C10Pred: A First Machine Learning Based Tool to Predict C10 Family Cysteine Peptidases Using Sequence-Derived Features
Abstract
:1. Introduction
2. Results
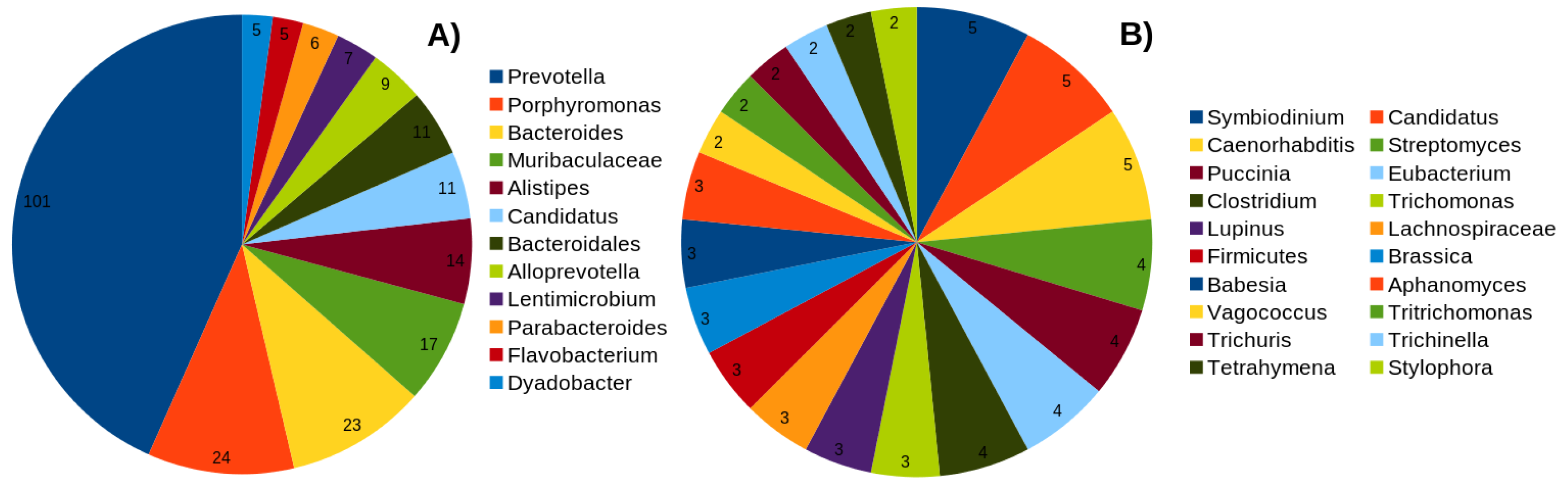
2.1. Overview of the Dataset
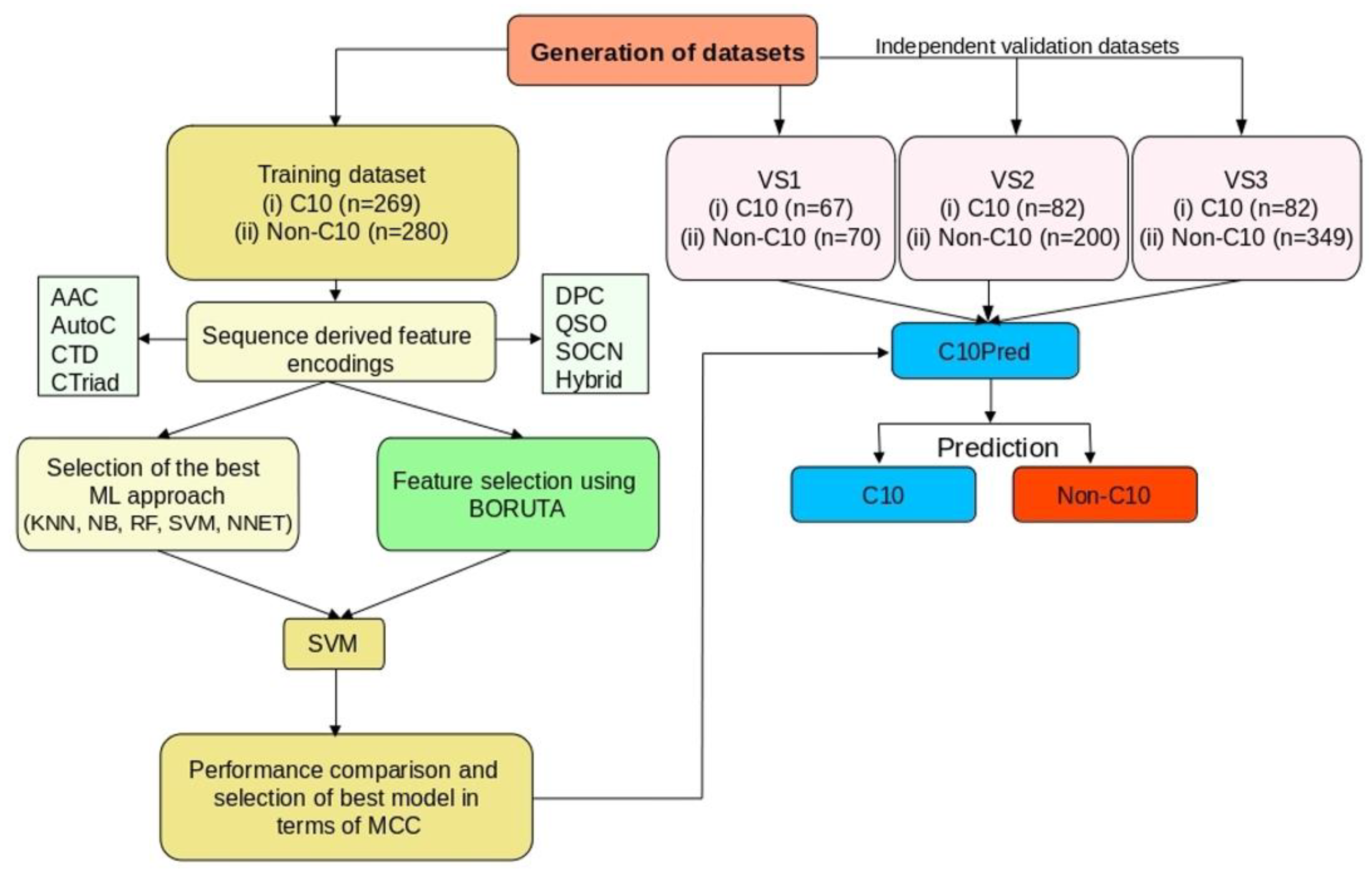
2.2. The Overall Framework of the Proposed Predictor
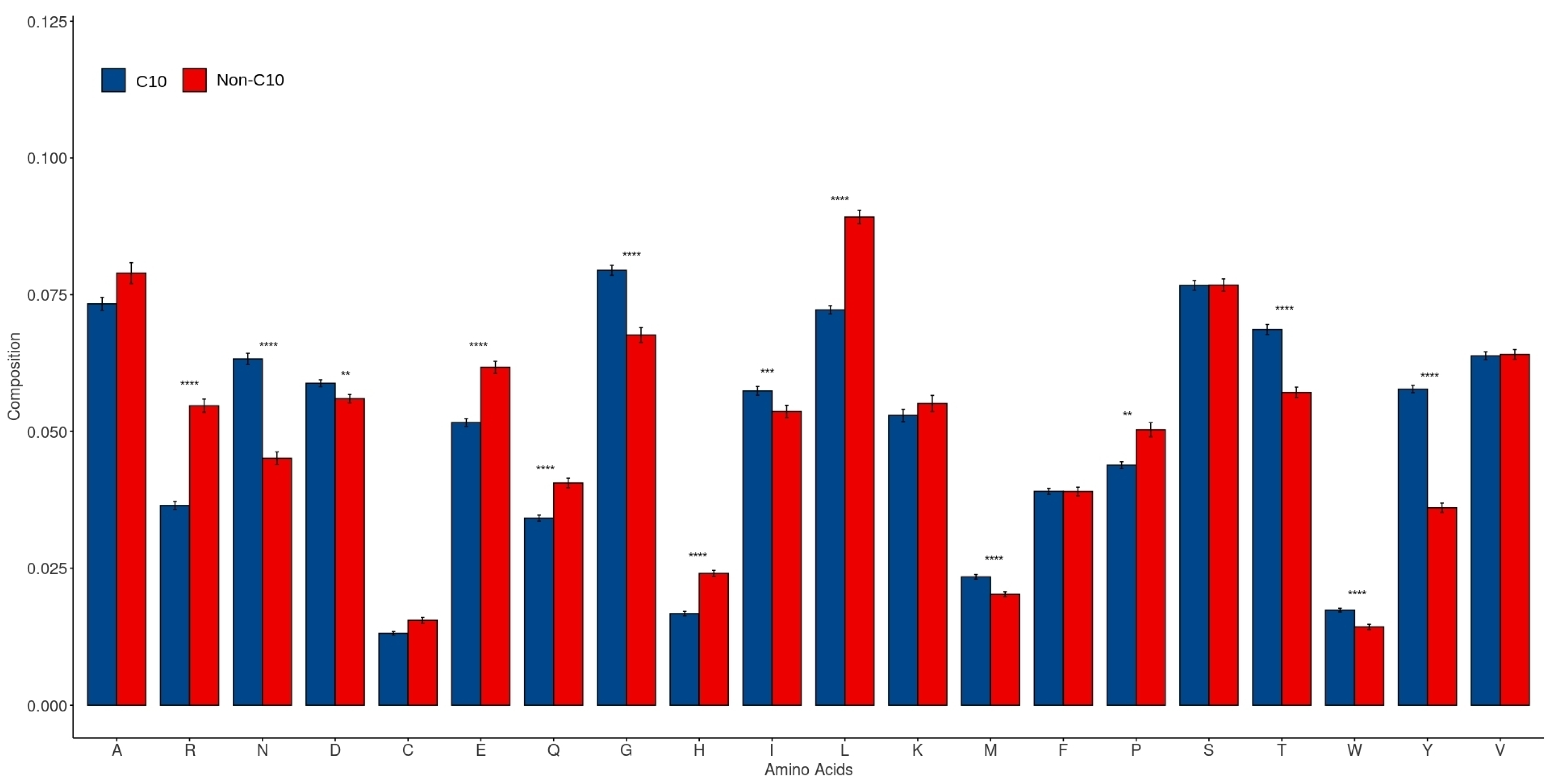
2.3. Amino Acid Composition in C10 and Non-C10 Sequences
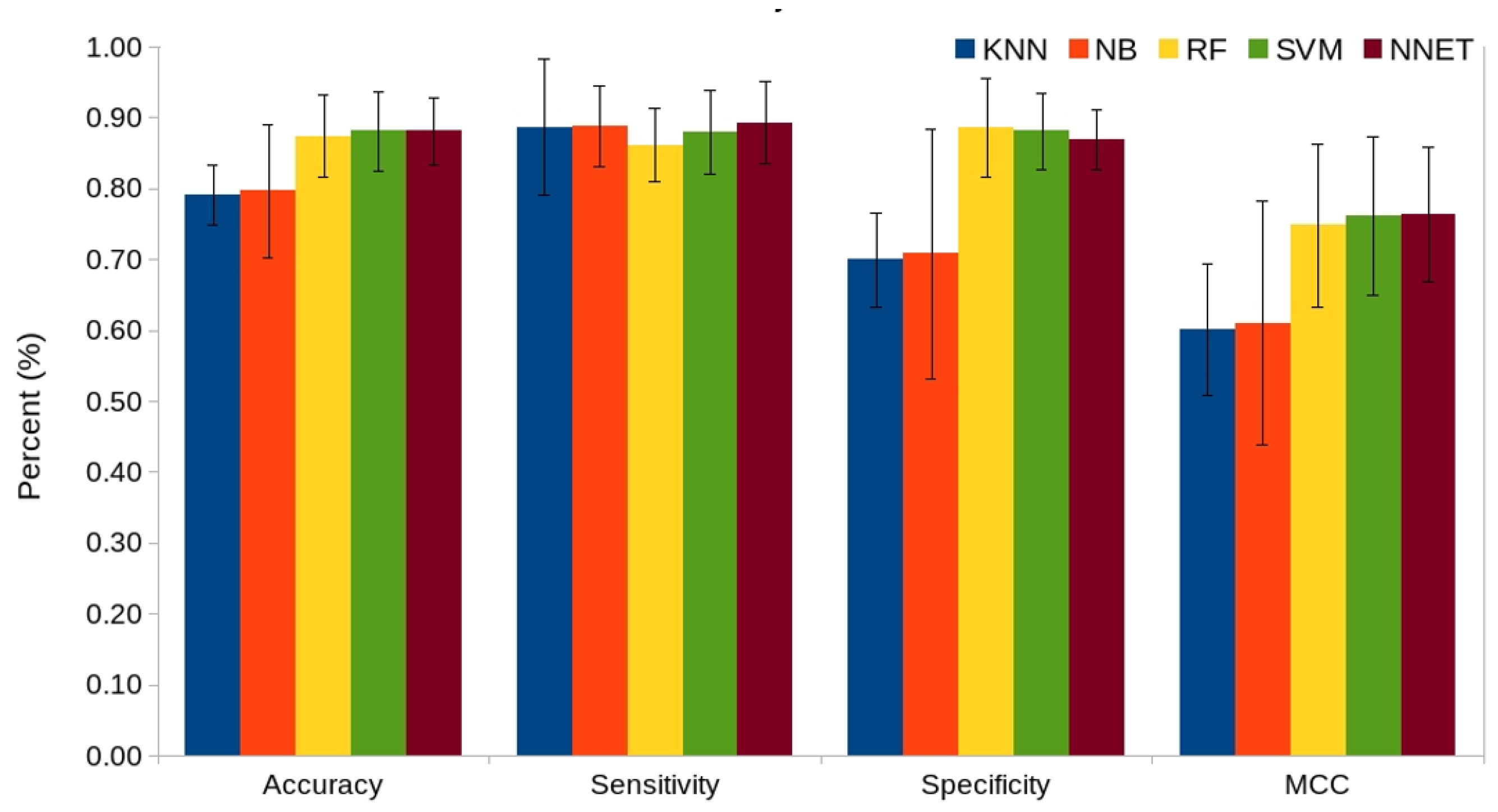
2.4. Comparison of Various Machine Learning Classifiers
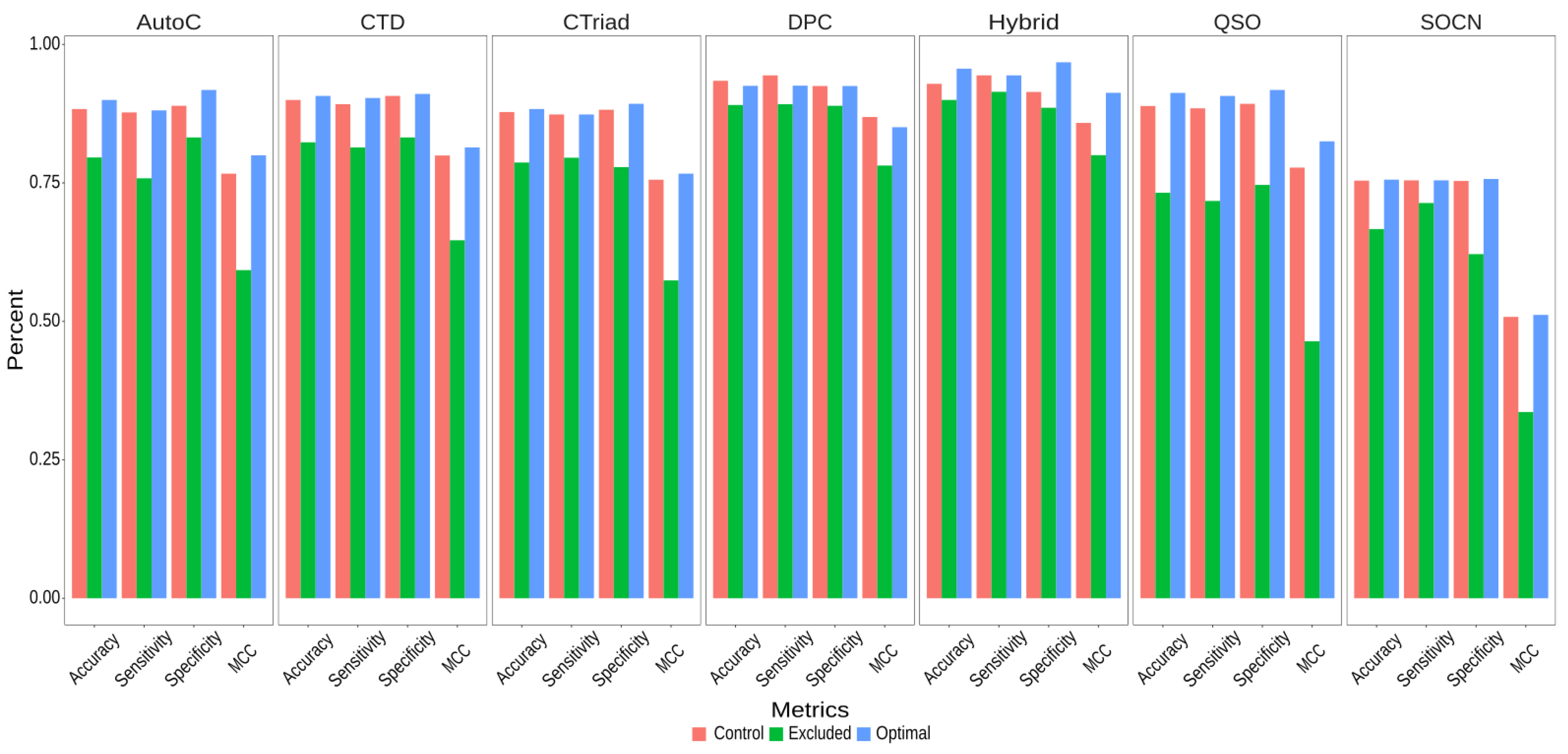
2.5. Performance Evaluation of Various Feature Encodings
2.6. Optimal Feature Selection for Each Encoding
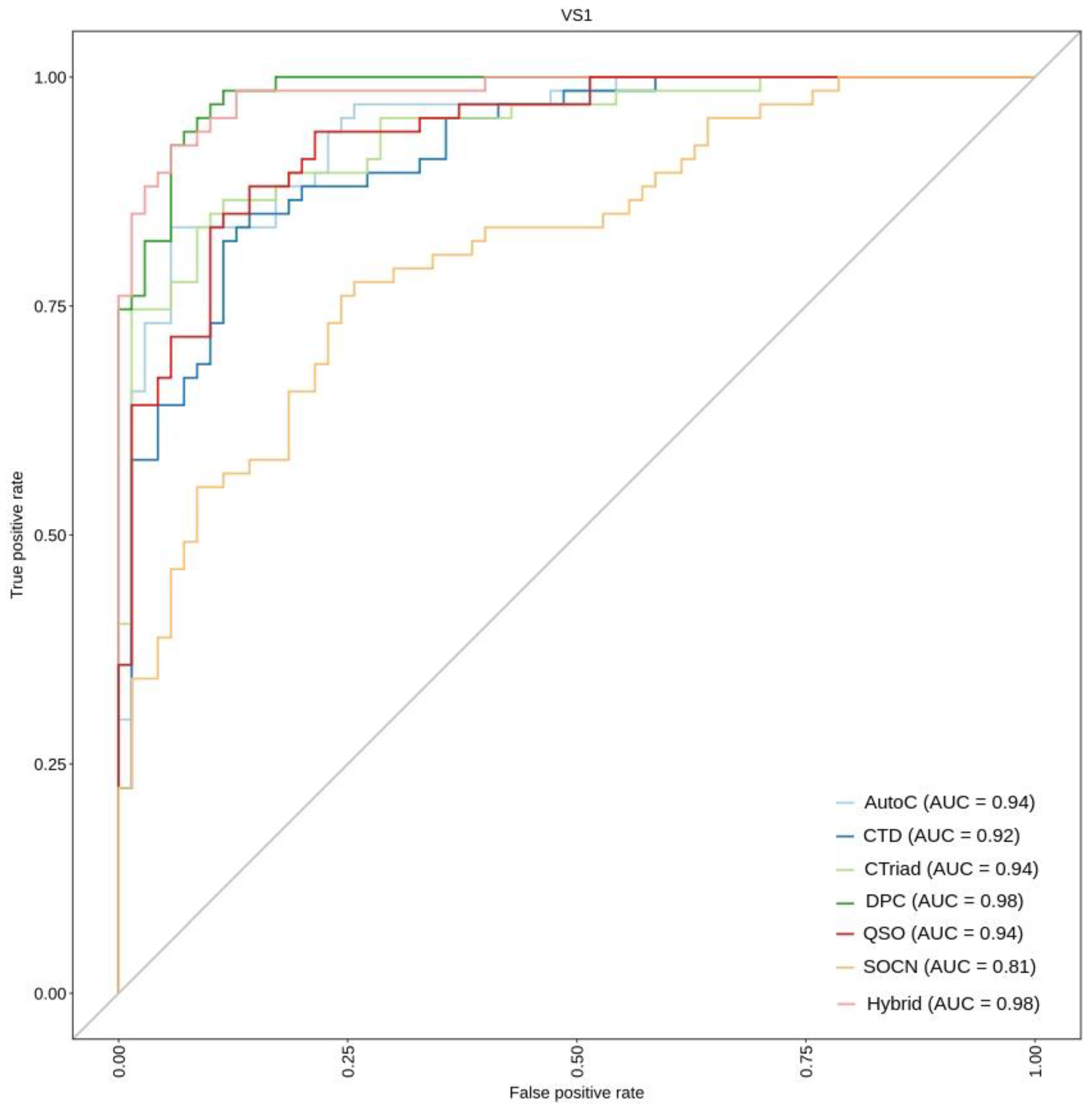
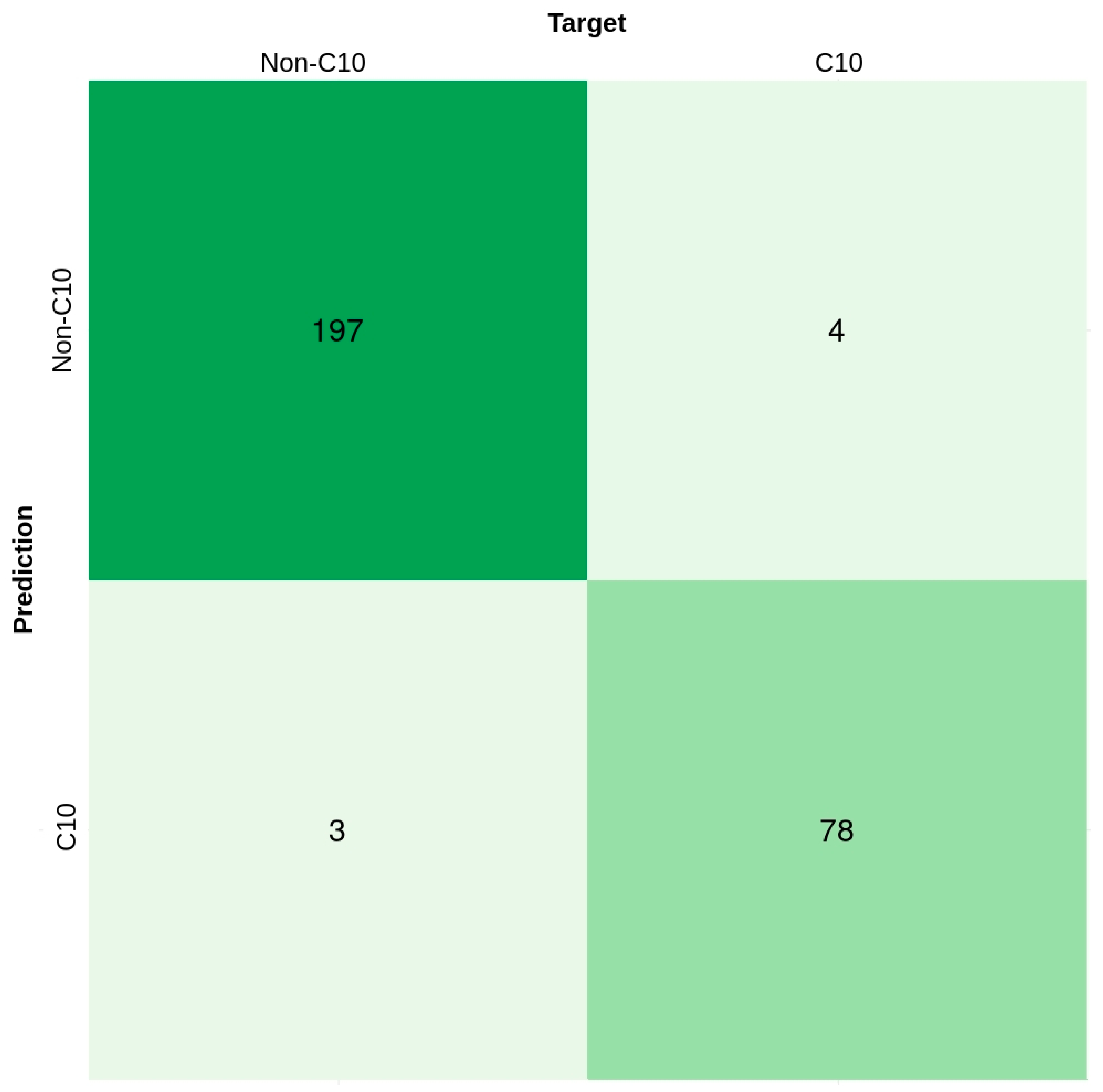
2.7. Performance Comparison on Independent Datasets
2.8. Software Availability
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Data Organization
4.2. Feature Encoding
4.2.1. Amino Acid Composition (AAC)
4.2.2. Autocorrelation (AutoC)
4.2.3. Composition (C), Transition (T), and Distribution (D) (CTD)
4.2.4. Conjoint Triad (CTriad)
4.2.5. Dipeptide Composition (DPC)
4.2.6. Quasi-Sequence Order (QSO)
4.2.7. Sequence Order Coupling Number (SOCN)
4.3. Machine Learning Models
4.4. Feature Selection
4.5. Performance Evaluation Metrics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cannon, J.W.; Zhung, J.; Bennett, J.; Moreland, N.J.; Baker, M.G.; Geelhoed, E.; Fraser, J.; Carapetis, J.R.; Jack, S. The economic and health burdens of diseases caused by group A Streptococcus in New Zealand. Int. J. Infect. Dis. 2021, 103, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.E.; Pondo, T.; Toews, K.-A.; Farley, M.M.; Lindegren, M.L.; Lynfield, R.; Aragon, D.; Zansky, S.M.; Watt, J.P.; Cieslak, P.R.; et al. Epidemiology of Invasive Group A Streptococcal Infections in the United States, 2005–2012. Clin. Infect. Dis. 2016, 63, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Carapetis, J.R.; Steer, A.C.; Mulholland, E.K.; Weber, M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005, 5, 685–694. [Google Scholar] [CrossRef]
- Bowen, A.C.; Tong, S.Y.C.; Chatfield, M.D.; Carapetis, J.R. The microbiology of impetigo in Indigenous children: Associations between Streptococcus pyogenes, Staphylococcus aureus, scabies, and nasal carriage. BMC Infect. Dis. 2014, 14, 727. [Google Scholar] [CrossRef] [Green Version]
- Bowen, A.C.; Mahé, A.; Hay, R.J.; Andrews, R.M.; Steer, A.C.; Tong, S.Y.C.; Carapetis, J.R. The Global Epidemiology of Impetigo: A Systematic Review of the Population Prevalence of Impetigo and Pyoderma. PLoS ONE 2015, 10, e0136789. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, M.W. Pathogenesis of Group A Streptococcal Infections. Clin. Microbiol. Rev. 2000, 13, 470–511. [Google Scholar] [CrossRef]
- Castro, S.A.; Dorfmueller, H.C. A brief review on Group A Streptococcus pathogenesis and vaccine development. R. Soc. Open Sci. 2021, 8, 201991. [Google Scholar] [CrossRef]
- Carapetis, J.R.; Beaton, A.; Cunningham, M.W.; Guilherme, L.; Karthikeyan, G.; Mayosi, B.M.; Sable, C.; Steer, A.; Wilson, N.; Wyber, R.; et al. Acute rheumatic fever and rheumatic heart disease. Nat. Rev. Dis. Prim. 2016, 2, 15084. [Google Scholar] [CrossRef] [Green Version]
- Hoy, W.E.; White, A.V.; Dowling, A.; Sharma, S.K.; Bloomfield, H.; Tipiloura, B.T.; Swanson, C.E.; Mathews, J.D.; McCredie, D.A. Post-streptococcal glomerulonephritis is a strong risk factor for chronic kidney disease in later life. Kidney Int. 2012, 81, 1026–1032. [Google Scholar] [CrossRef] [Green Version]
- Marshall, C.S.; Cheng, A.C.; Markey, P.G.; Towers, R.J.; Richardson, L.J.; Fagan, P.K.; Scott, L.; Krause, V.L.; Currie, B.J. Acute Post-Streptococcal Glomerulonephritis in the Northern Territory of Australia: A Review of 16 Years Data and Comparison with the Literature. Am. J. Trop. Med. Hyg. 2011, 85, 703–710. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J.; Pierse, N.; Williamson, D.A.; Baker, M.G. Estimating the likely true changes in rheumatic fever incidence using two data sources. Epidemiol. Infect. 2017, 146, 265–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, A.M.; Lennon, D.R.; van der Werf, B.; Diack, M.; Neutze, J.M.; Horsfall, M.; Emery, D.; Wong, W. Post-streptococcal glomerulonephritis: Some reduction in a disease of disparities. J. Paediatr. Child Health 2018, 55, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Stockmann, C.; Ampofo, K.; Hersh, A.L.; Blaschke, A.J.; Kendall, B.A.; Korgenski, K.; Daly, J.; Hill, H.R.; Byington, C.L.; Pavia, A.T. Evolving Epidemiologic Characteristics of Invasive Group A Streptococcal Disease in Utah, 2002–2010. Clin. Infect. Dis. 2012, 55, 479–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamagni, T.L.; Efstratiou, A.; Vuopio-Varkila, J.; Jasir, A.; Schalén, C.; Euro, S. The epidemiology of severe Streptococcus pyogenes associated disease in Europe. Eurosurveillance 2005, 10, 9–10. [Google Scholar] [CrossRef]
- Watkins, D.A.; Johnson, C.O.; Colquhoun, S.M.; Karthikeyan, G.; Beaton, A.; Bukhman, G.; Forouzanfar, M.H.; Longenecker, C.T.; Mayosi, B.M.; Mensah, G.A.; et al. Global, Regional, and National Burden of Rheumatic Heart Disease, 1990–2015. N. Engl. J. Med. 2017, 377, 713–722. [Google Scholar] [CrossRef]
- Gubba, S.; Low, D.E.; Musser, J.M. Expression and Characterization of Group A Streptococcus Extracellular Cysteine Protease Recombinant Mutant Proteins and Documentation of Seroconversion during Human Invasive Disease Episodes. Infect. Immun. 1998, 66, 765–770. [Google Scholar] [CrossRef] [Green Version]
- Gerlach, D.; Knöll, H.; Köhler, W.; Ozegowski, J.-H.; Hríbalova, V. Isolation and characterization of erythrogenic toxins V. Communication: Identity of erythrogenic toxin type B and Streptococcal proteinase precursor. Zentralbl. Bakteriol. Mikrobiol. Hyg. A Med. Mikrobiol. Infekt. Parasitol. 1983, 255, 221–233. [Google Scholar] [CrossRef]
- Hauser, A.R.; Schlievert, P.M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J. Bacteriol. 1990, 172, 4536–4542. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J. Introduction: The Clans and Families of Cysteine Peptidases. In Handbook of Proteolytic Enzymes; Academic Press: Cambridge, MA, USA, 2013; pp. 1743–1773. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Luo, S.-C.; Kuo, C.-F.; Lin, Y.-S.; Wu, J.-J.; Lin, M.T.; Liu, C.-C.; Jeng, W.-Y.; Chuang, W.-J. Maturation Processing and Characterization of Streptopain. J. Biol. Chem. 2003, 278, 17336–17343. [Google Scholar] [CrossRef] [Green Version]
- Kagawa, T.F.; Cooney, J.C.; Baker, H.M.; McSweeney, S.; Liu, M.; Gubba, S.; Musser, J.M.; Baker, E.N. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: An integrin-binding cysteine protease. Proc. Natl. Acad. Sci. USA 2000, 97, 2235–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musser, J.M.; Hauser, A.R.; Kim, M.H.; Schlievert, P.M.; Nelson, K.; Selander, R.K. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: Clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 1991, 88, 2668–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.Y.; Elliott, S.D. Streptococcal proteinase: The zymogen to enzyme transfromation. J. Biol. Chem. 1965, 240, 1138–1142. [Google Scholar] [CrossRef]
- Liu, T.Y.; Elliott, S.D. Activation of Streptococcal Proteinase and its Zymogen by Bacterial Cell Walls. Nature 1965, 206, 33–34. [Google Scholar] [CrossRef]
- Wang, C.-C.; Houng, H.-C.; Chen, C.-L.; Wang, P.-J.; Kuo, C.-F.; Lin, Y.-S.; Wu, J.-J.; Lin, M.T.; Liu, C.-C.; Huang, W.; et al. Solution structure and backbone dynamics of streptopain: Insight into diverse substrate specificity. J. Biol. Chem. 2009, 284, 10957–10967. [Google Scholar] [CrossRef] [Green Version]
- Walker, M.; Hollands, A.; Sanderson-Smith, M.; Cole, J.N.; Kirk, J.K.; Henningham, A.; McArthur, J.D.; Dinkla, K.; Aziz, R.; Kansal, R.G.; et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 2007, 13, 981–985. [Google Scholar] [CrossRef]
- Kapur, V.; Topouzis, S.; Majesky, M.W.; Li, L.-L.; Hamrick, M.R.; Hamill, R.J.; Patti, J.M.; Musser, J.M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 1993, 15, 327–346. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Mahajan, N.; Dhawan, V. Acknowledged Signatures of Matrix Metalloproteinases in Takayasu’s Arteritis. BioMed Res. Int. 2014, 2014, 827105. [Google Scholar] [CrossRef]
- Tamura, F.; Nakagawa, R.; Akuta, T.; Okamoto, S.; Hamada, S.; Maeda, H.; Kawabata, S.; Akaike, T. Proapoptotic Effect of Proteolytic Activation of Matrix Metalloproteinases by Streptococcus pyogenes Thiol Proteinase ( Streptococcus Pyrogenic Exotoxin B). Infect. Immun. 2004, 72, 4836–4847. [Google Scholar] [CrossRef] [Green Version]
- Stockbauer, K.E.; Magoun, L.; Liu, M.; Burns, E.H.; Gubba, S.; Renish, S.; Pan, X.; Bodary, S.C.; Baker, E.; Coburn, J.; et al. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins alphavbeta3 and alphaIIbbeta3. Proc. Natl. Acad. Sci. USA 1999, 96, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Byrne, D.P.; Wawrzonek, K.; Jaworska, A.; Birss, A.J.; Potempa, J.; Smalley, J.W. Role of the cysteine protease interpain A of Prevotella intermedia in breakdown and release of haem from haemoglobin. Biochem. J. 2009, 425, 257–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, D.; Potempa, J.; Kordula, T.; Travis, J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J. Biol. Chem. 1999, 274, 12245–12251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, E.; Alvarez, B.; Duchaud, E.; Guijarro, J.A. Development of a Markerless Deletion System for the Fish-Pathogenic Bacterium Flavobacterium psychrophilum. PLoS ONE 2015, 10, e0117969. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pascual, D.; Lunazzi, A.; Magdelenat, G.; Rouy, Z.; Roulet, A.; Lopez-Roques, C.; Larocque, R.; Barbeyron, T.; Gobet, A.; Michel, G.; et al. The Complete Genome Sequence of the Fish Pathogen Tenacibaculum maritimum Provides Insights into Virulence Mechanisms. Front. Microbiol. 2017, 8, 1542. [Google Scholar] [CrossRef]
- Tett, A.; Huang, K.D.; Asnicar, F.; Fehlner-Peach, H.; Pasolli, E.; Karcher, N.; Armanini, F.; Manghi, P.; Bonham, K.; Zolfo, M.; et al. The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell Host Microbe 2019, 26, 666–679.e7. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, M.; Subramanian, A.; Anishetty, S. Comparative pan genome analysis of oral Prevotella species implicated in periodontitis. Funct. Integr. Genom. 2017, 17, 513–536. [Google Scholar] [CrossRef]
- Patra, A.K.; Yu, Z. Genomic Insights into the Distribution of Peptidases and Proteolytic Capacity among Prevotella and Paraprevotella Species. Microbiol. Spectr. 2022, 10. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; López, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Štambuk, N.; Konjevoda, P. The Role of Independent Test Set in Modeling of Protein Folding Kinetics. Adv. Exp. Med. Biol. 2011, 696, 279–284. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.K.; Musser, J.M. From transcription to activation: How group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol. Microbiol. 2011, 81, 588–601. [Google Scholar] [CrossRef] [PubMed]
- Bridel, S.; Bourgeon, F.; Marie, A.; Saulnier, D.; Pasek, S.; Nicolas, P.; Bernardet, J.-F.; Duchaud, E. Genetic diversity and population structure of Tenacibaculum maritimum, a serious bacterial pathogen of marine fish: From genome comparisons to high throughput MALDI-TOF typing. Vet. Res. 2020, 51, 60. [Google Scholar] [CrossRef]
- Lithgow, K.V.; Buchholz, V.C.H.; Ku, E.; Konschuh, S.; D’Aubeterre, A.; Sycuro, L.K. Protease activities of vaginal Porphyromonas species disrupt coagulation and extracellular matrix in the cervicovaginal niche. NPJ Biofilms Microbiomes 2022, 8, 8. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Manavalan, B.; Subramaniyam, S.; Shin, T.H.; Kim, M.O.; Lee, G. Machine-Learning-Based Prediction of Cell-Penetrating Peptides and Their Uptake Efficiency with Improved Accuracy. J. Proteome Res. 2018, 17, 2715–2726. [Google Scholar] [CrossRef]
- Manavalan, B.; Shin, T.H.; Lee, G. PVP-SVM: Sequence-Based Prediction of Phage Virion Proteins Using a Support Vector Machine. Front. Microbiol. 2018, 9, 476. [Google Scholar] [CrossRef]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.-C. mACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef] [Green Version]
- Singh, O.; Hsu, W.-L.; Su, E.C.-Y. ILeukin10Pred: A Computational Approach for Predicting IL-10-Inducing Immunosuppressive Peptides Using Combinations of Amino Acid Global Features. Biology 2021, 11, 5. [Google Scholar] [CrossRef]
- Malik, A.; Subramaniyam, S.; Kim, C.-B.; Manavalan, B. SortPred: The first machine learning based predictor to identify bacterial sortases and their classes using sequence-derived information. Comput. Struct. Biotechnol. J. 2021, 20, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, X.-M.; Takemoto, K.; Xu, H.; Li, Y.; Akutsu, T.; Song, J. FunSAV: Predicting the Functional Effect of Single Amino Acid Variants Using a Two-Stage Random Forest Model. PLoS ONE 2012, 7, e43847. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, H.; Wang, J.; Leier, A.; Marquez-Lago, T.; Yang, B.; Zhang, Z.; Akutsu, T.; Webb, G.I.; Daly, R.J. PhosphoPredict: A bioinformatics tool for prediction of human kinase-specific phosphorylation substrates and sites by integrating heterogeneous feature selection. Sci. Rep. 2017, 7, 6862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; He, W.; Malik, A.; Su, R.; Cui, L.; Manavalan, B. Computational prediction and interpretation of cell-specific replication origin sites from multiple eukaryotes by exploiting stacking framework. Briefings Bioinform. 2021, 22, bbaa275. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Lee, G.; Manavalan, B. STALLION: A stacking-based ensemble learning framework for prokaryotic lysine acetylation site prediction. Briefings Bioinform. 2022, 23, bbab376. [Google Scholar] [CrossRef]
- Li, Z.; Guo, W.; Ding, S.; Chen, L.; Feng, K.; Huang, T.; Cai, Y.-D. Identifying Key MicroRNA Signatures for Neurodegenerative Diseases With Machine Learning Methods. Front. Genet. 2022, 13, 880997. [Google Scholar] [CrossRef]
- Uchida, Y.; Yoshida, S.; Arita, Y.; Shimoda, H.; Kimura, K.; Yamada, I.; Tanaka, H.; Yokoyama, M.; Matsuoka, Y.; Jinzaki, M.; et al. Apparent Diffusion Coefficient Map-Based Texture Analysis for the Differentiation of Chromophobe Renal Cell Carcinoma from Renal Oncocytoma. Diagnostics 2022, 12, 817. [Google Scholar] [CrossRef]
- Chieregato, M.; Frangiamore, F.; Morassi, M.; Baresi, C.; Nici, S.; Bassetti, C.; Bnà, C.; Galelli, M. A hybrid machine learning/deep learning COVID-19 severity predictive model from CT images and clinical data. Sci. Rep. 2022, 12, 4329. [Google Scholar] [CrossRef]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Xiao, N.; Cao, D.-S.; Zhu, M.-F.; Xu, Q.-S. protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics 2015, 31, 1857–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubchak, I.; Muchnik, I.; Holbrook, S.R.; Kim, S.H. Prediction of protein folding class using global description of amino acid sequence. Proc. Natl. Acad. Sci. USA 1995, 92, 8700–8704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubchak, I.; Muchnik, I.; Mayor, C.; Dralyuk, I.; Kim, S.H. Recognition of a protein fold in the context of the Structural Classification of Proteins (SCOP) classification. Proteins 1999, 35, 401–407. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.-C. Prediction of Protein Subcellular Locations by Incorporating Quasi-Sequence-Order Effect. Biochem. Biophys. Res. Commun. 2000, 278, 477–483. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Yang, B.; Xie, R.; Marquez-Lago, T.T.; Leier, A.; Hayashida, M.; Akutsu, T.; Zhang, Y.; Chou, K.-C.; et al. Bastion3: A two-layer ensemble predictor of type III secreted effectors. Bioinformatics 2019, 35, 2017–2028. [Google Scholar] [CrossRef] [Green Version]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Acharjee, A.; Larkman, J.; Xu, Y.; Cardoso, V.R.; Gkoutos, G.V. A random forest based biomarker discovery and power analysis framework for diagnostics research. BMC Med Genom. 2020, 13, 178. [Google Scholar] [CrossRef]
- Chen, R.C.; Dewi, C.; Huang, S.W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. J. Big Data 2022, 7. [Google Scholar] [CrossRef]
- Yang, Z.; Jin, M.; Zhang, Z.; Lu, J.; Hao, K. Classification Based on Feature Extraction For Hepatocellular Carcinoma Diagnosis Using High-throughput Dna Methylation Sequencing Data. Procedia Comput. Sci. 2017, 107, 412–417. [Google Scholar] [CrossRef]
- Honaas, L.; Hargarten, H.; Hadish, J.; Ficklin, S.P.; Serra, S.; Musacchi, S.; Wafula, E.; Mattheis, J.; dePamphilis, C.W.; Rudell, D. Transcriptomics of Differential Ripening in ‘d’Anjou’ Pear (Pyrus communis L.). Front. Plant Sci. 2021, 12, 609684. [Google Scholar] [CrossRef] [PubMed]
| Datasets | ||||
|---|---|---|---|---|
| Class | Training Set | Independent Validation Sets | ||
| VS1 | VS2 | VS3 | ||
| Positive (C10 family cysteine proteases) | 269 | 67 | 82 | 82 |
| Negative | 280 | 70 | 200 * | 349 ** |
| Features | Dimension Size | Accuracy | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| AAC | 20 | 0.880 | 0.874 | 0.886 | 0.759 |
| AutoC | 720 | 0.883 | 0.877 | 0.889 | 0.767 |
| CTD | 147 | 0.900 | 0.892 | 0.907 | 0.800 |
| CTriad | 343 | 0.878 | 0.874 | 0.882 | 0.756 |
| DPC | 400 | 0.934 | 0.944 | 0.925 | 0.869 |
| QSO | 100 | 0.889 | 0.885 | 0.893 | 0.778 |
| SOCN | 60 | 0.754 | 0.755 | 0.754 | 0.508 |
| Hybrid | 1790 | 0.929 | 0.944 | 0.914 | 0.858 |
| Features | Dimension Size | Accuracy | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| AutoC | 102 | 0.900 | 0.881 | 0.918 | 0.800 |
| CTD | 89 | 0.907 | 0.903 | 0.911 | 0.814 |
| CTriad | 67 | 0.883 | 0.874 | 0.893 | 0.767 |
| DPC | 79 | 0.925 | 0.926 | 0.925 | 0.851 |
| QSO | 45 | 0.913 | 0.907 | 0.918 | 0.825 |
| SOCN | 58 | 0.756 | 0.755 | 0.757 | 0.512 |
| Hybrid | 139 | 0.956 | 0.944 | 0.968 | 0.913 |
| Features | Accuracy | Sensitivity | Specificity | MCC |
|---|---|---|---|---|
| AutoC | 0.839 | 0.731 | 0.943 | 0.692 |
| CTD | 0.839 | 0.791 | 0.886 | 0.681 |
| CTriad | 0.861 | 0.776 | 0.943 | 0.731 |
| DPC | 0.891 | 0.836 | 0.943 | 0.785 |
| QSO | 0.869 | 0.836 | 0.900 | 0.738 |
| SOCN | 0.737 | 0.701 | 0.771 | 0.474 |
| Hybrid | 0.927 | 0.896 | 0.957 | 0.855 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malik, A.; Mahajan, N.; Dar, T.A.; Kim, C.-B. C10Pred: A First Machine Learning Based Tool to Predict C10 Family Cysteine Peptidases Using Sequence-Derived Features. Int. J. Mol. Sci. 2022, 23, 9518. https://doi.org/10.3390/ijms23179518
Malik A, Mahajan N, Dar TA, Kim C-B. C10Pred: A First Machine Learning Based Tool to Predict C10 Family Cysteine Peptidases Using Sequence-Derived Features. International Journal of Molecular Sciences. 2022; 23(17):9518. https://doi.org/10.3390/ijms23179518
Chicago/Turabian StyleMalik, Adeel, Nitin Mahajan, Tanveer Ali Dar, and Chang-Bae Kim. 2022. "C10Pred: A First Machine Learning Based Tool to Predict C10 Family Cysteine Peptidases Using Sequence-Derived Features" International Journal of Molecular Sciences 23, no. 17: 9518. https://doi.org/10.3390/ijms23179518
APA StyleMalik, A., Mahajan, N., Dar, T. A., & Kim, C.-B. (2022). C10Pred: A First Machine Learning Based Tool to Predict C10 Family Cysteine Peptidases Using Sequence-Derived Features. International Journal of Molecular Sciences, 23(17), 9518. https://doi.org/10.3390/ijms23179518







