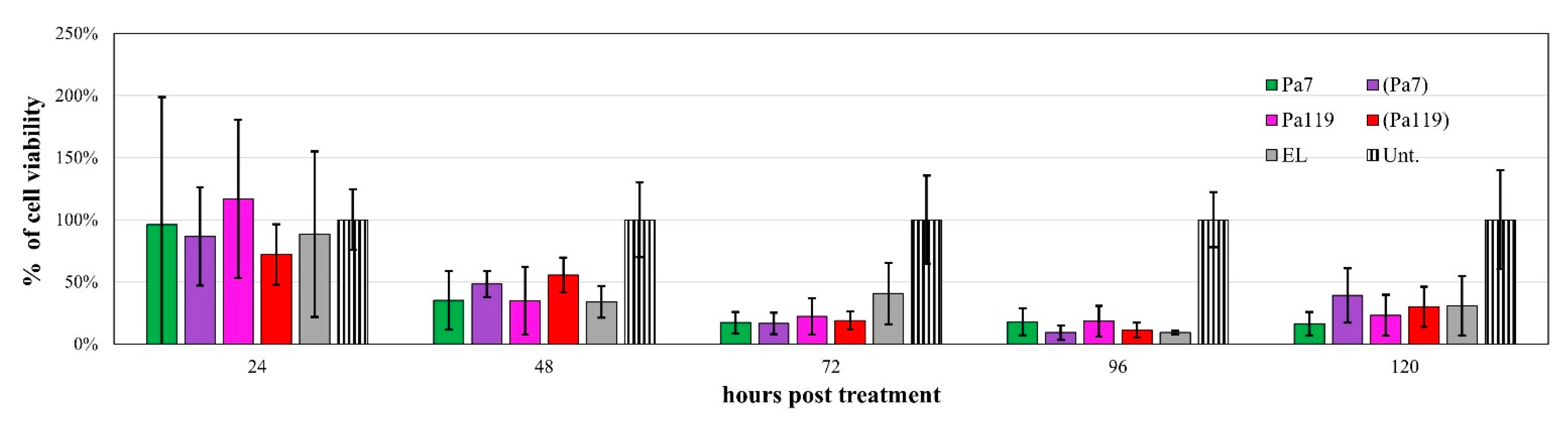
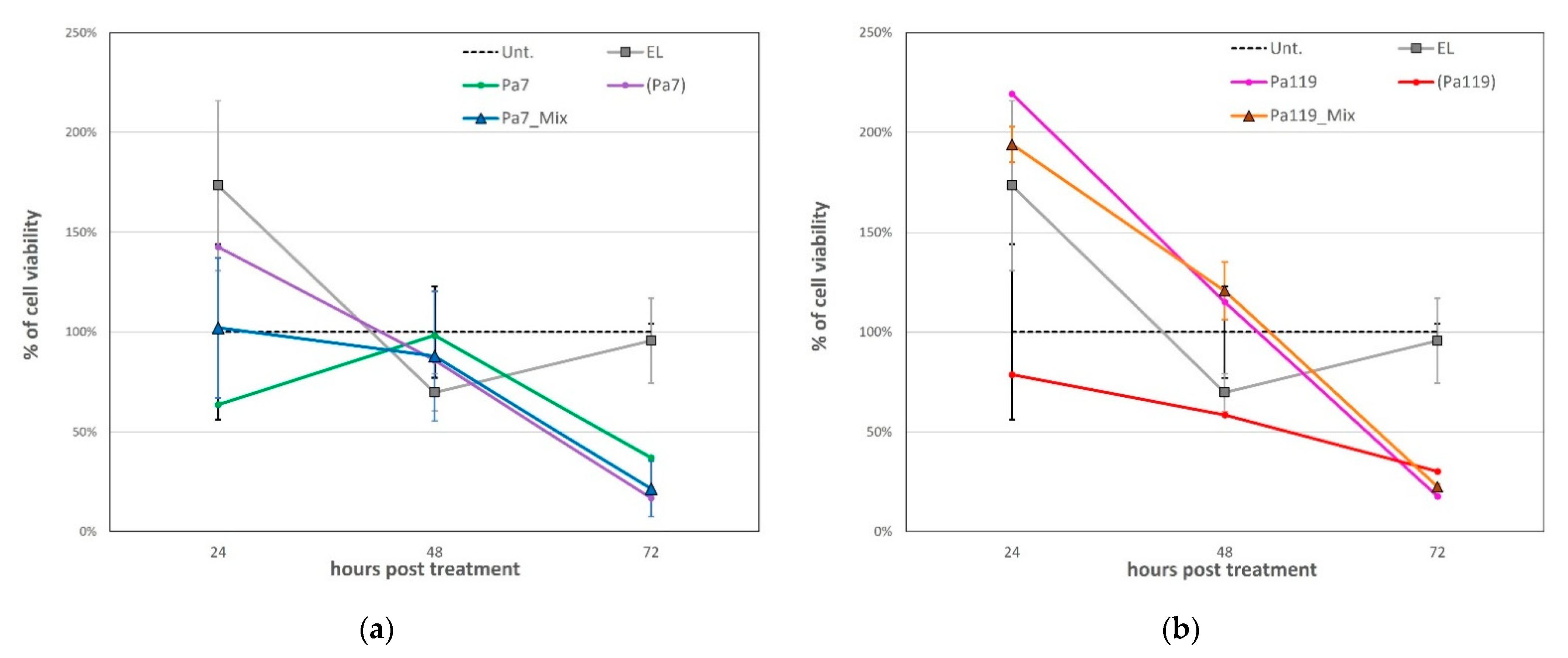
2.5. Liposome Deliver Phage Lysins to the Peptidoglycan of P. aeruginosa
3. Discussion
4. Materials and Methods
4.1. P. aeruginosa Genomes
4.2. Prophage Identification and Classification
4.3. Endolysin Identification
4.4. Lysin Genes Cloning
4.5. Protein Expression and Purification
4.6. Zymogram Analysis
4.7. Antimicrobial Activity of Endolysins
4.8. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration Determination (MBC)
4.9. Encapsulation of Selected Endolysins in Liposomes
4.10. Antimicrobial Activity of Encapsulated Endolysins
4.11. Cocktail Assays
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Wrafy, F.; Brzozowska, E.; Górska, S.; Gamian, A. Pathogenic Factors of Pseudomonas Aeruginosa-the Role of Biofilm in Pathogenicity and as a Target for Phage Therapy. Postepy Hig. I Med. Dosw. 2017, 71, 78–91. [Google Scholar] [CrossRef]
- Krylov, V.N. Chapter Five–Bacteriophages of Pseudomonas aeruginosa: Long-Term Prospects for Use in Phage Therapy. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2014; Volume 88, pp. 227–278. ISBN 9780128000984. [Google Scholar]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-garbajosa, P.; Cantón, R. Epidemiology of Antibiotic Resistance in Pseudomonas aeruginosa. Implications for Empiric and Definitive Therapy. Rev. Esp. Quimioter. 2017, 30, 8–12. [Google Scholar] [PubMed]
- Hancock, R.E.W.; Speert, D.P. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Impact on Treatment. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2000, 3, 247–255. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Heath Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Roxo-Rosa, M.; Oleastro, M.; Vale, F.F. Helicobacter pylori Eradication-the Alternatives beyond Antibiotics. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Formatex Research Center: Badajoz, Spain, 2013; pp. 1656–1667. [Google Scholar]
- Cisek, A.A.; Dabrowska, I.; Gregorczyk, K.P.; Wyzewski, Z. Phage Therapy in Bacterial Infections Treatment: One Hundred Years After the Discovery of Bacteriophages. Curr. Microbiol. 2017, 74, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, B.; Harper, D.R.; Anderson, J.; McConville, M.; Enright, M.C. Bacteriophage Therapy: Potential Uses in the Control of Antibiotic-Resistant Pathogens. Expert Rev. Anti-Infect. Ther. 2011, 9, 775–785. [Google Scholar] [CrossRef]
- Brussow, H.; Kutter, E. Phage Ecology. In Bacteriophages: Biology and Applications; CRC Press: Boca Raton, FL, USA, 2005; pp. 129–163. [Google Scholar]
- Vale, F.F.; Nunes, A.; Oleastro, M.; Gomes, J.P.; Sampaio, D.A.; Rocha, R.; Vítor, J.M.B.; Engstrand, L.; Pascoe, B.; Berthenet, E.; et al. Genomic Structure and Insertion Sites of Helicobacter pylori Prophages from Various Geographical Origins. Sci. Rep. 2017, 7, 42471. [Google Scholar] [CrossRef]
- Tanoeiro, L.; Oleastro, M.; Nunes, A.; Marques, A.T.; Duarte, S.V.; Gomes, J.P.; Matos, A.P.A.; Vítor, J.M.B.; Vale, F.F. Cryptic Prophages Contribution for Campylobacter Jejuni and Campylobacter Coli Introgression. Microorganisms 2022, 10, 516. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan Structure and Architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef]
- Vázquez, R.; García, E.; García, P. Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Front. Immunol. 2018, 9, 2252. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, N.M.D.; Parisien, A.; Lan, C.Q. Production and Application of Bacteriophage and Bacteriophage-Encoded Lysins. Recent Pat. Biotechnol. 2009, 3, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M. Agents That Increase the Permeability of the Outer Membrane. Microbiol. Rev. 1992, 56, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.-P.; et al. Engineered Endolysin-Based “Artilysins” to Combat Multidrug-Resistant Gram-Negative Pathogens. mBio 2014, 5, e01379-14. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.E.M.; Corvo, M.L.; Martins, M.B.; Simões, S.; Gaspar, M.M. Liposomes as Tools to Improve Therapeutic Enzyme Performance. Pharmaceutics 2022, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Ogren, M.; Dias, J.N.R.; Silva, M.; Gil, S.; Tavares, L.; Aires-da-Silva, F.; Gaspar, M.M.; Aguiar, S.I. Liposomes as Antibiotic Delivery Systems: A Promising Nanotechnological Strategy against Antimicrobial Resistance. Molecules 2021, 26, 2047. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, M.M.; Cruz, A.; Penha, A.F.; Reymao, J.; Sousa, A.C.; Eleuterio, C.V.; Domingues, S.A.; Fraga, A.G.; Filho, A.L.; Cruz, M.E.; et al. Rifabutin Encapsulated in Liposomes Exhibits Increased Therapeutic Activity in a Model of Disseminated Tuberculosis. Int. J. Antimicrob. Agents 2008, 31, 37–45. [Google Scholar] [CrossRef]
- Gaspar, M.M.; Calado, S.; Pereira, J.; Ferronha, H.; Correia, I.; Castro, H.; Tomas, A.M.; Cruz, M.E. Targeted Delivery of Paromomycin in Murine Infectious Diseases through Association to Nano Lipid Systems. Nanomedicine 2015, 11, 1851–1860. [Google Scholar] [CrossRef]
- Ferreira, M.; Pinto, S.N.; Aires-da-Silva, F.; Bettencourt, A.; Aguiar, S.I.; Gaspar, M.M. Liposomes as a Nanoplatform to Improve the Delivery of Antibiotics into Staphylococcus aureus Biofilms. Pharmaceutics 2021, 13, 321. [Google Scholar] [CrossRef]
- Nicolosi, D.; Scalia, M.; Nicolosi, V.M.; Pignatello, R. Encapsulation in Fusogenic Liposomes Broadens the Spectrum of Action of Vancomycin against Gram-Negative Bacteria. Int. J. Antimicrob. Agents 2010, 35, 553–558. [Google Scholar] [CrossRef] [Green Version]
- Nicolosi, D.; Cupri, S.; Genovese, C.; Tempera, G.; Mattina, R.; Pignatello, R. Nanotechnology Approaches for Antibacterial Drug Delivery: Preparation and Microbiological Evaluation of Fusogenic Liposomes Carrying Fusidic Acid. Int. J. Antimicrob. Agents 2015, 45, 622–626. [Google Scholar] [CrossRef]
- Lopes, A.; Tavares, P.; Petit, M.; Guérois, R.; Zinn-justin, S. Automated Classification of Tailed Bacteriophages According to Their Neck Organization. BMC Biotechnol. 2014, 15, 1027. [Google Scholar] [CrossRef] [PubMed]
- Mageeney, C.M.; Sinha, A.; Mosesso, R.A.; Medlin, D.L.; Lau, B.Y.; Rokes, A.B.; Lane, T.W.; Branda, S.S.; Williams, K.P. Computational Basis for On-Demand Production of Diversified Therapeutic Phage Cocktails. mSystems 2020, 5, e00659-20. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Marques, A.T.; Tanoeiro, L.; Duarte, A.; Gonçalves, L.; Vítor, J.M.B.; Vale, F.F. Genomic Analysis of Prophages from Klebsiella Pneumoniae Clinical Isolates. Microorganisms 2021, 9, 2252. [Google Scholar] [CrossRef]
- Krishnamurthy, S.R.; Wang, D. Origins and Challenges of Viral Dark Matter. Virus Res. 2017, 239, 136–142. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A More Efficient Report with Usability Improvements. Nucleic Acids Res. 2013, 41, 29–33. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Robertus, J.D.; Monzingo, A.F.; Marcotte, E.M.; Hart, P.J. Structural Analysis Shows Five Glycohydrolase Families Diverged from a Common Ancestor. J. Exp. Zool 1998, 282, 127–132. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–608. [Google Scholar] [CrossRef]
- Grela, E.; Kozłowska, J.; Grabowiecka, A. Current Methodology of MTT Assay in Bacteria—A Review. Acta Histochem. 2018, 120, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, A.B.; Stepanian, J.; Trespalacios, A.A.; Vale, F.F. Bacteriophages of Helicobacter pylori. Front. Microbiol. 2020, 11, 549084. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.; Banerjee, S.; Putonti, C. Diversity of Pseudomonas aeruginosa Temperate Phages. mSphere 2022, 7, e0101521. [Google Scholar] [CrossRef]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brussow, H. Prophage Genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef]
- Cahill, J.; Young, R. Phage Lysis: Multiple Genes for Multiple Barriers. Adv. Virus Res. 2019, 103, 33–70. [Google Scholar] [CrossRef]
- Oliveira, H.; Melo, L.D.R.; Santos, S.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular Aspects and Comparative Genomics of Bacteriophage Endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [Green Version]
- Glazko, G.; Makarenkov, V.; Liu, J.; Mushegian, A. Evolutionary History of Bacteriophages with Double-Stranded DNA Genomes. Biol. Direct 2007, 2, 36. [Google Scholar] [CrossRef]
- Yahara, K.; Lehours, P.; Vale, F.F. Analysis of Genetic Recombination and the Pan-Genome of a Highly Recombinogenic Bacteriophage Species. Microb. Genom. 2019, 5, e000282. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Lysis from Without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- O’Mahony, J.; Fenton, M.; Henry, M.; Sleator, R.D.; Coffey, A. Lysins to Kill-a Tale of Viral Weapons of Mass Destruction. Bioeng. Bugs 2011, 2, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Feng, C.; Ren, J.; Zhuang, X.; Zhang, Y.; Zhu, Y.; Dong, K.; He, P.; Guo, X.; Qin, J. A Novel Antimicrobial Endolysin, LysPA26, against Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 293. [Google Scholar] [CrossRef]
- Briers, Y.; Walmagh, M.; Lavigne, R. Use of Bacteriophage Endolysin EL188 and Outer Membrane Permeabilizers against Pseudomonas Aeruginosa. J. Appl. Microbiol. 2011, 110, 778–785. [Google Scholar] [CrossRef]
- Vázquez, R.; Blanco-Gañán, S.; Ruiz, S.; García, P. Mining of Gram-Negative Surface-Active Enzybiotic Candidates by Sequence-Based Calculation of Physicochemical Properties. Front. Microbiol. 2021, 12, 1009. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Le, S.; Shen, W.; Chen, Q.; Huang, Y.; Lu, S.; Tan, Y.; Li, M.; Hu, F.; Li, Y. Antibacterial Activity of a Lytic Enzyme Encoded by Pseudomonas aeruginosa Double Stranded RNA Bacteriophage PhiYY. Front. Microbiol. 2018, 9, 1778. [Google Scholar] [CrossRef]
- George, T.; Brady, M.F. EDTA. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Dos Santos Ramos, M.A.; Dos Santos, K.C.; da Silva, P.B.; de Toledo, L.G.; Marena, G.D.; Rodero, C.F.; de Camargo, B.A.F.; Fortunato, G.C.; Bauab, T.M.; Chorilli, M. Nanotechnological Strategies for Systemic Microbial Infections Treatment: A Review. Int. J. Pharm. 2020, 589, 119780. [Google Scholar] [CrossRef]
- Bai, J.; Yang, E.; Chang, P.-S.; Ryu, S. Preparation and Characterization of Endolysin-Containing Liposomes and Evaluation of Their Antimicrobial Activities against Gram-Negative Bacteria. Enzym. Microb. Technol. 2019, 128, 40–48. [Google Scholar] [CrossRef]
- Silva, M.D.; Paris, J.L.; Gama, F.M.; Silva, B.F.B.; Sillankorva, S. Sustained Release of a Streptococcus Pneumoniae Endolysin from Liposomes for Potential Otitis Media Treatment. ACS Infect. Dis. 2021, 7, 2127–2137. [Google Scholar] [CrossRef]
- Chirgwin, M.E.; Dedloff, M.R.; Holban, A.M.; Gestal, M.C. Novel Therapeutic Strategies Applied to Pseudomonas aeruginosa Infections in Cystic Fibrosis. Materials 2019, 12, 4093. [Google Scholar] [CrossRef] [PubMed]
- Okusanya, O.O.; Bhavnani, S.M.; Hammel, J.; Minic, P.; Dupont, L.J.; Forrest, A.; Mulder, G.-J.; Mackinson, C.; Ambrose, P.G.; Gupta, R. Pharmacokinetic and Pharmacodynamic Evaluation of Liposomal Amikacin for Inhalation in Cystic Fibrosis Patients with Chronic Pseudomonal Infection. Antimicrob. Agents Chemother. 2009, 53, 3847–3854. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Vale, F.F.; Lehours, P. Relating Phage Genomes to Helicobacter pylori Population Structure: General Steps Using Whole-Genome Sequencing Data. Int. J. Mol. Sci. 2018, 19, 1831. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.-X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An Integrative Hunting Tool for Active Prophages. Nucleic Acids Re.s 2019, 47, W74–W80. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol.Biol Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, R.; Serra, F.; Tarraga, J.; Medina, I.; Carbonell, J.; Pulido, L.; de Maria, A.; Capella-Gutierrez, S.; Huerta-Cepas, J.; Gabaldon, T.; et al. Phylemon 2.0: A Suite of Web-Tools for Molecular Evolution, Phylogenetics, Phylogenomics and Hypotheses Testing. Nucleic Acids Res. 2011, 39, W470–W474. [Google Scholar] [CrossRef]
- Froger, A.; Hall, J.E. Transformation of Plasmid DNA into E. Coli Using the Heat Shock Method. J. Vis. Exp. 2007, 6, e253. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.R. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE). In Current Protocols Essential Laboratory Techniques; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Yang, P.-C.; Mahmood, T. Western Blot: Technique, Theory, and Trouble Shooting. N. Am. J. Med. Sci. 2012, 4, 429. [Google Scholar] [CrossRef] [PubMed]
- Catalão, M.J.; Milho, C.; Gil, F.; Moniz-Pereira, J.; Pimentel, M. A Second Endolysin Gene Is Fully Embedded In-Frame with the LysA Gene of Mycobacteriophage Ms6. PLoS ONE 2011, 6, e20515. [Google Scholar] [CrossRef]
- Rodríguez-Tudela, J.L.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Cuenca-Estrella, M.; Denning, D.; Donnelly, J.P.; Dupont, B.; Fegeler, W.; Moore, C.; et al. Method for the Determination of Minimum Inhibitory Concentration (MIC) by Broth Dilution of Fermentative Yeasts. Clin. Microbiol. Infect. 2003, 9, i. [Google Scholar] [CrossRef]
- Mimoso, I.M.; Francisco, A.P.G.; Cruz, M.E.M. Liposomal Formulation of Netilmicin. Int. J. Pharm. 1997, 147, 109–117. [Google Scholar] [CrossRef]
- Lasch, J.; Weissig, V.B.M. Preparation of Liposomes. In Liposomes A Practical Approach; Torchilin, V., Weissig, V., Eds.; Oxford University Press Inc.: New York, NY, USA, 2003; pp. 3–30. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Wang, C.; Smith, R.L. Lowry Determination of Protein in the Presence of Triton X-100. Anal. Biochem. 1975, 63, 414–417. [Google Scholar] [CrossRef]
- Vale, F.F.; Vadivelu, J.; Oleastro, M.; Breurec, S.; Engstrand, L.; Perets, T.T.; Mégraud, F.; Lehours, P. Dormant Phages of Helicobacter pylori Reveal Distinct Populations in Europe. Sci. Rep. 2015, 5, 14333. [Google Scholar] [CrossRef] [Green Version]
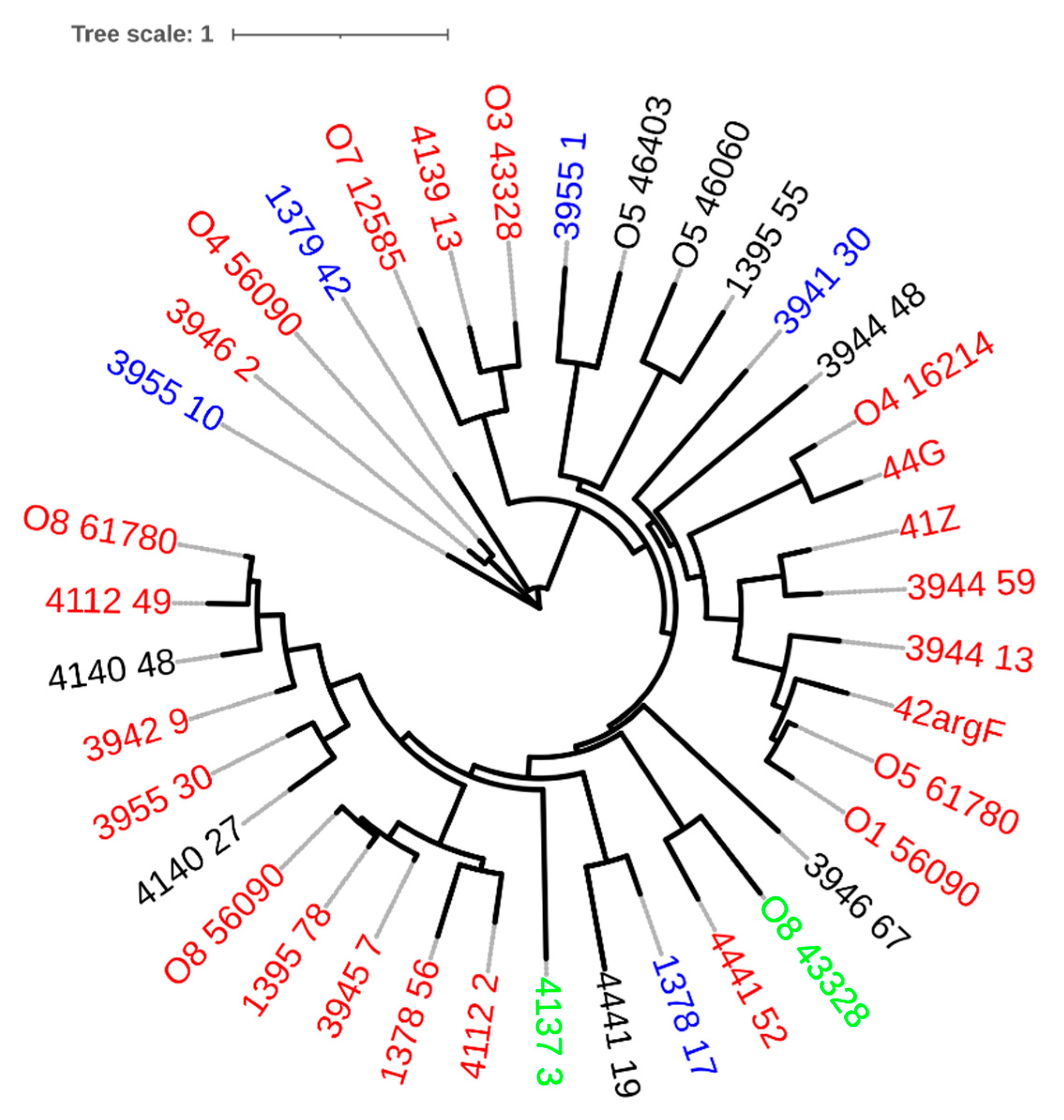
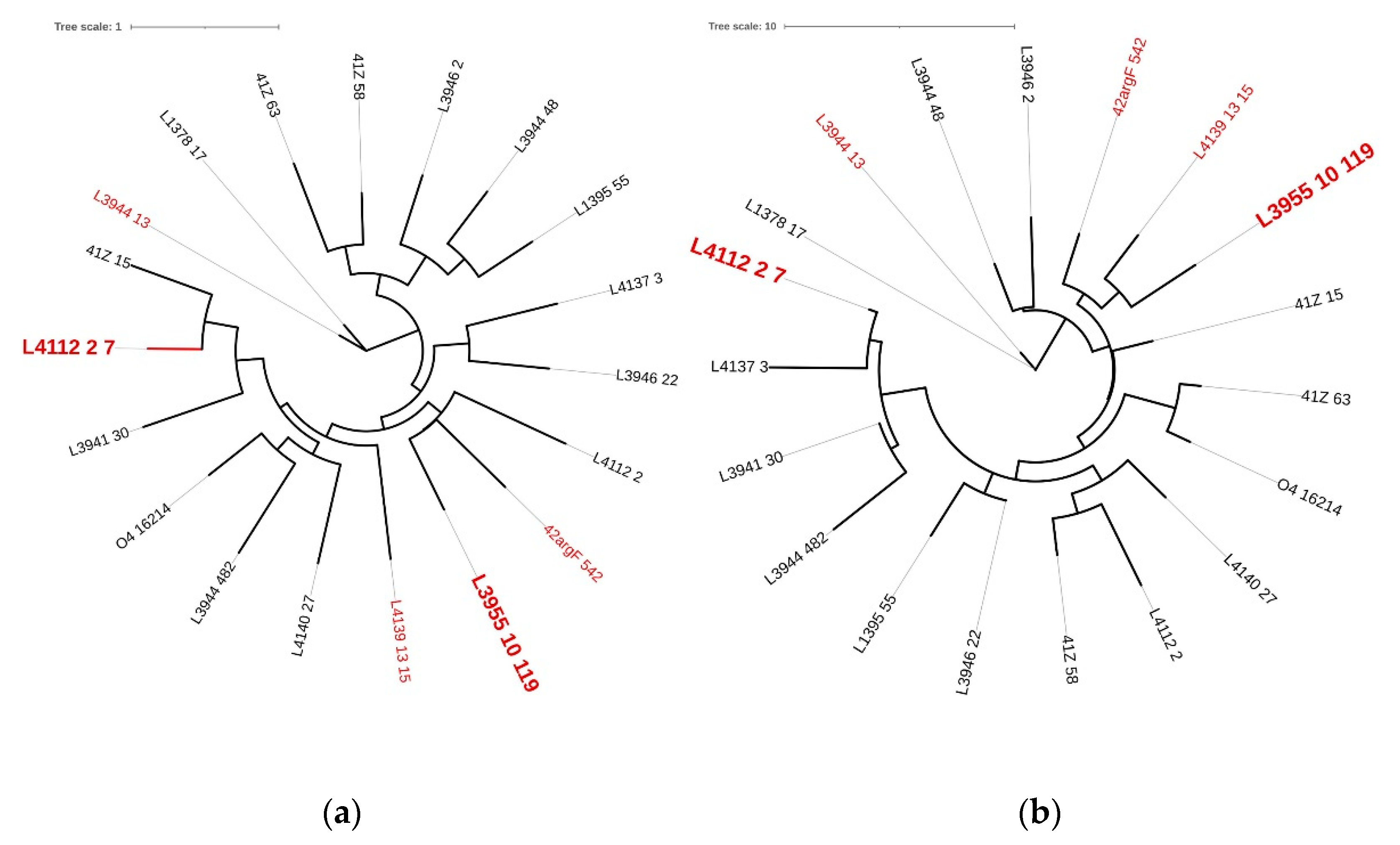
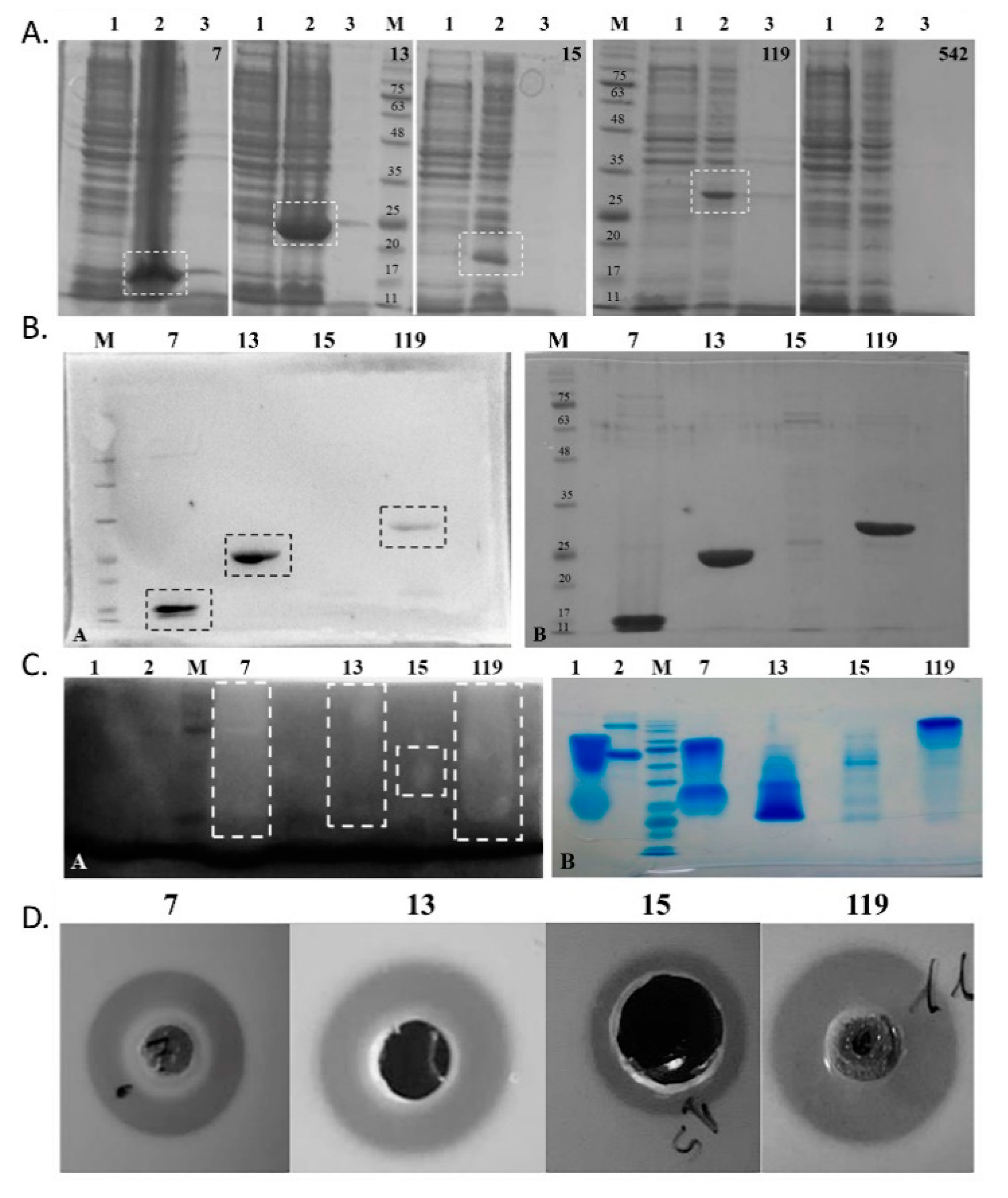
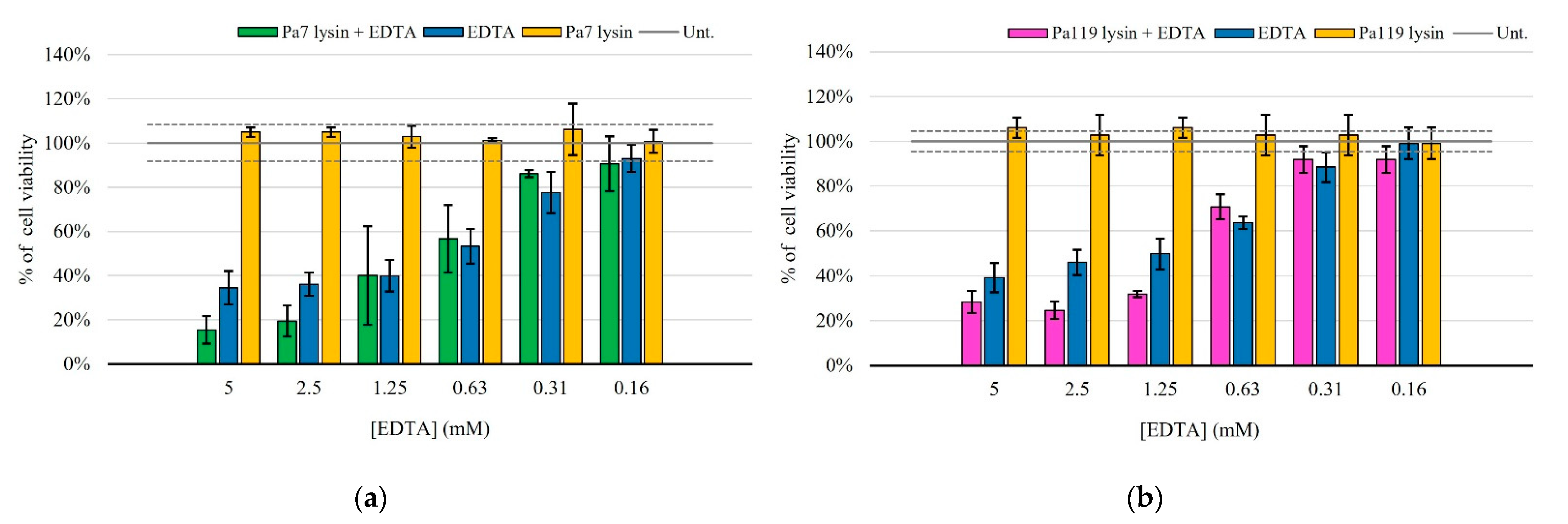


| Prophage | GC(%) | Size (kb) | CDS | Coding Region (%) | tRNA | Family | Reference |
|---|---|---|---|---|---|---|---|
| 41Z | 60.60 | 40.94 | 70 | 92.57 | 0 | Siphoviridae | [27] |
| 42argF | 61.80 | 41.59 | 67 | 92.92 | 0 | Siphoviridae | [27] |
| 44G | 63.00 | 43.67 | 54 | 90.81 | 0 | Siphoviridae | [27] |
| L1378_17 | 64.20 | 28.58 | 36 | 91.26 | 0 | Myoviridae | This study |
| L1378_56 | 57.30 | 38.16 | 50 | 86.99 | 3 | Siphoviridae | This study |
| L1379_42 | 62.00 | 37.57 | 51 | 90.39 | 0 | Myoviridae | This study |
| L1395_55 | 58.70 | 35.27 | 60 | 81.40 | 2 | ND | This study |
| L1395_78 | 60.00 | 21.74 | 25 | 84.85 | 0 | Siphoviridae | This study |
| L3941_30 | 62.50 | 31.45 | 38 | 89.85 | 0 | Myoviridae | This study |
| L3942_9 | 61.10 | 62.09 | 84 | 82.46 | 0 | Siphoviridae | This study |
| L3944_13 | 60.80 | 34.57 | 50 | 91.42 | 0 | Siphoviridae | This study |
| L3944_48 | 58.80 | 56.82 | 101 | 77.15 | 0 | ND | This study |
| L3944_59 | 59.90 | 45.34 | 68 | 88.79 | 0 | Siphoviridae | This study |
| L3945_7 | 58.90 | 33.76 | 42 | 77.92 | 0 | Siphoviridae | This study |
| L3946_2 | 64.40 | 37.21 | 57 | 95.86 | 0 | Siphoviridae | This study |
| L3946_67 | 57.30 | 30.85 | 42 | 89.90 | 3 | ND | This study |
| L3955_1 | 60.20 | 37.76 | 48 | 87.82 | 0 | Myoviridae | This study |
| L3955_10 | 62.70 | 36.34 | 45 | 88.47 | 0 | Myoviridae | This study |
| L3955_30 | 58.90 | 53.78 | 87 | 86.32 | 1 | Siphoviridae | This study |
| L4112_2 | 57.80 | 49.05 | 72 | 85.93 | 2 | Siphoviridae | This study |
| L4112_49 | 62.70 | 42.67 | 53 | 87.04 | 0 | Siphoviridae | This study |
| L4137_3 | 62.00 | 66.16 | 65 | 90.81 | 0 | Podoviridae | This study |
| L4139_13 | 62.70 | 39.72 | 55 | 95.00 | 0 | Siphoviridae | This study |
| L4140_27 | 58.70 | 28.55 | 53 | 82.15 | 1 | ND | This study |
| L4140_48 | 63.90 | 29.22 | 44 | 88.65 | 0 | ND | This study |
| L4441_19 | 60.80 | 74.64 | 77 | 84.36 | 0 | ND | This study |
| L4441_52 | 62.20 | 38.28 | 46 | 87.18 | 0 | Siphoviridae | This study |
| O4_56090 | 64.20 | 37.18 | 55 | 95.37 | 0 | Siphoviridae | [28] |
| O5_46060 | 59.80 | 58.78 | 45 | 86.48 | 0 | ND | [28] |
| O5_46403 | 55.40 | 35.8 | 34 | 88.88 | 0 | ND | [28] |
| O8_56090 | 59.00 | 60.54 | 99 | 85.63 | 2 | Siphoviridae | [28] |
| O1_56090 | 61.70 | 40.52 | 62 | 93.48 | 0 | Siphoviridae | [28] |
| O4_16214 | 60.30 | 48.74 | 53 | 74.68 | 1 | Siphoviridae | [28] |
| O3_43328 | 63.40 | 38.77 | 53 | 95.02 | 0 | Siphoviridae | [28] |
| O5_61780 | 62.30 | 39.78 | 60 | 93.77 | 0 | Siphoviridae | [28] |
| O7_12585 | 61.90 | 72.9 | 69 | 93.46 | 0 | Siphoviridae | [28] |
| O8_43328 | 64.60 | 57.23 | 66 | 92.92 | 0 | Podoviridae | [28] |
| O8_61780 | 62.10 | 53.39 | 72 | 85.91 | 0 | Siphoviridae | [28] |
| Putative Lysin | InterPro [34] | Phyre [35] | |
|---|---|---|---|
| Family | Description | Description | |
| 41Z_15 | Repressor Cro | Helix-turn-helix | Chitinase |
| 41Z_58 | - | - | Endolysin |
| 41Z_63 | - | - | Endolysin |
| 42argF_542 * | Phage lysis | Phage lytic protein Rz | Endopeptidase |
| L4112_2_7 * | Phage lysozyme | Lysozyme | Lysozyme |
| L3944_13 * | Glycosidic hydrolase family 19 | Chitinase class I | Endolysin |
| L4139_13_15 * | Transglycosylase | Transglycosylase | Transglycosylase |
| L3955_10_119 * | Muramidase | N-acetylmuramidase | Peptidoglycan binding domain |
| L1378_17 | Phage lysozyme | Lysozyme | Endolysin |
| L1395_55 | - | - | Endolysin |
| L3941_30 | Phage lysozyme | Lysozyme | Endolysin |
| L3944_48 | - | - | Endolysin |
| L3944_482 | - | - | Endolysin |
| L3946_2 | - | - | Endolysin |
| L3946_22 | - | - | Endolysin |
| L4112_2 | Endopeptidase | Endopeptidase | Lysozyme |
| L4137_3 | Phage lysozyme | Lysozyme | Lysozyme |
| L4140_27 | - | - | Transglycosylase |
| O4_16214 | Peptidase | Endopeptidase | Hydrolase |
| Lysin | BLASTp [37] | Molecular Weight (kDa) | Theoretical pI [38] | Number of Aminoacids | Zymogram (Peptidoglycan Hydrolyzing Activity) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Classification | Query Cover (%) | Identity (%) | E-Value | Predicted Protparam [38] | Experimental SDS-PAGE | M. luteus | P. aeruginosa | |||
| Pa7 | Lysozyme | 100 | 99.31 | 1.00 × 10−101 | 16.20 | 16.5 | 9.69 | 144 | yes | yes |
| Pa13 | Chitinase | 100 | 100 | 3.00 × 10−151 | 22.86 | 25.0 | 9.41 | 205 | yes | yes |
| Pa15 | Transglycosylase | 100 | 98.56 | 6.00 × 10−150 | 23.50 | 18.5 | 9.54 | 209 | yes | yes |
| Pa119 | N-acetylmuramidase | 99 | 99.25 | 0.00 | 29.30 | 30.0 | 7.13 | 268 | yes | yes |
| Pa542 | Rz lytic protein | 99 | 95.97 | 8.00 × 10−99 | 16.06 | ND | 9.23 | 150 | ND | ND |
| Lipid Composition (Molar Ratio) | Lysin | (Prot/Lip)i (μg/μmol) | (Prot/Lip)f (μg/μmol) | E.E. (%) | Mean Size (nm) | P. I. | Zeta Pot (mV) |
|---|---|---|---|---|---|---|---|
| DMPC:DOPE:CHEMS (4:4:2) | Pa7 | 16 ± 5 | 5 ± 2 | 33 ± 7 | 151 ± 6 | 0.111 ± 0.011 | −21 ± 2 |
| DMPC:DOPE:CHEMS (4:4:2) | Pa119 | 10 ± 3 | 3 ± 1 | 32 ±9 | 149 ± 4 | 0.106 ± 0.012 | −22 ± 2 |
| DMPC:DOPE:CHEMS (4:4:2) | EL | NA | NA | NA | 142 ± 2 | 0.125 ± 0.015 | −22 ± 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morais, D.; Tanoeiro, L.; Marques, A.T.; Gonçalves, T.; Duarte, A.; Matos, A.P.A.; Vital, J.S.; Cruz, M.E.M.; Carvalheiro, M.C.; Anes, E.; et al. Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model. Int. J. Mol. Sci. 2022, 23, 10143. https://doi.org/10.3390/ijms231710143
Morais D, Tanoeiro L, Marques AT, Gonçalves T, Duarte A, Matos APA, Vital JS, Cruz MEM, Carvalheiro MC, Anes E, et al. Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model. International Journal of Molecular Sciences. 2022; 23(17):10143. https://doi.org/10.3390/ijms231710143
Chicago/Turabian StyleMorais, Diana, Luís Tanoeiro, Andreia T. Marques, Tiago Gonçalves, Aida Duarte, António Pedro Alves Matos, Joana S. Vital, Maria Eugénia Meirinhos Cruz, Manuela Colla Carvalheiro, Elsa Anes, and et al. 2022. "Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model" International Journal of Molecular Sciences 23, no. 17: 10143. https://doi.org/10.3390/ijms231710143
APA StyleMorais, D., Tanoeiro, L., Marques, A. T., Gonçalves, T., Duarte, A., Matos, A. P. A., Vital, J. S., Cruz, M. E. M., Carvalheiro, M. C., Anes, E., Vítor, J. M. B., Gaspar, M. M., & Vale, F. F. (2022). Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model. International Journal of Molecular Sciences, 23(17), 10143. https://doi.org/10.3390/ijms231710143








