Serum Proteome Signatures of Anti-SARS-CoV-2 Vaccinated Healthcare Workers in Greece Associated with Their Prior Infection Status †
Abstract
:1. Introduction
2. Results
2.1. Anti-SARS-CoV-2-Induced Antibody Response
2.2. Global Proteomic Analysis of Post-COVID-19 Patients and Non-Infected Individuals Following Anti-SARS-CoV-2 Vaccination
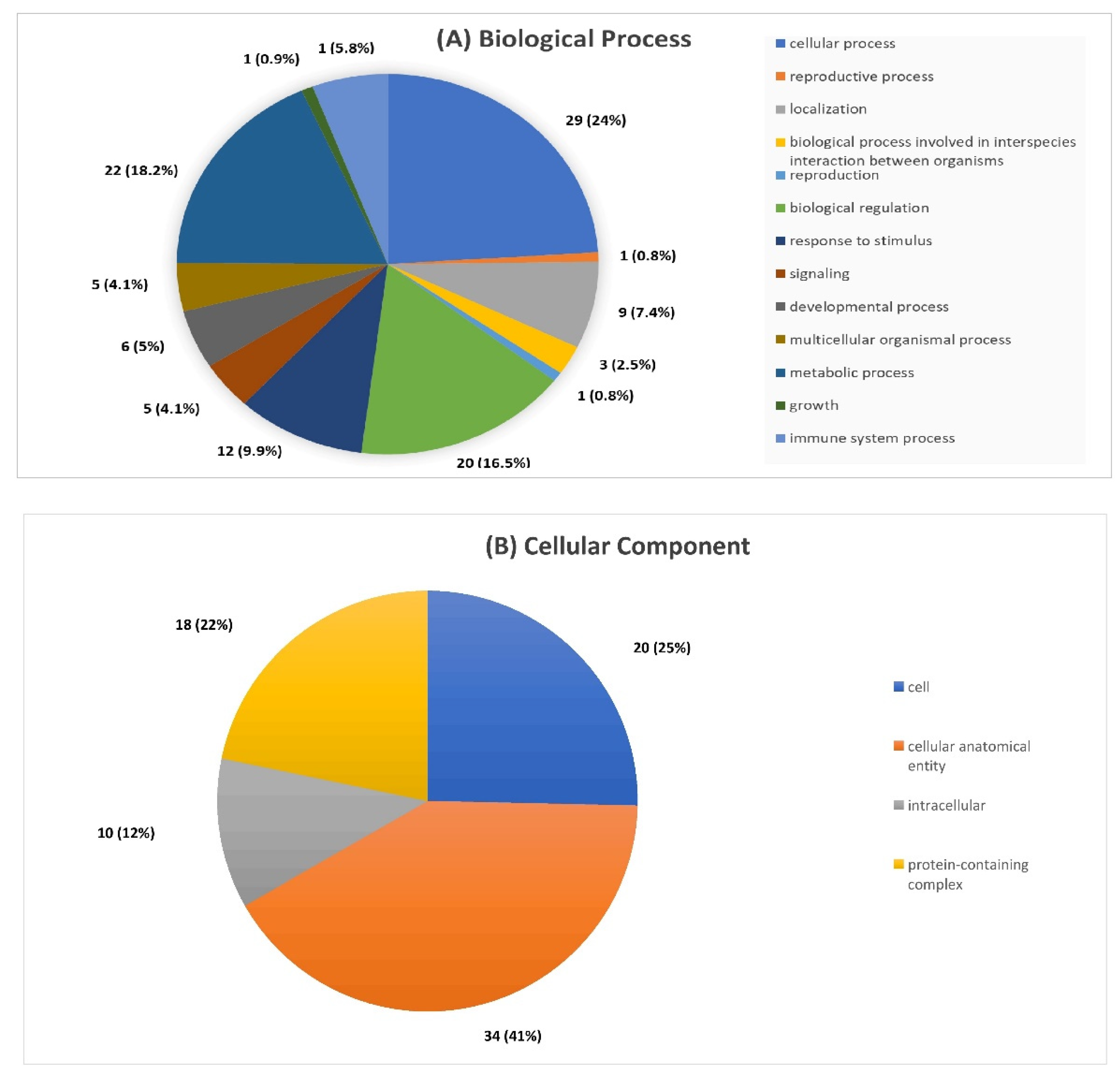
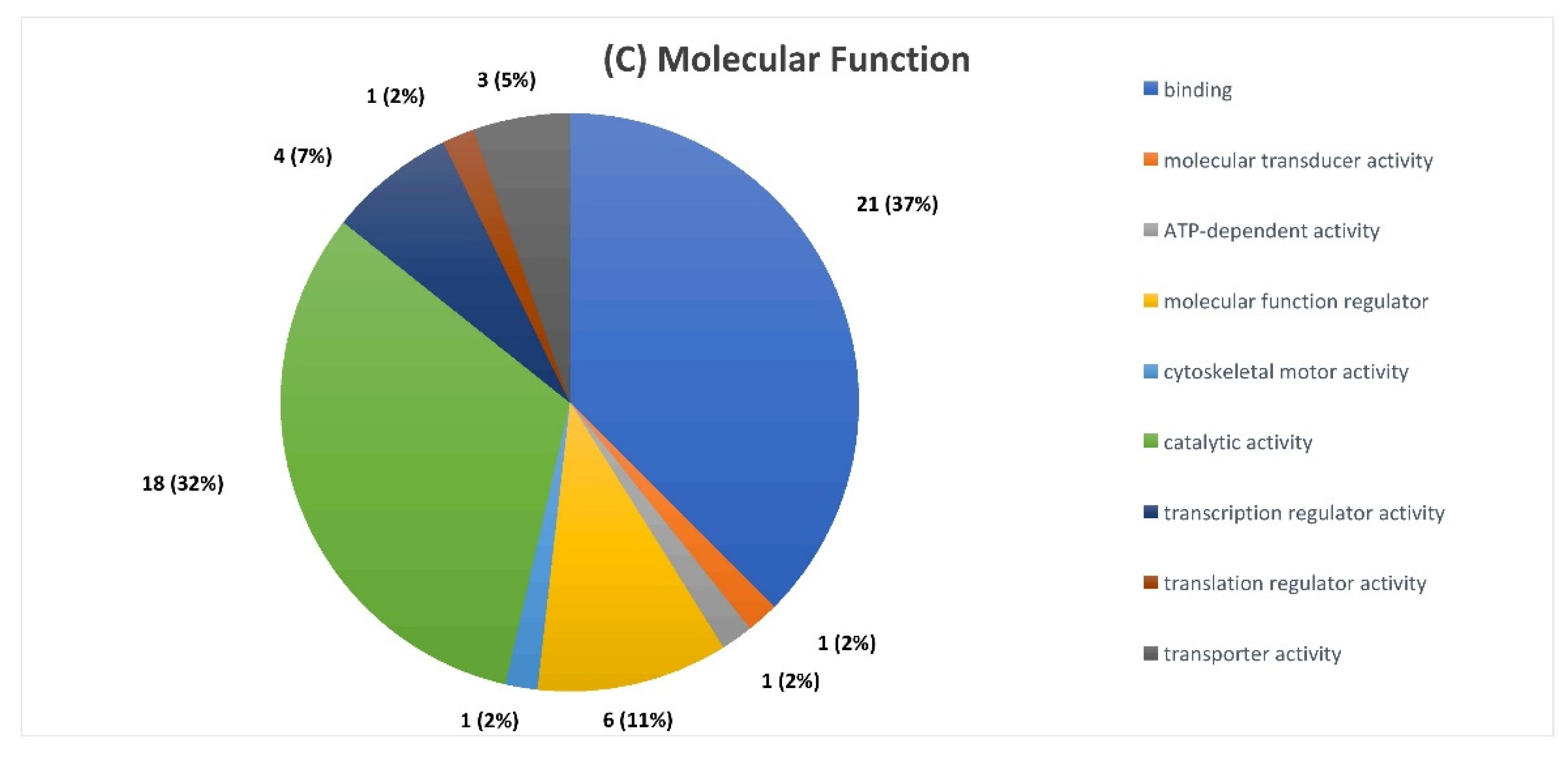
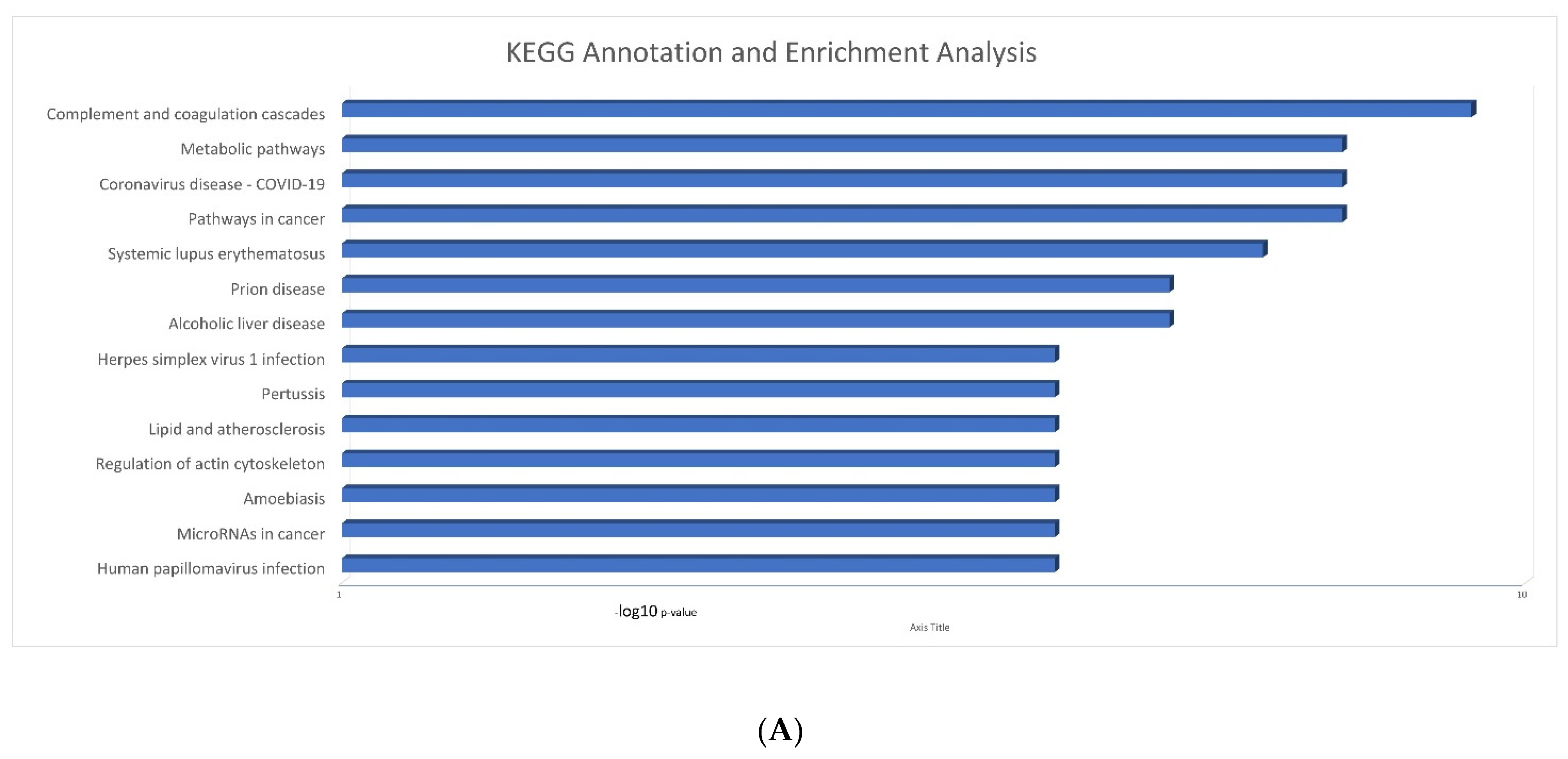
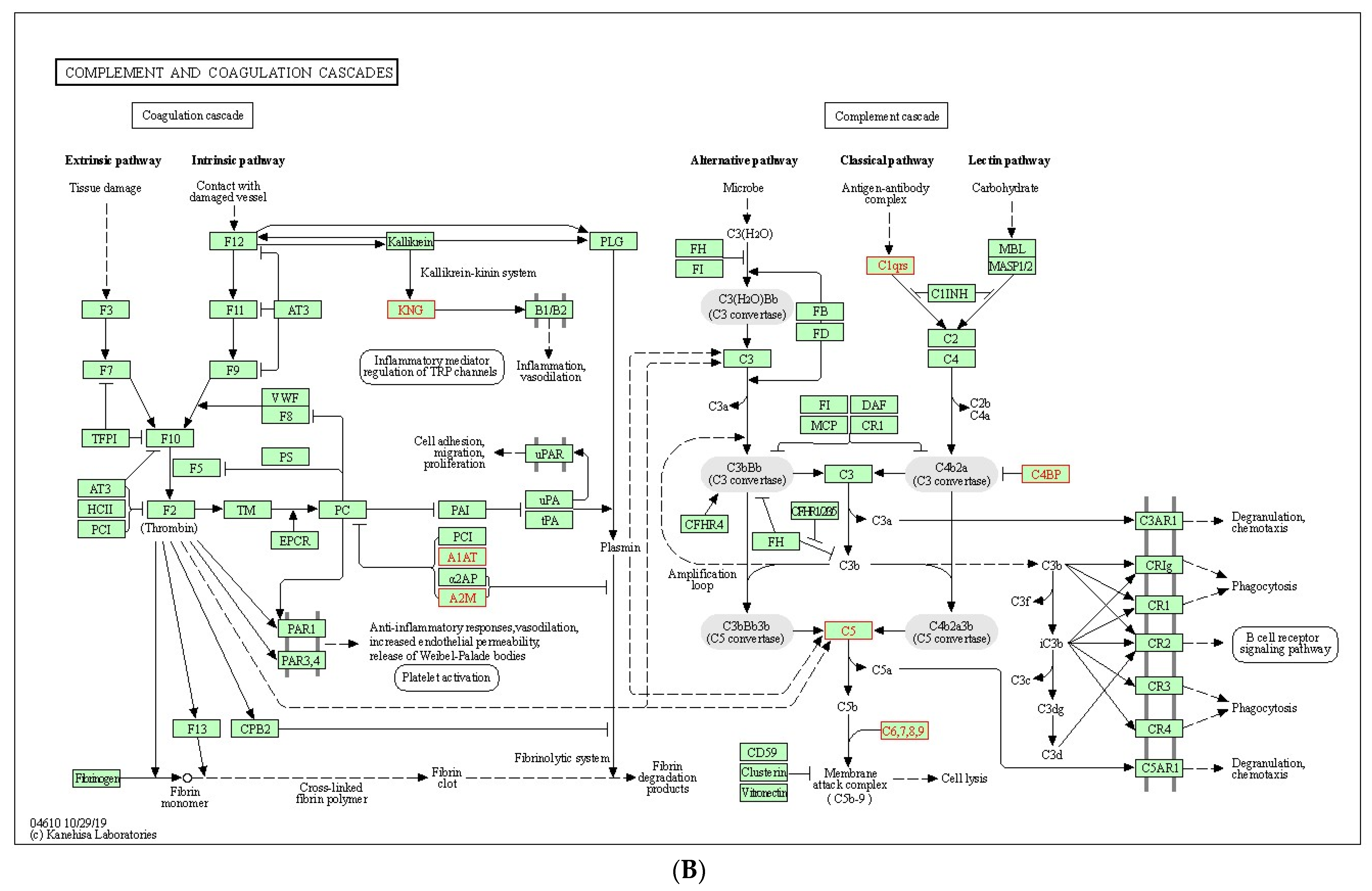
2.3. Serum Proteomic Signatures of Anti-SARS-CoV-2 Vaccination in Post-COVID-19 Patients
3. Discussion
4. Materials and Methods
4.1. Patient Samples and Experimental Design
4.2. COVID-19-Specific IgG Quantitative Determination
4.3. Proteomic Investigation
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clemente-Suárez, V.J.; Navarro-Jiménez, E.; Moreno-Luna, L.; Saavedra-Serrano, M.C.; Jimenez, M.; Simón, J.A.; Tornero-Aguilera, J.F. The impact of the COVID-19 pandemic on social, health, and economy. Sustainability 2021, 13, 6314. [Google Scholar] [CrossRef]
- Taribagil, P.; Creer, D.; Tahir, H. ‘Long COVID’ syndrome. BMJ Case Rep. 2021, 14, e241485. [Google Scholar] [CrossRef] [PubMed]
- Crook, H.; Raza, S.; Nowell, J.; Young, M.; Edison, P. Long covid—mechanisms, risk factors, and management. BMJ Case Rep. 2021, 374, n1648. [Google Scholar] [CrossRef] [PubMed]
- Iida, S.; Arashiro, T.; Suzuki, T. Insights into Pathology and Pathogenesis of Coronavirus Disease 2019 from a Histopathological and Immunological Perspective. JMA J. 2021, 4, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Chiba, T.; Kaneko, T.; Ao, J.; Kan, M.; Muroyama, R.; Nakamoto, S.; Kanda, T.; Maruyama, H.; Kato, J.; et al. The RNA-Binding Protein ELAVL1 Regulates Hepatitis B Virus Replication and Growth of Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2022, 23, 7878. [Google Scholar] [CrossRef] [PubMed]
- Lachén-Montes, M.; Corrales, F.J.; Fernández-Irigoyen, J.; Santamaría, E. Proteomics insights into the molecular basis of SARS-CoV-2 infection: What we can learn from the human olfactory Axis. Front. Microbiol. 2020, 11, 2101. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Caterino, M.; Fedele, R.; Cevenini, A.; Pontillo, M.; Barra, L.; Ruoppolo, M. COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19. Int. J. Mol. Sci. 2022, 23, 2414. [Google Scholar] [CrossRef]
- Claverie, J.M. A putative role of de-mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 Nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses 2020, 12, 646. [Google Scholar] [CrossRef]
- Kumar, V. Can proteomics-based approaches further help COVID-19 prevention and therapy? Expert Rev. Proteom. 2021, 18, 241–245. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 2020, 182, 59–72. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Deutsche COVID-19 OMICS Initiative (DeCOI). Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 2020, 182, 1419–1440. [Google Scholar] [CrossRef]
- Wu, Z.; Hu, Y.; Xu, M.; Chen, Z.; Yang, W.; Jiang, Z.; Li, M.; Jin, H.; Cui, G.; Chen, P.; et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet. Infect. Dis. 2021, 21, 803–812. [Google Scholar] [CrossRef]
- Skendros, P.; Mitsios, A.; Chrysanthopoulou, A.; Mastellos, D.C.; Metallidis, S.; Rafailidis, P.; Ntinopoulou, M.; Sertaridou, E.; Tsironidou, V.; Tsigalou, C.; et al. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Investig. 2020, 130, 6151–6157. [Google Scholar] [CrossRef]
- Holter, J.C.; Pischke, S.E.; Boer, E.; Lind, A.; Jenum, S.; Holten, A.R.; Tonby, K.; Barratt-Due, A.; Sokolova, M.; Schjalm, C.; et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl. Acad. Sci. USA 2020, 117, 25018–25025. [Google Scholar] [CrossRef]
- Yu, J.; Gerber, G.F.; Chen, H.; Yuan, X.; Chaturvedi, S.; Braunstein, E.M.; Brodsky, R.A. Complement dysregulation is associated with severe COVID-19 illness. Haematologica 2022, 107, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Alosaimi, B.; Mubarak, A.; Hamed, M.E.; Almutairi, A.Z.; Alrashed, A.A.; AlJuryyan, A.; Enani, M.; Alenzi, F.; Alturaiki, W. Complement Anaphylatoxins and Inflammatory Cytokines as Prognostic Markers for COVID-19 Severity and In-Hospital Mortality. Front. Immunol. 2021, 12, 668725. [Google Scholar] [CrossRef]
- Gauchel, N.; Rieder, M.; Krauel, K.; Goller, I.; Jeserich, M.; Salzer, U.; Venhoff, A.C.; Baldus, N.; Pollmeier, L.; Wirth, L.; et al. Complement system component dysregulation is a distinctive feature of COVID-19 disease: A prospective and comparative analysis of patients admitted to the emergency department for suspected COVID-19 disease. J. Thromb. Thrombolysis 2022, 53, 788–797. [Google Scholar] [CrossRef]
- Magro, C.; Mulvey, J.J.; Berlin, D.; Nuovo, G.; Salvatore, S.; Harp, J.; Baxter-Stoltzfus, A.; Laurence, J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 2020, 220, 1–13. [Google Scholar] [CrossRef]
- Degroote, R.L.; Uhl, P.B.; Amann, B.; Krackhardt, A.M.; Ueffing, M.; Hauck, S.M.; Deeg, C. Formin like 1 expression is increased on CD4+ T lymphocytes in spontaneous autoimmune uveitis. J. Proteom. 2017, 154, 102–108. [Google Scholar] [CrossRef]
- Thompson, S.B.; Sandor, A.M.; Lui, V.; Chung, J.W.; Waldman, M.M.; Long, R.A.; Estin, M.L.; Matsuda, J.; Friedman, R.; Jacobelli, J. Formin-like 1 mediates effector T cell trafficking to inflammatory sites to enable T cell-mediated autoimmunity. Elife 2020, 9, e58046. [Google Scholar] [CrossRef]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, Å.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, 107–113. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Idid, S.Z. Can Zn Be a Critical Element in COVID-19 Treatment? Biol. Trace Elem. Res. 2021, 199, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Arnesano, F.; Rosato, A. The Zinc Proteome of SARS-CoV-2. Metallomics 2022, 14, mfac047. [Google Scholar] [CrossRef]
- Chasapis, C.T.; Georgiopoulou, A.K.; Perlepes, S.P.; Bjørklund, G.; Peana, M. A SARS-CoV-2 -human metalloproteome interaction map. J. Inorg. Biochem. 2021, 219, 111423. [Google Scholar] [CrossRef]
- Esposito, S.; D’Abrosca, G.; Antolak, A.; Pedone, P.V.; Isernia, C.; Malgieri, G. Host and Viral Zinc-Finger Proteins in COVID-19. Int. J. Mol. Sci. 2022, 23, 3711. [Google Scholar] [CrossRef]
- Prasher, P.; Sharma, M. Mucoadhesive nanoformulations and their potential for combating COVID-19. Nanomed 2021, 16, 2497–2501. [Google Scholar] [CrossRef]
- Prasher, P.; Sharma, M. Targeting mucin hypersecretion in COVID-19 therapy. Future Med. Chem. 2021, 14, 681–684. [Google Scholar] [CrossRef]
- Kousathanas, A.; Pairo-Castineira, E.; Rawlik, K.; Stuckey, A.; Odhams, C.A.; Walker, S.; Russell, C.D.; Malinauskas, T.; Wu, Y.; Millar, J.; et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 2022, 607, 97–103. [Google Scholar] [CrossRef]
- Devadoss, D.; Acharya, A.; Manevski, M.; Houserova, D.; Cioffi, M.D.; Pandey, K.; Nair, M.; Chapagain, P.; Mirsaeidi, M.; Borchert, G.M.; et al. Immunomodulatory LncRNA on antisense strand of ICAM-1 augments SARS-CoV-2 infection-associated airway mucoinflammatory phenotype. Iscience 2022, 25, 104685. [Google Scholar] [CrossRef]
- Lee, S.; Na, H.G.; Choi, Y.S.; Bae, C.H.; Song, S.Y.; Kim, Y.D. SARS-CoV-2 Induces Expression of Cytokine and MUC5AC/5B in Human Nasal Epithelial Cell through ACE 2 Receptor. BioMed. Res. Int. 2022, 2022, 2743046. [Google Scholar] [CrossRef]
- Hafez, W.; Saleh, H.; Arya, A.; Alzouhbi, M.; Fdl, A.O.; Lal, K.; Kishk, S.; Ali, S.; Raghu, S.; Elgaili, W.; et al. Vitamin D Status in Relation to the Clinical Outcome of Hospitalized COVID-19 Patients. Front. Med. 2022, 9, 843737. [Google Scholar] [CrossRef]
- Speeckaert, M.M.; Delanghe, J.R. Commentary: Vitamin D Status in Relation to the Clinical Outcome of Hospitalized COVID-19 Patients. Front. Med. 2022, 9, 922820. [Google Scholar] [CrossRef]
- Dancer, R.C.A.; Parekh, D.; Lax, S.; D’Souza, V.; Zheng, S.; Bassford, C.R.; Park, D.; Bartis, D.G.; Mahida, R.; Turner, A.M.; et al. Vitamin D deficiency contributes directly to the acute respiratory distress syndrome (ARDS). Thorax 2015, 70, 617–624. [Google Scholar] [CrossRef]
- Kelsh, R.; You, R.; Horzempa, C.; Zheng, M.; McKeown-Longo, P.J. Regulation of the innate immune response by fibronectin: Synergism between the III-1 and EDA domains. PLoS ONE 2014, 9, e102974. [Google Scholar] [CrossRef]
- Lemańska-Perek, A.; Krzyżanowska-Gołąb, D.; Dragan, B.; Tyszko, M.; Adamik, B. Fibronectin as a Marker of Disease Severity in Critically Ill COVID-19 Patients. Cells 2022, 11, 1566. [Google Scholar] [CrossRef]
- Duan, Q.; Xia, S.; Jiao, F.; Wang, Q.; Wang, R.; Lu, L.; Jiang, S.; Xu, W. A Modified Fibronectin Type III Domain-Conjugated, Long-Acting Pan-Coronavirus Fusion Inhibitor with Extended Half-Life. Viruses 2022, 14, 655. [Google Scholar] [CrossRef]
- Nezametdinova, V.Z.; Yunes, R.A.; Dukhinova, M.S.; Alekseeva, M.G.; Danilenko, V.N. The Role of the PFNA Operon of Bifidobacteria in the Recognition of Host’s Immune Signals: Prospects for the Use of the FN3 Protein in the Treatment of COVID-19. Int. J. Mol. Sci. 2021, 22, 9219. [Google Scholar] [CrossRef]
- Plaper, T.; Aupič, J.; Dekleva, P.; Lapenta, F.; Keber, M.M.; Jerala, R.; Benčina, M. Coiled-coil heterodimers with increased stability for cellular regulation and sensing SARS-CoV-2 spike protein-mediated cell fusion. Sci. Rep. 2021, 11, 9136. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.A.; Akhter, M.S.; Kubra, K.T.; Barabutis, N. Induction of the NEK family of kinases in the lungs of mice subjected to cecal ligation and puncture model of sepsis. Tissue Barrier. 2021, 9, 1929787. [Google Scholar] [CrossRef]
- Chatterjee, B.; Thakur, S.S. SARS-CoV-2 Infection Triggers Phosphorylation: Potential Target for Anti-COVID-19 Therapeutics. Front. Immunol. 2022, 13, 829474. [Google Scholar] [CrossRef] [PubMed]
- Dimou, S.; Georgiou, X.; Sarantidi, E.; Diallinas, G.; Anagnostopoulos, A.K. Profile of Membrane Cargo Trafficking Proteins and Transporters Expressed under N Source Derepressing Conditions in Aspergillus nidulans. J. Fungi 2021, 7, 560. [Google Scholar] [CrossRef] [PubMed]



| N | % | Age | 6 Months GMC | 95% CI | ||
|---|---|---|---|---|---|---|
| 252 | 100 | 22–65 | 980.956 | (878.954–1094.794) | ||
| Male | 83 | 33 | 47.84337 | 912.77 | (730.801–1140.045) | |
| Female | 169 | 67 | 48.00599 | 1016.29 | (898.312–1149.754) | |
| Prior Status | No | 217 | 86 | 825.98 | (745.958–914.591) | |
| YES | 35 | 14 | 2848.78 | (2120.774–3826.677) |
| Num. of Total Quant. | Regulation Type | Fold-Change | ||||
|---|---|---|---|---|---|---|
| >0 | >1.2 | >1.3 | >1.5 | >2.0 | ||
| 47 | Up-regulated | 1 | 0 | 1 | 0 | 4 |
| Down-regulated | 23 | 2 | 2 | 5 | 8 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stamoula, E.; Sarantidi, E.; Dimakopoulos, V.; Ainatzoglou, A.; Dardalas, I.; Papazisis, G.; Kontopoulou, K.; Anagnostopoulos, A.K. Serum Proteome Signatures of Anti-SARS-CoV-2 Vaccinated Healthcare Workers in Greece Associated with Their Prior Infection Status. Int. J. Mol. Sci. 2022, 23, 10153. https://doi.org/10.3390/ijms231710153
Stamoula E, Sarantidi E, Dimakopoulos V, Ainatzoglou A, Dardalas I, Papazisis G, Kontopoulou K, Anagnostopoulos AK. Serum Proteome Signatures of Anti-SARS-CoV-2 Vaccinated Healthcare Workers in Greece Associated with Their Prior Infection Status. International Journal of Molecular Sciences. 2022; 23(17):10153. https://doi.org/10.3390/ijms231710153
Chicago/Turabian StyleStamoula, Eleni, Eleana Sarantidi, Vasilis Dimakopoulos, Alexandra Ainatzoglou, Ioannis Dardalas, Georgios Papazisis, Konstantina Kontopoulou, and Athanasios K. Anagnostopoulos. 2022. "Serum Proteome Signatures of Anti-SARS-CoV-2 Vaccinated Healthcare Workers in Greece Associated with Their Prior Infection Status" International Journal of Molecular Sciences 23, no. 17: 10153. https://doi.org/10.3390/ijms231710153
APA StyleStamoula, E., Sarantidi, E., Dimakopoulos, V., Ainatzoglou, A., Dardalas, I., Papazisis, G., Kontopoulou, K., & Anagnostopoulos, A. K. (2022). Serum Proteome Signatures of Anti-SARS-CoV-2 Vaccinated Healthcare Workers in Greece Associated with Their Prior Infection Status. International Journal of Molecular Sciences, 23(17), 10153. https://doi.org/10.3390/ijms231710153








