Novel Roles of MT1-MMP and MMP-2: Beyond the Extracellular Milieu
Abstract
:1. Introduction
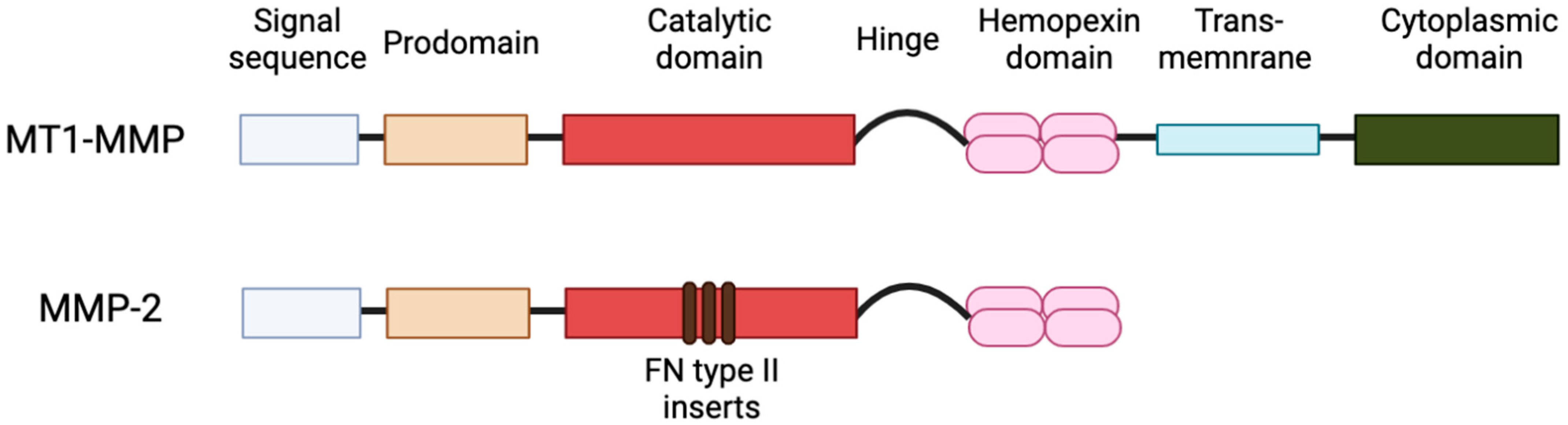
2. Subcellular Localization of MT1-MMP
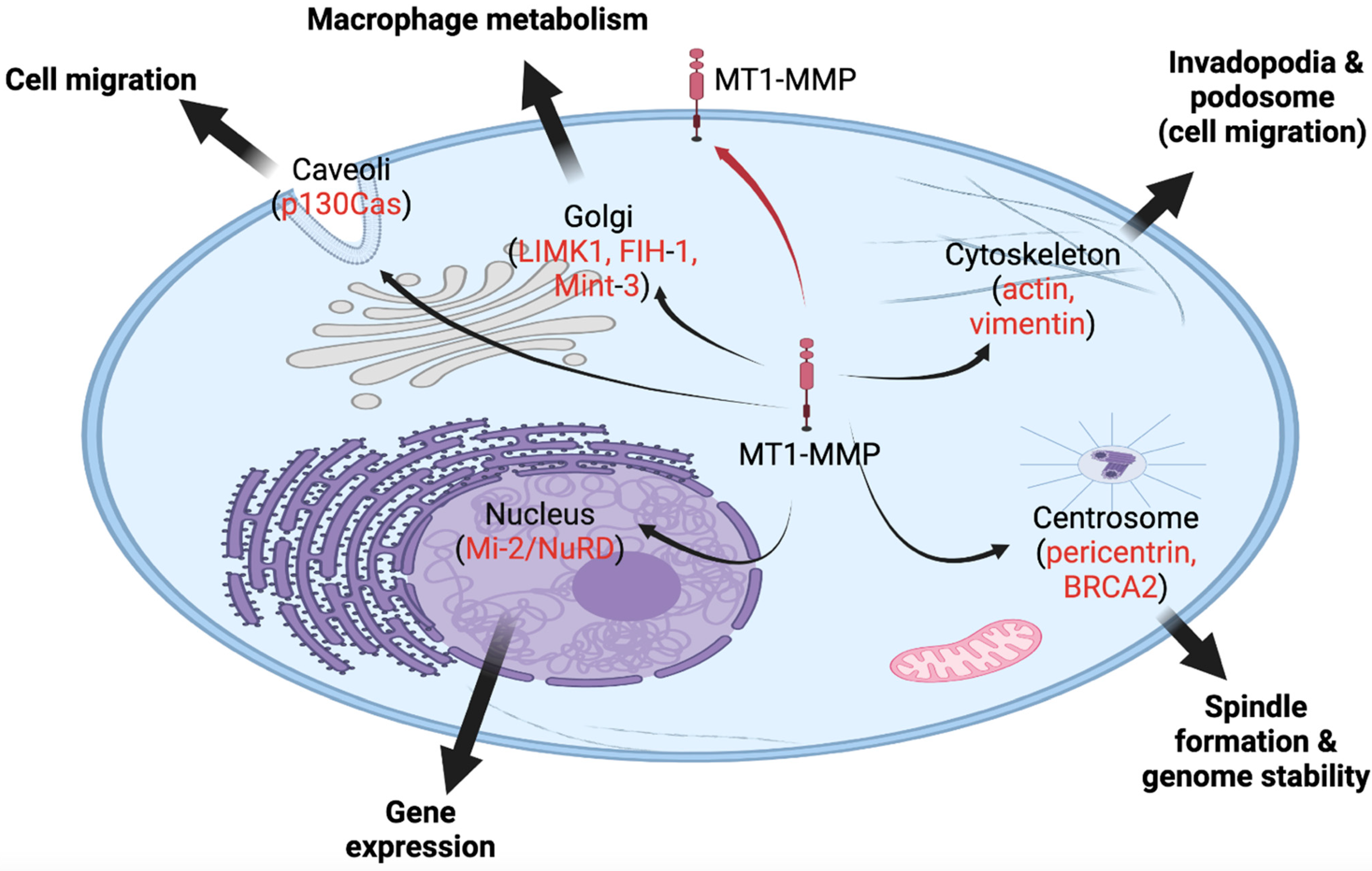
3. Novel Roles of MT1-MMP inside Cells
4. Novel Role in Centrosome Function
5. Novel Role in Migration
6. MT1-MMP Interacts with Tumor Suppressors
7. Novel Role of Caveolae in MT1-MMP Recycling
8. Novel Role of MT1-MMP in Invadopodia and Podosome Formation
9. Non-Proteolytic Role in Macrophage Regulation and Metabolism
10. Novel Role in Gene Regulation
11. Subcellular Localizations of MMP-2
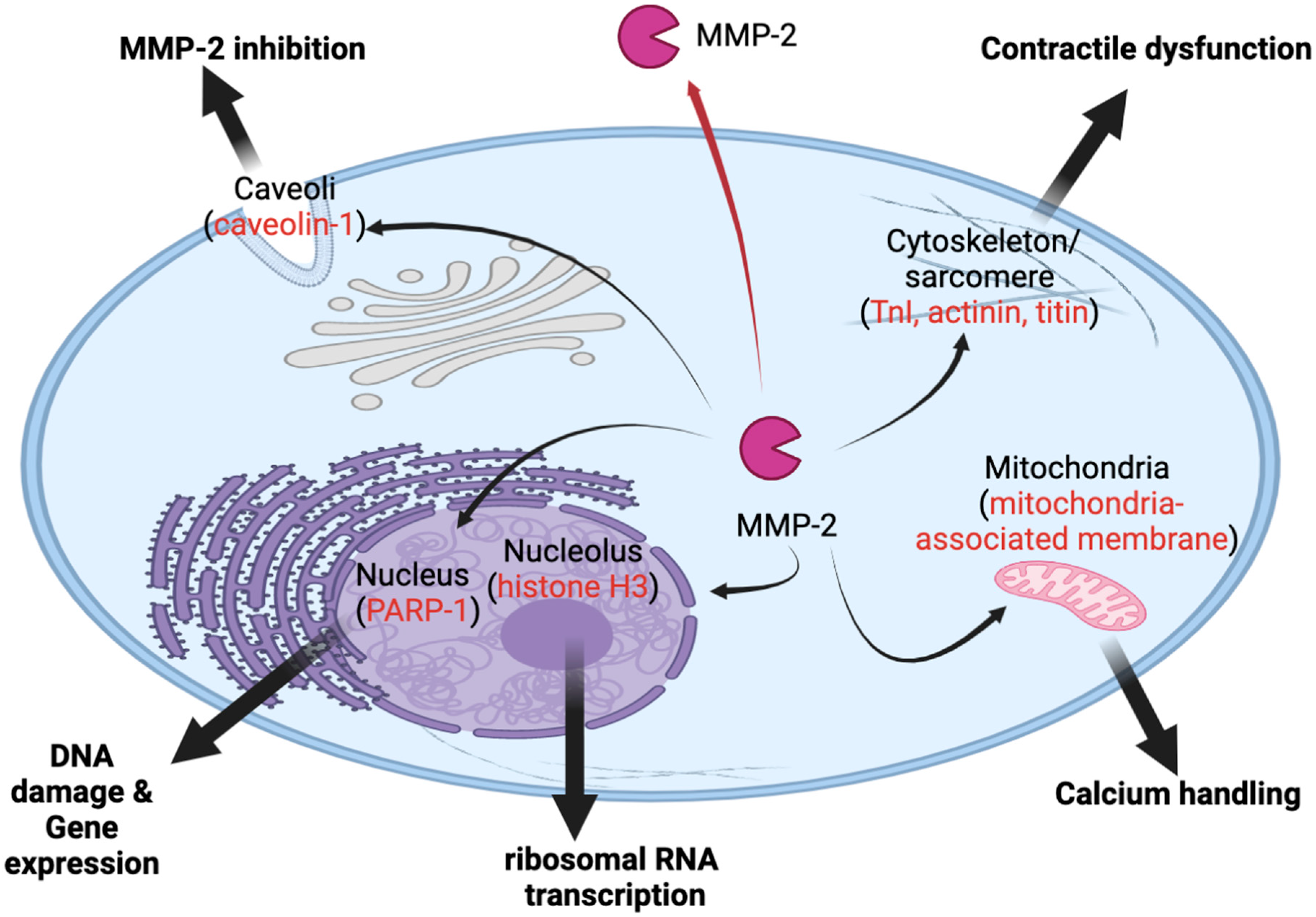
12. Novel Roles of MMP-2 inside Cells
13. Novel Role in Cardiac Dysfunction
14. Novel Roles of MMP-2 inside the Nucleus
15. Novel Role in Ribosomal RNA Transcription
16. Potential Interaction of MT1-MMP and MMP-2 inside the Cell
17. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bassiouni, W.; Ali, M.A.M.; Schulz, R. Multifunctional Intracellular Matrix Metalloproteinases: Implications in Disease. FEBS J. 2021, 288, 7162–7182. [Google Scholar] [CrossRef] [PubMed]
- Woessner, J.F.; Taplin, C.J. Purification and Properties of a Small Latent Matrix Metalloproteinase of the Rat Uterus. J. Biol. Chem. 1988, 263, 16918–16925. [Google Scholar] [CrossRef]
- Cross, J.B.; Duca, J.S.; Kaminski, J.J.; Madison, V.S. The Active Site of a Zinc-Dependent Metalloproteinase Influences the Computed pKa of Ligands Coordinated to the Catalytic Zinc Ion. J. Am. Chem. Soc. 2002, 124, 11004–11007. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.P.; Patterson, N.L.; Fields, G.B.; Lindsey, M.L. The History of Matrix Metalloproteinases: Milestones, Myths, and Misperceptions. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H919–H930. [Google Scholar] [CrossRef] [Green Version]
- Konttinen, Y.T.; Ainola, M.; Valleala, H.; Ma, J.; Ida, H.; Mandelin, J.; Kinne, R.W.; Santavirta, S.; Sorsa, T.; Lopez-Otin, C.; et al. Analysis of 16 Different Matrix Metalloproteinases (MMP-1 to MMP-20) in the Synovial Membrane: Different Profiles in Trauma and Rheumatoid Arthritis. Ann. Rheum. Dis. 1999, 58, 691–697. [Google Scholar] [CrossRef]
- Rozanov, D.V.; Ghebrehiwet, B.; Postnova, T.I.; Eichinger, A.; Deryugina, E.I.; Strongin, A.Y. The Hemopexin-like C-Terminal Domain of Membrane Type 1 Matrix Metalloproteinase Regulates Proteolysis of a Multifunctional Protein, GC1qR. J. Biol. Chem. 2002, 277, 9318–9325. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.-H.; Huang, Y.-H.; Cunningham, C.M.; Han, K.-Y.; Chang, M.; Seiki, M.; Zhou, Z.; Azar, D.T. Matrix Metalloproteinase 14 Modulates Signal Transduction and Angiogenesis in the Cornea. Surv. Ophthalmol. 2016, 61, 478–497. [Google Scholar] [CrossRef] [PubMed]
- Snyman, C.; Niesler, C.U. MMP-14 in Skeletal Muscle Repair. J. Muscle Res. Cell Motil. 2015, 36, 215–225. [Google Scholar] [CrossRef]
- Davis, M.E.; Gumucio, J.P.; Sugg, K.B.; Bedi, A.; Mendias, C.L. MMP Inhibition as a Potential Method to Augment the Healing of Skeletal Muscle and Tendon Extracellular Matrix. J. Appl. Physiol. 2013, 115, 884–891. [Google Scholar] [CrossRef] [Green Version]
- Stevens, L.J.; Page-McCaw, A. A Secreted MMP Is Required for Reepithelialization during Wound Healing. MBoC 2012, 23, 1068–1079. [Google Scholar] [CrossRef]
- Thompson, M.M.; Squire, I.B. Matrix Metalloproteinase-9 Expression after Myocardial Infarction: Physiological or Pathological? Cardiovasc. Res. 2002, 54, 495–498. [Google Scholar] [CrossRef]
- Jobin, P.G.; Butler, G.S.; Overall, C.M. New Intracellular Activities of Matrix Metalloproteinases Shine in the Moonlight. Biochim. Biophys. Acta. Mol. Cell Res. 2017, 1864, 2043–2055. [Google Scholar] [CrossRef] [PubMed]
- Laronha, H.; Caldeira, J. Structure and Function of Human Matrix Metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef]
- Krampert, M.; Bloch, W.; Sasaki, T.; Bugnon, P.; Rülicke, T.; Wolf, E.; Aumailley, M.; Parks, W.C.; Werner, S. Activities of the Matrix Metalloproteinase Stromelysin-2 (MMP-10) in Matrix Degradation and Keratinocyte Organization in Wounded Skin. MBoC 2004, 15, 5242–5254. [Google Scholar] [CrossRef] [Green Version]
- Forsyth, P.A.; Wong, H.; Laing, T.D.; Rewcastle, N.B.; Morris, D.G.; Muzik, H.; Leco, K.J.; Johnston, R.N.; Brasher, P.M.A.; Sutherland, G.; et al. Gelatinase-A (MMP-2), Gelatinase-B (MMP-9) and Membrane Type Matrix Metalloproteinase-1 (MT1-MMP) Are Involved in Different Aspects of the Pathophysiology of Malignant Gliomas. Br. J. Cancer 1999, 79, 1828–1835. [Google Scholar] [CrossRef]
- Dean, R.A.; Cox, J.H.; Bellac, C.L.; Doucet, A.; Starr, A.E.; Overall, C.M. Macrophage-Specific Metalloelastase (MMP-12) Truncates and Inactivates ELR+ CXC Chemokines and Generates CCL2, -7, -8, and -13 Antagonists: Potential Role of the Macrophage in Terminating Polymorphonuclear Leukocyte Influx. Blood 2008, 112, 3455–3464. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, D.; Kirkbride, K.C.; Costello, K.; Clark, E.S.; Sinha, S.; Grega-Larson, N.; Tyska, M.J.; Weaver, A.M. Exosome Secretion Is Enhanced by Invadopodia and Drives Invasive Behavior. Cell Rep. 2013, 5, 1159–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limb, G.A.; Matter, K.; Murphy, G.; Cambrey, A.D.; Bishop, P.N.; Morris, G.E.; Khaw, P.T. Matrix Metalloproteinase-1 Associates with Intracellular Organelles and Confers Resistance to Lamin A/C Degradation during Apoptosis. Am. J. Pathol. 2005, 166, 1555–1563. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Schulze, C.J.; Suarez-Pinzon, W.L.; Dyck, J.R.B.; Sawicki, G.; Schulz, R. Intracellular Action of Matrix Metalloproteinase-2 Accounts for Acute Myocardial Ischemia and Reperfusion Injury. Circulation 2002, 106, 1543–1549. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.H.; Kim, E.-M.; Son, H.J.; Joh, T.H.; Kim, Y.S.; Kim, D.; Flint Beal, M.; Hwang, O. A Novel Intracellular Role of Matrix Metalloproteinase-3 during Apoptosis of Dopaminergic Cells. J. Neurochem. 2008, 106, 405–415. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, J.; Li, X.; Meng, X.; Fang, X. Identification of the Endoplasmic Reticulum Localization Sequence and N -Glycosylation of Matrix Metalloproteinase 26. RSC Adv. 2019, 9, 23053–23060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.A.M.; Chow, A.K.; Kandasamy, A.D.; Fan, X.; West, L.J.; Crawford, B.D.; Simmen, T.; Schulz, R. Mechanisms of Cytosolic Targeting of Matrix Metalloproteinase-2. J. Cell Physiol. 2012, 227, 3397–3404. [Google Scholar] [CrossRef] [PubMed]
- Lovett, D.H.; Mahimkar, R.; Raffai, R.L.; Cape, L.; Maklashina, E.; Cecchini, G.; Karliner, J.S. A Novel Intracellular Isoform of Matrix Metalloproteinase-2 Induced by Oxidative Stress Activates Innate Immunity. PLoS ONE 2012, 7, e34177. [Google Scholar] [CrossRef] [PubMed]
- Arai, A.L.; Migliorini, M.; Au, D.T.; Hahn-Dantona, E.; Peeney, D.; Stetler-Stevenson, W.G.; Muratoglu, S.C.; Strickland, D.K. High-Affinity Binding of LDL Receptor-Related Protein 1 to Matrix Metalloprotease 1 Requires Protease:Inhibitor Complex Formation. Biochemistry 2020, 59, 2922–2933. [Google Scholar] [CrossRef]
- Eshaq, R.S.; Harris, N.R. The Role of Matrix Metalloproteinase 2 (MMP2) In Diabetes-induced Loss of PECAM-1 in The Retina: Direct and Indirect Mechanisms. FASEB J. 2018, 32, 706.2. [Google Scholar] [CrossRef]
- Shimizu-Hirota, R.; Xiong, W.; Baxter, B.T.; Kunkel, S.L.; Maillard, I.; Chen, X.-W.; Sabeh, F.; Liu, R.; Li, X.-Y.; Weiss, S.J. MT1-MMP Regulates the PI3Kδ·Mi-2/NuRD-Dependent Control of Macrophage Immune Function. Genes Dev. 2012, 26, 395–413. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.A.M.; Garcia-Vilas, J.A.; Cromwell, C.R.; Hubbard, B.P.; Hendzel, M.J.; Schulz, R. Matrix Metalloproteinase-2 Mediates Ribosomal RNA Transcription by Cleaving Nucleolar Histones. FEBS J. 2021, 288, 6736–6751. [Google Scholar] [CrossRef]
- Gonzalez-Avila, G.; Sommer, B.; Mendoza-Posada, D.A.; Ramos, C.; Garcia-Hernandez, A.A.; Falfan-Valencia, R. Matrix Metalloproteinases Participation in the Metastatic Process and Their Diagnostic and Therapeutic Applications in Cancer. Crit. Rev. Oncol. Hematol. 2019, 137, 57–83. [Google Scholar] [CrossRef]
- Koziol, A.; Martín-Alonso, M.; Clemente, C.; Gonzalo, P.; Arroyo, A.G. Site-Specific Cellular Functions of MT1-MMP. Eur. J. Cell Biol. 2012, 91, 889–895. [Google Scholar] [CrossRef]
- Knapinska, A.M.; Fields, G.B. The Expanding Role of MT1-MMP in Cancer Progression. Pharmaceuticals 2019, 12, 77. [Google Scholar] [CrossRef] [Green Version]
- Gingras, D.; Béliveau, R. Emerging Concepts in the Regulation of Membrane-Type 1 Matrix Metalloproteinase Activity. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 142–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozanov, D.V.; Deryugina, E.I.; Monosov, E.Z.; Marchenko, N.D.; Strongin, A.Y. Aberrant, Persistent Inclusion into Lipid Rafts Limits the Tumorigenic Function of Membrane Type-1 Matrix Metalloproteinase in Malignant Cells. Exp. Cell Res. 2004, 293, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Golubkov, V.S.; Boyd, S.; Savinov, A.Y.; Chekanov, A.V.; Osterman, A.L.; Remacle, A.; Rozanov, D.V.; Doxsey, S.J.; Strongin, A.Y. Membrane Type-1 Matrix Metalloproteinase (MT1-MMP) Exhibits an Important Intracellular Cleavage Function and Causes Chromosome Instability. J. Biol. Chem. 2005, 280, 25079–25086. [Google Scholar] [CrossRef] [Green Version]
- Golubkov, V.S.; Strongin, A.Y. Proteolysis-Driven Oncogenesis. Cell Cycle 2007, 6, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.; Lehti, K.; Wang, X.; Weiss, S.J.; Keski-Oja, J.; Pei, D. Regulation of Membrane-Type Matrix Metalloproteinase 1 Activity by Dynamin-Mediated Endocytosis. Proc. Natl. Acad. Sci. USA 2001, 98, 13693–13698. [Google Scholar] [CrossRef] [Green Version]
- Uekita, T.; Itoh, Y.; Yana, I.; Ohno, H.; Seiki, M. Cytoplasmic Tail-Dependent Internalization of Membrane-Type 1 Matrix Metalloproteinase Is Important for Its Invasion-Promoting Activity. J. Cell Biol. 2001, 155, 1345–1356. [Google Scholar] [CrossRef]
- Urena, J.M.; Merlos-Suarez, A.; Baselga, J.; Arribas, J. The Cytoplasmic Carboxy-Terminal Amino Acid Determines the Subcellular Localization of ProTGF-(Alpha) and Membrane Type Matrix Metalloprotease (MT1-MMP). J. Cell Sci. 1999, 112, 773–784. [Google Scholar] [CrossRef]
- Yana, I.; Weiss, S.J. Regulation of Membrane Type-1 Matrix Metalloproteinase Activation by Proprotein Convertases. MBoC 2000, 11, 2387–2401. [Google Scholar] [CrossRef] [Green Version]
- Strongin, A.Y. Proteolytic and Non-Proteolytic Roles of Membrane Type-1 Matrix Metalloproteinase in Malignancy. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y.; Seiki, M. MT1-MMP: A Potent Modifier of Pericellular Microenvironment. J. Cell Physiol. 2006, 206, 1–8. [Google Scholar] [CrossRef]
- Annabi, B.; Lachambre, M.; Bousquet-Gagnon, N.; Pagé, M.; Gingras, D.; Béliveau, R. Localization of Membrane-Type 1 Matrix Metalloproteinase in Caveolae Membrane Domains. Biochem. J 2001, 353, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, B.G.; Matías-Román, S.; Yáñez-Mó, M.; Vicente-Manzanares, M.; Sánchez-Madrid, F.; Arroyo, A.G. Caveolae Are a Novel Pathway for Membrane-Type 1 Matrix Metalloproteinase Traffic in Human Endothelial Cells. Mol. Biol. Cell 2004, 15, 678–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remacle, A.; Murphy, G.; Roghi, C. Membrane Type I-Matrix Metalloproteinase (MT1-MMP) Is Internalised by Two Different Pathways and Is Recycled to the Cell Surface. J. Cell Sci. 2003, 116, 3905–3916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frittoli, E.; Palamidessi, A.; Disanza, A.; Scita, G. Secretory and Endo/Exocytic Trafficking in Invadopodia Formation: The MT1-MMP Paradigm. Eur. J. Cell Biol. 2011, 90, 108–114. [Google Scholar] [CrossRef]
- Jacob, A.; Prekeris, R. The Regulation of MMP Targeting to Invadopodia during Cancer Metastasis. Front. Cell Dev. Biol. 2015, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Yu, H.; Liu, S.-Y.; Xiao, X.-S.; Dong, W.-H.; Chen, Y.-N.; Xu, W.; Zhu, T. Prognostic Value of Tissue Inhibitor of Metalloproteinase-2 Expression in Patients with Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0124230. [Google Scholar] [CrossRef] [Green Version]
- Castro-Castro, A.; Marchesin, V.; Monteiro, P.; Lodillinsky, C.; Rossé, C.; Chavrier, P. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu. Rev. Cell Dev. Biol. 2016, 32, 555–576. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Takeo, Y.; Yoshida, S.; Kouchi, Z.; Nakamura, Y.; Fukami, K. Lipid Rafts and Caveolin-1 Are Required for Invadopodia Formation and Extracellular Matrix Degradation by Human Breast Cancer Cells. Cancer Res. 2009, 69, 8594–8602. [Google Scholar] [CrossRef] [Green Version]
- Labrecque, L.; Nyalendo, C.; Langlois, S.; Durocher, Y.; Roghi, C.; Murphy, G.; Gingras, D.; Béliveau, R. Src-Mediated Tyrosine Phosphorylation of Caveolin-1 Induces Its Association with Membrane Type 1 Matrix Metalloproteinase. J. Biol. Chem. 2004, 279, 52132–52140. [Google Scholar] [CrossRef] [Green Version]
- Poincloux, R.; Lizárraga, F.; Chavrier, P. Matrix Invasion by Tumour Cells: A Focus on MT1-MMP Trafficking to Invadopodia. J. Cell Sci. 2009, 122, 3015–3024. [Google Scholar] [CrossRef] [Green Version]
- Tapia, T.; Ottman, R.; Chakrabarti, R. LIM Kinase1 Modulates Function of Membrane Type Matrix Metalloproteinase 1: Implication in Invasion of Prostate Cancer Cells. Mol. Cancer 2011, 10, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ip, Y.C.; Cheung, S.T.; Fan, S.T. Atypical Localization of Membrane Type 1-Matrix Metalloproteinase in the Nucleus Is Associated with Aggressive Features of Hepatocellular Carcinoma. Mol. Carcinog. 2007, 46, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Si-Tayeb, K.; Monvoisin, A.; Mazzocco, C.; Lepreux, S.; Decossas, M.; Cubel, G.; Taras, D.; Blanc, J.-F.; Robinson, D.R.; Rosenbaum, J. Matrix Metalloproteinase 3 Is Present in the Cell Nucleus and Is Involved in Apoptosis. Am. J. Pathol. 2006, 169, 1390–1401. [Google Scholar] [CrossRef] [Green Version]
- Benmerah, A.; Scott, M.; Poupon, V.; Marullo, S. Nuclear Functions for Plasma Membrane-Associated Proteins?: Traffic Between Plasma Membrane and Nucleus. Traffic 2003, 4, 503–511. [Google Scholar] [CrossRef]
- Lee, K.-W.; Liu, B.; Ma, L.; Li, H.; Bang, P.; Koeffler, H.P.; Cohen, P. Cellular Internalization of Insulin-like Growth Factor Binding Protein-3: Distinct Endocytic Pathways Facilitate Re-Uptake and Nuclear Localization. J. Biol. Chem. 2004, 279, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Seiki, M. Membrane-Type 1 Matrix Metalloproteinase: A Key Enzyme for Tumor Invasion. Cancer Lett. 2003, 194, 1–11. [Google Scholar] [CrossRef]
- Genís, L.; Gálvez, B.G.; Gonzalo, P.; Arroyo, A.G. MT1-MMP: Universal or Particular Player in Angiogenesis? Cancer Metastasis Rev. 2006, 25, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Sabeh, F.; Ota, I.; Holmbeck, K.; Birkedal-Hansen, H.; Soloway, P.; Balbin, M.; Lopez-Otin, C.; Shapiro, S.; Inada, M.; Krane, S.; et al. Tumor Cell Traffic through the Extracellular Matrix Is Controlled by the Membrane-Anchored Collagenase MT1-MMP. J. Cell Biol. 2004, 167, 769–781. [Google Scholar] [CrossRef] [Green Version]
- Hotary, K.B.; Allen, E.D.; Brooks, P.C.; Datta, N.S.; Long, M.W.; Weiss, S.J. Membrane Type I Matrix Metalloproteinase Usurps Tumor Growth Control Imposed by the Three-Dimensional Extracellular Matrix. Cell 2003, 114, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Roghi, C.; Jones, L.; Gratian, M.; English, W.R.; Murphy, G. Golgi Reassembly Stacking Protein 55 Interacts with Membrane-Type (MT) 1-Matrix Metalloprotease (MMP) and Furin and Plays a Role in the Activation of the MT1-MMP Zymogen: Role of GRASP55 in MT1-MMP Activation. FEBS J. 2010, 277, 3158–3175. [Google Scholar] [CrossRef]
- Xie, Y.; Mustafa, A.; Yerzhan, A.; Merzhakupova, D.; Yerlan, P.; N Orakov, A.; Wang, X.; Huang, Y.; Miao, L. Nuclear Matrix Metalloproteinases: Functions Resemble the Evolution from the Intracellular to the Extracellular Compartment. Cell Death Discov. 2017, 3, 17036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golubkov, V.S.; Chekanov, A.V.; Savinov, A.Y.; Rozanov, D.V.; Golubkova, N.V.; Strongin, A.Y. Membrane Type-1 Matrix Metalloproteinase Confers Aneuploidy and Tumorigenicity on Mammary Epithelial Cells. Cancer Res. 2006, 66, 10460–10465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wali, N.; Hosokawa, K.; Malik, S.; Saito, H.; Miyaguchi, K.; Imajoh-Ohmi, S.; Miki, Y.; Nakanishi, A. Centrosomal BRCA2 Is a Target Protein of Membrane Type-1 Matrix Metalloproteinase (MT1-MMP). Biochem. Biophys. Res. Commun. 2014, 443, 1148–1154. [Google Scholar] [CrossRef]
- Lehti, K.; Valtanen, H.; Wickström, S.A.; Lohi, J.; Keski-Oja, J. Regulation of Membrane-Type-1 Matrix Metalloproteinase Activity by Its Cytoplasmic Domain. J. Biol. Chem. 2000, 275, 15006–15013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gingras, D.; Bousquet-Gagnon, N.; Langlois, S.; Lachambre, M.-P.; Annabi, B.; Béliveau, R. Activation of the Extracellular Signal-Regulated Protein Kinase (ERK) Cascade by Membrane-Type-1 Matrix Metalloproteinase (MT1-MMP). FEBS Lett. 2001, 507, 231–236. [Google Scholar] [CrossRef] [Green Version]
- Sounni, N.E.; Devy, L.; Hajitou, A.; Frankenne, F.; Munaut, C.; Gilles, C.; Deroanne, C.; Thompson, E.W.; Foidart, J.M.; Noel, A. MT1-MMP Expression Promotes Tumor Growth and Angiogenesis through an up-Regulation of Vascular Endothelial Growth Factor Expression. FASEB J. 2002, 16, 555–564. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, S.; Ferrari, G.; Cinnante, K.; Scheerer, W.; Galloway, A.C.; Roses, D.F.; Rozanov, D.V.; Remacle, A.G.; Oh, E.-S.; Shiryaev, S.A.; et al. Tissue Inhibitor of Metalloproteinases-2 Binding to Membrane-Type 1 Matrix Metalloproteinase Induces MAPK Activation and Cell Growth by a Non-Proteolytic Mechanism. J. Biol. Chem. 2008, 283, 87–99. [Google Scholar] [CrossRef]
- Rozanov, D.V.; Savinov, A.Y.; Williams, R.; Liu, K.; Golubkov, V.S.; Krajewski, S.; Strongin, A.Y. Molecular Signature of MT1-MMP: Transactivation of the Downstream Universal Gene Network in Cancer. Cancer Res. 2008, 68, 4086–4096. [Google Scholar] [CrossRef] [Green Version]
- Nyalendo, C.; Michaud, M.; Beaulieu, E.; Roghi, C.; Murphy, G.; Gingras, D.; Béliveau, R. Src-Dependent Phosphorylation of Membrane Type I Matrix Metalloproteinase on Cytoplasmic Tyrosine 573: Role in Endothelial and Tumor Cell Migration. J. Biol. Chem. 2007, 282, 15690–15699. [Google Scholar] [CrossRef] [Green Version]
- Gingras, D.; Michaud, M.; Di Tomasso, G.; Béliveau, E.; Nyalendo, C.; Béliveau, R. Sphingosine-1-Phosphate Induces the Association of Membrane-Type 1 Matrix Metalloproteinase with P130Cas in Endothelial Cells. FEBS Lett. 2008, 582, 399–404. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Guan, L.; Li, S.; Jiang, Y.; Xiong, N.; Li, L.; Wu, C.; Zeng, H.; Liu, Y. Mechanosensitive Caveolin-1 Activation-Induced PI3K/Akt/MTOR Signaling Pathway Promotes Breast Cancer Motility, Invadopodia Formation and Metastasis in Vivo. Oncotarget 2016, 7, 16227–16247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.P.X.; Nichols, B.J. Caveolae: One Function or Many? Trends Cell Biol. 2016, 26, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; del Pozo, M.A. Caveolae as Plasma Membrane Sensors, Protectors and Organizers. Nat. Rev. Mol. Cell Biol. 2013, 14, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.C.; Coppolino, M.G. Phosphorylation of Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) and Its Vesicle-Associated Membrane Protein 7 (VAMP7)-Dependent Trafficking Facilitate Cell Invasion and Migration. J. Biol. Chem. 2011, 286, 43405–43416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, D.; Cambier, S.; Fjellbirkeland, L.; Baron, J.L.; Munger, J.S.; Kawakatsu, H.; Sheppard, D.; Broaddus, V.C.; Nishimura, S.L. The Integrin Alpha(v)Beta8 Mediates Epithelial Homeostasis through MT1-MMP-Dependent Activation of TGF-Beta1. J. Cell Biol. 2002, 157, 493–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalo, P.; Moreno, V.; Gálvez, B.G.; Arroyo, A.G. MT1-MMP and Integrins: Hand-to-Hand in Cell Communication. BioFactors 2010, 36, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, N.G.; Shapiro, S.D. A Protean Protease: MMP-12 Fights Viruses as a Protease and a Transcription Factor. Nat. Med. 2014, 20, 470–472. [Google Scholar] [CrossRef]
- Marchant, D.J.; Bellac, C.L.; Moraes, T.J.; Wadsworth, S.J.; Dufour, A.; Butler, G.S.; Bilawchuk, L.M.; Hendry, R.G.; Robertson, A.G.; Cheung, C.T.; et al. A New Transcriptional Role for Matrix Metalloproteinase-12 in Antiviral Immunity. Nat. Med. 2014, 20, 493–502. [Google Scholar] [CrossRef]
- Shaverdashvili, K.; Zhang, K.; Osman, I.; Honda, K.; Jobava, R.; Bedogni, B. MT1-MMP Dependent Repression of the Tumor Suppressor SPRY4 Contributes to MT1-MMP Driven Melanoma Cell Motility. Oncotarget 2015, 6, 33512–33522. [Google Scholar] [CrossRef] [Green Version]
- Uekita, T.; Gotoh, I.; Kinoshita, T.; Itoh, Y.; Sato, H.; Shiomi, T.; Okada, Y.; Seiki, M. Membrane-Type 1 Matrix Metalloproteinase Cytoplasmic Tail-Binding Protein-1 Is a New Member of the Cupin Superfamily. A Possible Multifunctional Protein Acting as an Invasion Suppressor down-Regulated in Tumors. J. Biol. Chem. 2004, 279, 12734–12743. [Google Scholar] [CrossRef] [Green Version]
- Pratt, J.; Iddir, M.; Bourgault, S.; Annabi, B. Evidence of MTCBP-1 Interaction with the Cytoplasmic Domain of MT1-MMP: Implications in the Autophagy Cell Index of High-Grade Glioblastoma: MTCBP-1 INHIBITS MT1-MMP-MEDIATED AUTOPHAGY. Mol. Carcinog. 2016, 55, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Goetz, J.G.; Minguet, S.; Navarro-Lérida, I.; Lazcano, J.J.; Samaniego, R.; Calvo, E.; Tello, M.; Osteso-Ibáñez, T.; Pellinen, T.; Echarri, A.; et al. Biomechanical Remodeling of the Microenvironment by Stromal Caveolin-1 Favors Tumor Invasion and Metastasis. Cell 2011, 146, 148–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafleur, M.A.; Xu, D.; Hemler, M.E. Tetraspanin Proteins Regulate Membrane Type-1 Matrix Metalloproteinase-Dependent Pericellular Proteolysis. MBoC 2009, 20, 2030–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo-Cordero, J.J.; Marrero-Diaz, R.; Megías, D.; Genís, L.; García-Grande, A.; García, M.A.; Arroyo, A.G.; Montoya, M.C. MT1-MMP Proinvasive Activity Is Regulated by a Novel Rab8-Dependent Exocytic Pathway. EMBO J. 2007, 26, 1499–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez, H.; López-Martín, S.; Toribio, V.; Zamai, M.; Hernández-Riquer, M.V.; Genís, L.; Arroyo, A.G.; Yáñez-Mó, M. Regulation of MT1-MMP Activity through Its Association with ERMs. Cells 2020, 9, 348. [Google Scholar] [CrossRef] [Green Version]
- Clark, E.S.; Weaver, A.M. A New Role for Cortactin in Invadopodia: Regulation of Protease Secretion. Eur. J. Cell Biol. 2008, 87, 581–590. [Google Scholar] [CrossRef] [Green Version]
- Clark, E.S.; Whigham, A.S.; Yarbrough, W.G.; Weaver, A.M. Cortactin Is an Essential Regulator of Matrix Metalloproteinase Secretion and Extracellular Matrix Degradation in Invadopodia. Cancer Res. 2007, 67, 4227–4235. [Google Scholar] [CrossRef] [Green Version]
- Schoumacher, M.; Goldman, R.D.; Louvard, D.; Vignjevic, D.M. Actin, Microtubules, and Vimentin Intermediate Filaments Cooperate for Elongation of Invadopodia. J. Cell Biol. 2010, 189, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Kwak, H.-I.; Kang, H.; Dave, J.M.; Mendoza, E.A.; Su, S.-C.; Maxwell, S.A.; Bayless, K.J. Calpain-Mediated Vimentin Cleavage Occurs Upstream of MT1-MMP Membrane Translocation to Facilitate Endothelial Sprout Initiation. Angiogenesis 2012, 15, 287–303. [Google Scholar] [CrossRef] [Green Version]
- Baldassarre, M.; Ayala, I.; Beznoussenko, G.; Giacchetti, G.; Machesky, L.M.; Luini, A.; Buccione, R. Actin Dynamics at Sites of Extracellular Matrix Degradation. Eur. J. Cell Biol. 2006, 85, 1217–1231. [Google Scholar] [CrossRef]
- Linder, S.; Higgs, H.; Hüfner, K.; Schwarz, K.; Pannicke, U.; Aepfelbacher, M. The Polarization Defect of Wiskott-Aldrich Syndrome Macrophages Is Linked to Dislocalization of the Arp2/3 Complex. J. Immunol. 2000, 165, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Linder, S.; Aepfelbacher, M. Podosomes: Adhesion Hot-Spots of Invasive Cells. Trends Cell Biol. 2003, 13, 376–385. [Google Scholar] [CrossRef]
- El Azzouzi, K.; Wiesner, C.; Linder, S. Metalloproteinase MT1-MMP Islets Act as Memory Devices for Podosome Reemergence. J. Cell Biol. 2016, 213, 109–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, T.; Seiki, M. Cytoplasmic Tail of MT1-MMP Regulates Macrophage Motility Independently from Its Protease Activity. Genes Cells 2009, 14, 617–626. [Google Scholar] [CrossRef]
- Sounni, N.E.; Roghi, C.; Chabottaux, V.; Janssen, M.; Munaut, C.; Maquoi, E.; Galvez, B.G.; Gilles, C.; Frankenne, F.; Murphy, G.; et al. Up-Regulation of Vascular Endothelial Growth Factor-A by Active Membrane-Type 1 Matrix Metalloproteinase through Activation of Src-Tyrosine Kinases. J. Biol. Chem. 2004, 279, 13564–13574. [Google Scholar] [CrossRef] [Green Version]
- Deryugina, E.I.; Ratnikov, B.I.; Postnova, T.I.; Rozanov, D.V.; Strongin, A.Y. Processing of Integrin Alpha(v) Subunit by Membrane Type 1 Matrix Metalloproteinase Stimulates Migration of Breast Carcinoma Cells on Vitronectin and Enhances Tyrosine Phosphorylation of Focal Adhesion Kinase. J. Biol. Chem. 2002, 277, 9749–9756. [Google Scholar] [CrossRef] [Green Version]
- Eisenach, P.A.; Roghi, C.; Fogarasi, M.; Murphy, G.; English, W.R. MT1-MMP Regulates VEGF-A Expression through a Complex with VEGFR-2 and Src. J. Cell Sci. 2010, 123, 4182–4193. [Google Scholar] [CrossRef] [Green Version]
- Mannello, F.; Medda, V. Nuclear Localization of Matrix Metalloproteinases. Prog. Histochem. Cytochem. 2012, 47, 27–58. [Google Scholar] [CrossRef]
- Cho, W.J.; Chow, A.K.; Schulz, R.; Daniel, E.E. Matrix Metalloproteinase-2, Caveolins, Focal Adhesion Kinase and c-Kit in Cells of the Mouse Myocardium. J. Cell. Mol. Med. 2007, 11, 1069–1086. [Google Scholar] [CrossRef] [Green Version]
- Hughes, B.G.; Fan, X.; Cho, W.J.; Schulz, R. MMP-2 Is Localized to the Mitochondria-Associated Membrane of the Heart. Am. J. Physiol.-Heart Circ. Physiol. 2014, 306, H764–H770. [Google Scholar] [CrossRef] [Green Version]
- Kuittinen, O.; Soini, Y.; Turpeenniemi-Hujanen, T. Diverse Role of MMP-2 and MMP-9 in the Clinicopathological Behavior of Hodgkin’s Lymphoma. Eur. J. Haematol. 2002, 69, 205–212. [Google Scholar] [CrossRef]
- Fallata, A.M.; Wyatt, R.A.; Levesque, J.M.; Dufour, A.; Overall, C.M.; Crawford, B.D. Intracellular Localization in Zebrafish Muscle and Conserved Sequence Features Suggest Roles for Gelatinase A Moonlighting in Sarcomere Maintenance. Biomedicines 2019, 7, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwan, J.A.; Schulze, C.J.; Wang, W.; Leon, H.; Sariahmetoglu, M.; Sung, M.; Sawicka, J.; Sims, D.E.; Sawicki, G.; Schulz, R. Matrix Metalloproteinase-2 (MMP-2) Is Present in the Nucleus of Cardiac Myocytes and Is Capable of Cleaving Poly (ADP-ribose) Polymerase (PARP) in Vitro. FASEB J. 2004, 18, 690–692. [Google Scholar] [CrossRef]
- Ruta, A.; Mark, B.; Edward, B.; Jawaharlal, P.; Jianliang, Z. Nuclear Localization of Active Matrix Metalloproteinase-2 in Cigarette Smoke-Exposed Apoptotic Endothelial Cells. Exp. Lung Res. 2009, 35, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, A.D.; Chow, A.K.; Ali, M.A.M.; Schulz, R. Matrix Metalloproteinase-2 and Myocardial Oxidative Stress Injury: Beyond the Matrix. Cardiovasc. Res. 2010, 85, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.-F.; Chen, F.-H.; Lu, M.-H.; Hong, J.-H.; Chiang, C.-S. Dual Roles of Tumour Cells-Derived Matrix Metalloproteinase 2 on Brain Tumour Growth and Invasion. Br. J. Cancer 2017, 117, 1828–1836. [Google Scholar] [CrossRef] [Green Version]
- Sinha, S.K.; Asotra, K.; Uzui, H.; Nagwani, S.; Mishra, V.; Rajavashisth, T.B. Nuclear Localization of Catalytically Active MMP-2 in Endothelial Cells and Neurons. Am. J. Transl. Res. 2014, 6, 155–162. [Google Scholar]
- Frolova, A.S.; Petushkova, A.I.; Makarov, V.A.; Soond, S.M.; Zamyatnin, A.A. Unravelling the Network of Nuclear Matrix Metalloproteinases for Targeted Drug Design. Biology 2020, 9, 480. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin Alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Mittal, R.; Patel, A.P.; Debs, L.H.; Nguyen, D.; Patel, K.; Grati, M.; Mittal, J.; Yan, D.; Chapagain, P.; Liu, X.Z. Intricate Functions of Matrix Metalloproteinases in Physiological and Pathological Conditions: ROLE OF MMPs IN HEALTH AND DISEASE. J. Cell. Physiol. 2016, 231, 2599–2621. [Google Scholar] [CrossRef]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and Regulation of Nuclear Transport Processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Jang, H.S.; Watson, G.W.; Wellappili, D.P.; Tyler, B.M. Distinctive Nuclear Localization Signals in the Oomycete Phytophthora Sojae. Front. Microbiol. 2017, 8, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdukhakimova, D.; Xie, Y. Comparative Analysis of NLS Sequence Suggests the Evolutionary Origin of Nuclear Matrix Metalloproteinase 7 during Cancer Evolution. IJPMBS 2016, 5, 206–210. [Google Scholar]
- Eguchi, T.; Kubota, S.; Kawata, K.; Mukudai, Y.; Uehara, J.; Ohgawara, T.; Ibaragi, S.; Sasaki, A.; Kuboki, T.; Takigawa, M. Novel Transcription Factor-Like Function of Human Matrix Metalloproteinase 3 Regulating the CTGF/CCN2 Gene. Mol. Cell Biol. 2008, 28, 2391–2413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madzharova, E.; Kastl, P.; Sabino, F.; Auf dem Keller, U. Post-Translational Modification-Dependent Activity of Matrix Metalloproteinases. Int. J. Mol. Sci. 2019, 20, 3077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löffek, S.; Schilling, O.; Franzke, C.-W. Series “Matrix Metalloproteinases in Lung Health and Disease”: Biological Role of Matrix Metalloproteinases: A Critical Balance. Eur. Respir. J. 2011, 38, 191–208. [Google Scholar] [CrossRef] [Green Version]
- Surgucheva, I.; Chidambaram, K.; Willoughby, D.A.; Surguchov, A. Matrix Metalloproteinase 9 Expression: New Regulatory Elements. J. Ocul. Biol. Dis. Inform. 2010, 3, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Ramos-DeSimone, N.; Hahn-Dantona, E.; Sipley, J.; Nagase, H.; French, D.L.; Quigley, J.P. Activation of Matrix Metalloproteinase-9 (MMP-9) via a Converging Plasmin/Stromelysin-1 Cascade Enhances Tumor Cell Invasion. J. Biol. Chem. 1999, 274, 13066–13076. [Google Scholar] [CrossRef] [Green Version]
- Sariahmetoglu, M.; Crawford, B.D.; Leon, H.; Sawicka, J.; Li, L.; Ballermann, B.J.; Holmes, C.; Berthiaume, L.G.; Holt, A.; Sawicki, G.; et al. Regulation of Matrix Metalloproteinase-2 (MMP-2) Activity by Phosphorylation. FASEB J. 2007, 21, 2486–2495. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Rehemtulla, A.; Pavlaki, M.; Kozarekar, P.; Chiarelli, C. Furin Directly Cleaves ProMMP-2 in the Trans-Golgi Network Resulting in a Nonfunctioning Proteinase. J. Biol. Chem. 2005, 280, 10974–10980. [Google Scholar] [CrossRef] [Green Version]
- Pereira, A.M.M.; Strasberg-Rieber, M.; Rieber, M. Invasion-Associated MMP-2 and MMP-9 Are up-Regulated Intracellularly in Concert with Apoptosis Linked to Melanoma Cell Detachment. Clin. Exp. Metastasis 2005, 22, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Nagase, H. Progress in Matrix Metalloproteinase Research. Mol. Asp. Med. 2008, 29, 290–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.A.M.; Cho, W.J.; Hudson, B.; Kassiri, Z.; Granzier, H.; Schulz, R. Titin Is a Target of Matrix Metalloproteinase-2: Implications in Myocardial Ischemia/Reperfusion Injury. Circulation 2010, 122, 2039–2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeCoux, A.; Lindsey, M.L.; Villarreal, F.; Garcia, R.A.; Schulz, R. Myocardial Matrix Metalloproteinase-2: Inside out and Upside Down. J. Mol. Cell. Cardiol. 2014, 77, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandasamy, A.D.; Schulz, R. Glycogen Synthase Kinase-3 Is Activated by Matrix Metalloproteinase-2 Mediated Proteolysis in Cardiomyoblasts. Cardiovasc. Res. 2009, 83, 698–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gömöri, K.; Szabados, T.; Kenyeres, É.; Pipis, J.; Földesi, I.; Siska, A.; Dormán, G.; Ferdinandy, P.; Görbe, A.; Bencsik, P. Cardioprotective Effect of Novel Matrix Metalloproteinase Inhibitors. Int. J. Mol. Sci. 2020, 21, 6990. [Google Scholar] [CrossRef]
- Skrzypiec-Spring, M.; Urbaniak, J.; Sapa-Wojciechowska, A.; Pietkiewicz, J.; Orda, A.; Karolko, B.; Danielewicz, R.; Bil-Lula, I.; Woźniak, M.; Schulz, R.; et al. Matrix Metalloproteinase-2 Inhibition in Acute Ischemia-Reperfusion Heart Injury—Cardioprotective Properties of Carvedilol. Pharmaceuticals 2021, 14, 1276. [Google Scholar] [CrossRef]
- Koskivirta, I.; Kassiri, Z.; Rahkonen, O.; Kiviranta, R.; Oudit, G.Y.; McKee, T.D.; Kytö, V.; Saraste, A.; Jokinen, E.; Liu, P.P.; et al. Mice with Tissue Inhibitor of Metalloproteinases 4 (Timp4) Deletion Succumb to Induced Myocardial Infarction but Not to Cardiac Pressure Overload. J. Biol. Chem. 2010, 285, 24487–24493. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Jin, L.; Tan, Y. Different Roles of Matrix Metalloproteinase 2 in Osteolysis of Skeletal Dysplasia and Bone Metastasis (Review). Mol. Med. Rep. 2020, 23, 70. [Google Scholar] [CrossRef]
- Okamoto, T.; Akaike, T.; Nagano, T.; Miyajima, S.; Suga, M.; Ando, M.; Ichimori, K.; Maeda, H. Activation of Human Neutrophil Procollagenase by Nitrogen Dioxide and Peroxynitrite: A Novel Mechanism for Procollagenase Activation Involving Nitric Oxide. Arch. Biochem. Biophys. 1997, 342, 261–274. [Google Scholar] [CrossRef]
- Mashimo, M.; Onishi, M.; Uno, A.; Tanimichi, A.; Nobeyama, A.; Mori, M.; Yamada, S.; Negi, S.; Bu, X.; Kato, J.; et al. The 89-KDa PARP1 Cleavage Fragment Serves as a Cytoplasmic PAR Carrier to Induce AIF-Mediated Apoptosis. J. Biol. Chem. 2021, 296, 100046. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Kim, S.; Ghate, N.B.; Rhie, S.K.; An, W. MMP-9 Drives the Melanomagenic Transcription Program through Histone H3 Tail Proteolysis. Oncogene 2022, 41, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Morgunova, E.; Tuuttila, A.; Bergmann, U.; Isupov, M.; Lindqvist, Y.; Schneider, G.; Tryggvason, K. Structure of Human Pro-Matrix Metalloproteinase-2: Activation Mechanism Revealed. Science 1999, 284, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Lehti, K.; Lohi, J.; Valtanen, H.; Keski-Oja, J. Proteolytic Processing of Membrane-Type-1 Matrix Metalloproteinase Is Associated with Gelatinase A Activation at the Cell Surface. Biochem. J. 1998, 334, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanton, H.; Gavrilovic, J.; Atkinson, S.J.; d’Ortho, M.P.; Yamada, K.M.; Zardi, L.; Murphy, G. The Activation of ProMMP-2 (Gelatinase A) by HT1080 Fibrosarcoma Cells Is Promoted by Culture on a Fibronectin Substrate and Is Concomitant with an Increase in Processing of MT1-MMP (MMP-14) to a 45 KDa Form. J. Cell Sci. 1998, 111 Pt 18, 2789–2798. [Google Scholar] [CrossRef]
- Stamenkovic, I. Matrix Metalloproteinases in Tumor Invasion and Metastasis. Semin. Cancer Biol. 2000, 10, 415–433. [Google Scholar] [CrossRef]
- D’ortho, M.-P.; Will, H.; Atkinson, S.; Butler, G.; Messent, A.; Gavrilovic, J.; Smith, B.; Timpl, R.; Zardi, L.; Murphy, G. Membrane-Type Matrix Metalloproteinases 1 and 2 Exhibit Broad-Spectrum Proteolytic Capacities Comparable to Many Matrix Metalloproteinases. Eur. J. Biochem. 1997, 250, 751–757. [Google Scholar] [CrossRef] [Green Version]
- Hariton-Gazal, E.; Feder, R.; Mor, A.; Graessmann, A.; Brack-Werner, R.; Jans, D.; Gilon, C.; Loyter, A. Targeting of Nonkaryophilic Cell-Permeable Peptides into the Nuclei of Intact Cells by Covalently Attached Nuclear Localization Signals. Biochemistry 2002, 41, 9208–9214. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Tai, L.; Jiang, K.; Xie, C.; Li, Z.; Lin, Y.-Z.; Wei, G.; Lu, W.; Pan, W. Functionalized Cell Nucleus-Penetrating Peptide Combined with Doxorubicin for Synergistic Treatment of Glioma. Acta Biomater. 2016, 42, 90–101. [Google Scholar] [CrossRef]
- Li, H.K.; Hasegawa, S. Favorable Tumor Uptake and Nuclear Transport of Auger Electrons by Nuclear Targeting with 111In-Trastuzumab in an Intraperitoneal Tumor Mouse Model. Nucl. Med. Commun. 2022, 43, 763–769. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maybee, D.V.; Ink, N.L.; Ali, M.A.M. Novel Roles of MT1-MMP and MMP-2: Beyond the Extracellular Milieu. Int. J. Mol. Sci. 2022, 23, 9513. https://doi.org/10.3390/ijms23179513
Maybee DV, Ink NL, Ali MAM. Novel Roles of MT1-MMP and MMP-2: Beyond the Extracellular Milieu. International Journal of Molecular Sciences. 2022; 23(17):9513. https://doi.org/10.3390/ijms23179513
Chicago/Turabian StyleMaybee, Deanna V., Nicole L. Ink, and Mohammad A. M. Ali. 2022. "Novel Roles of MT1-MMP and MMP-2: Beyond the Extracellular Milieu" International Journal of Molecular Sciences 23, no. 17: 9513. https://doi.org/10.3390/ijms23179513






