Vitelline Membrane Protein 26 Mutagenesis, Using CRISPR/Cas9, Results in Egg Collapse in Plutella xylostella
Abstract
1. Introduction
2. Results
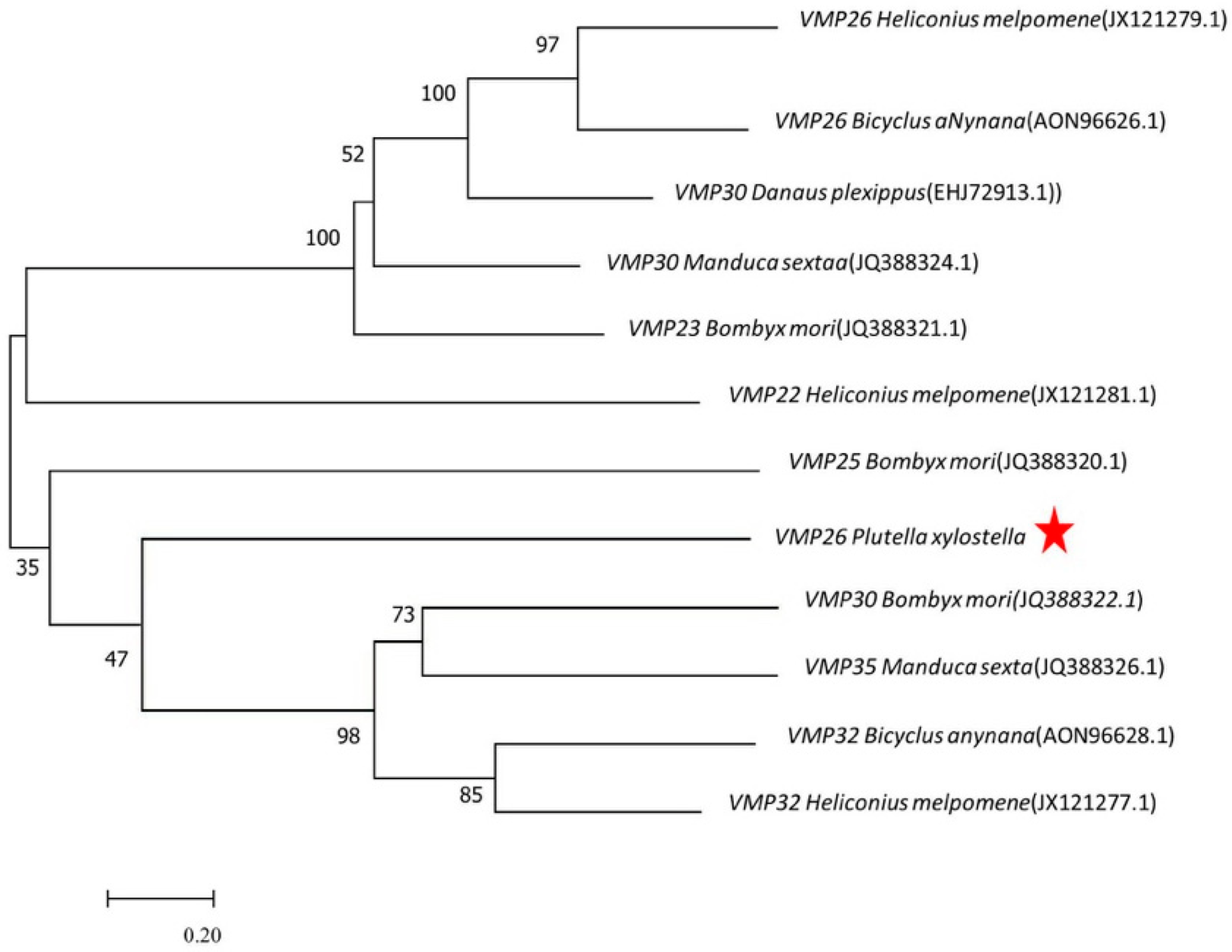
2.1. Identification and Analysis of PxVMP26
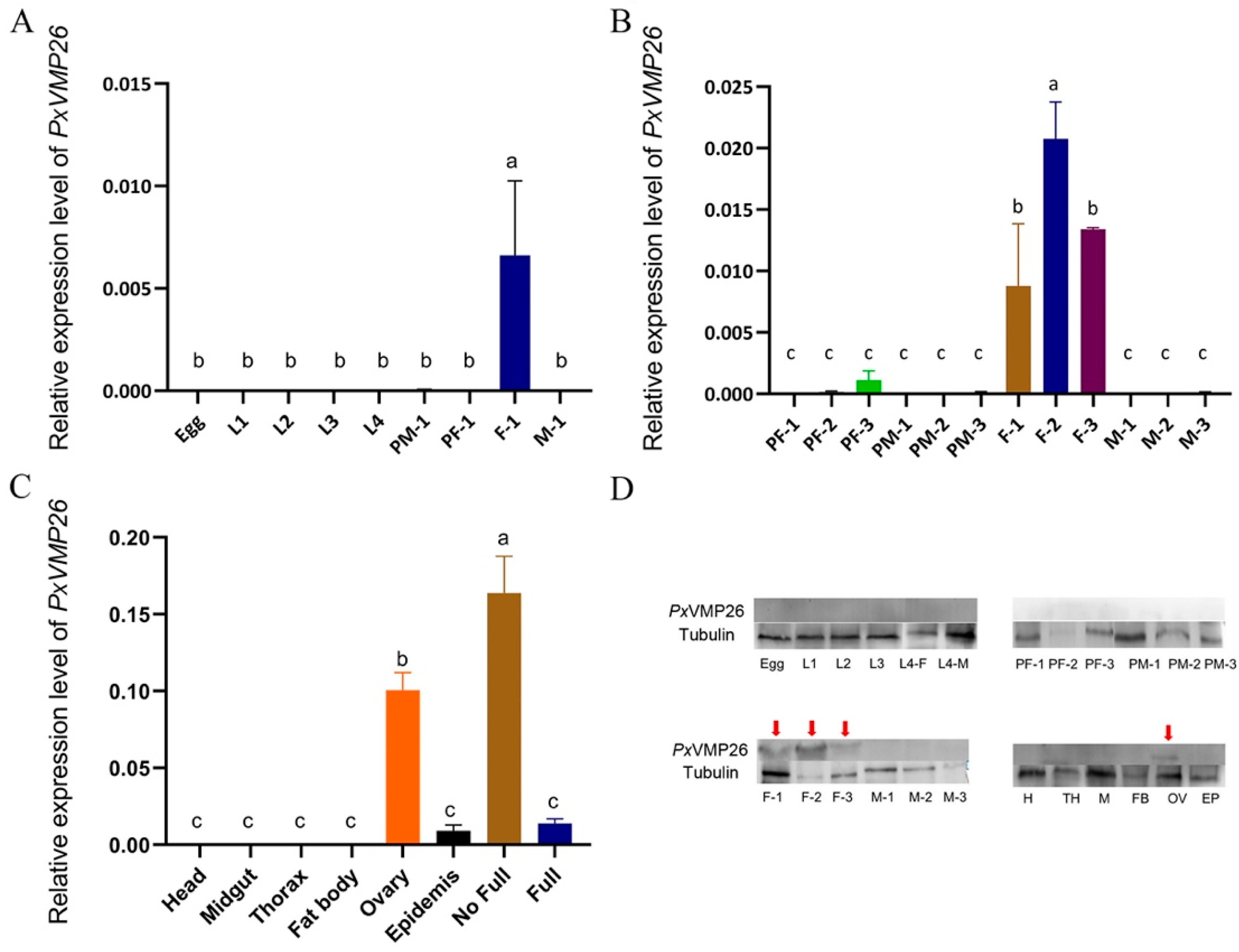
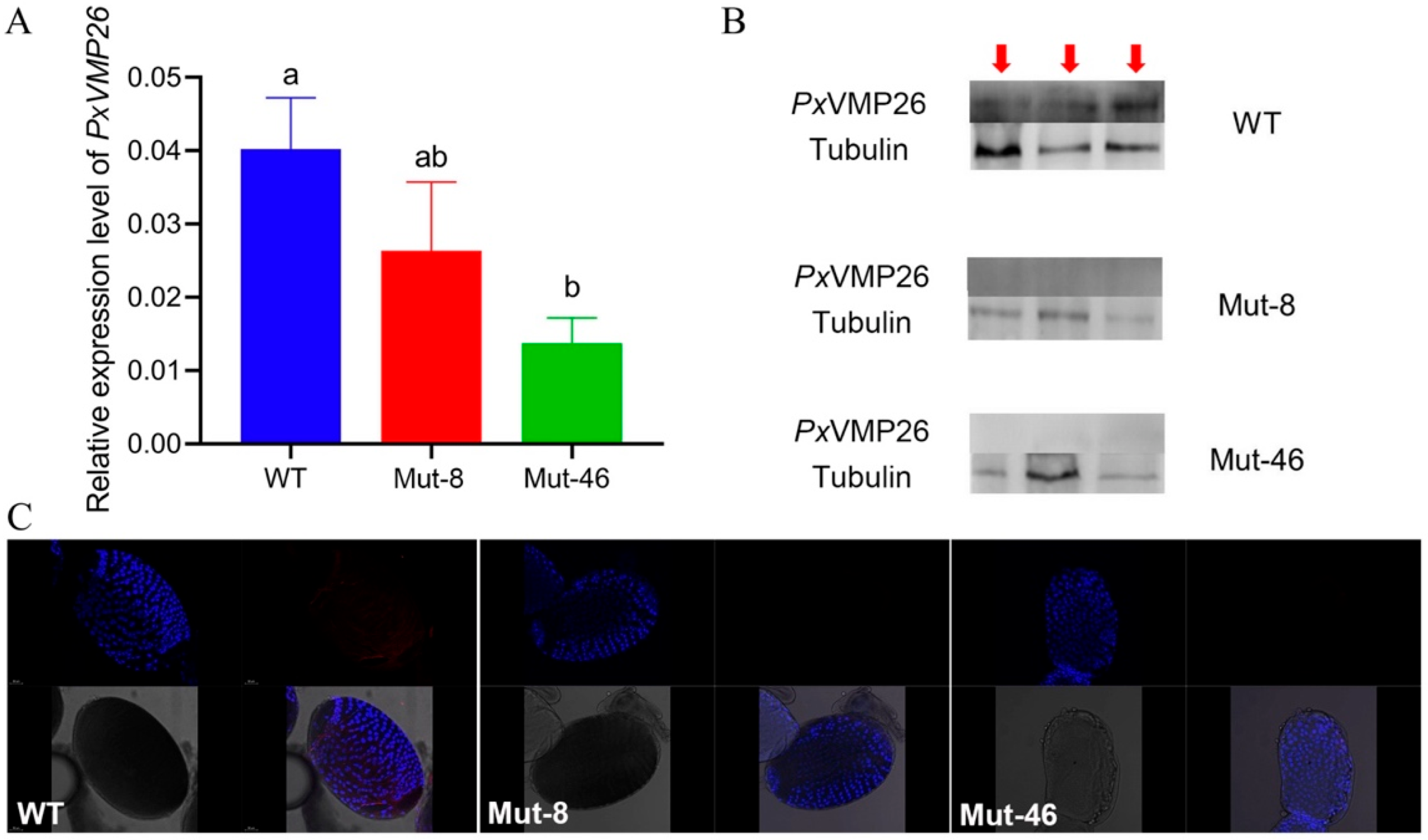
2.2. Expression Profile of PxVMP26
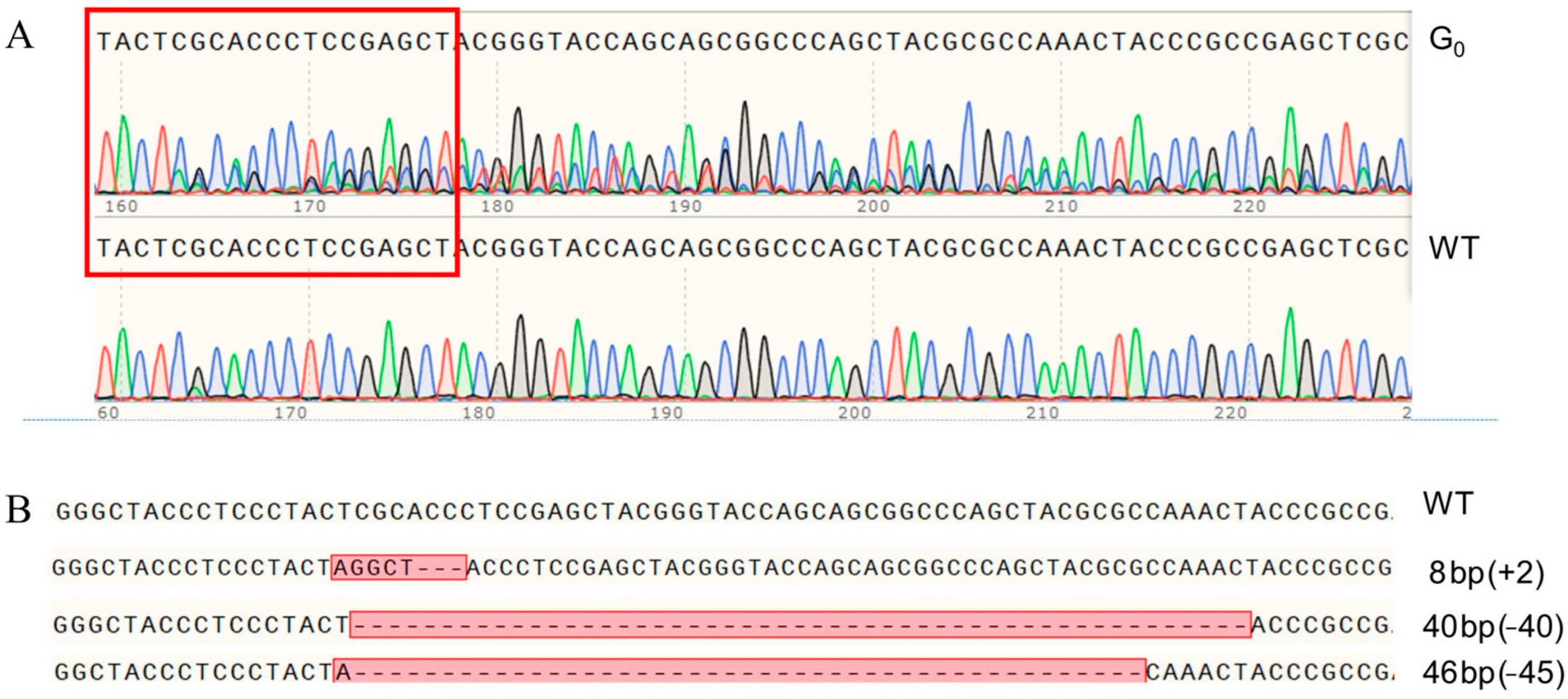
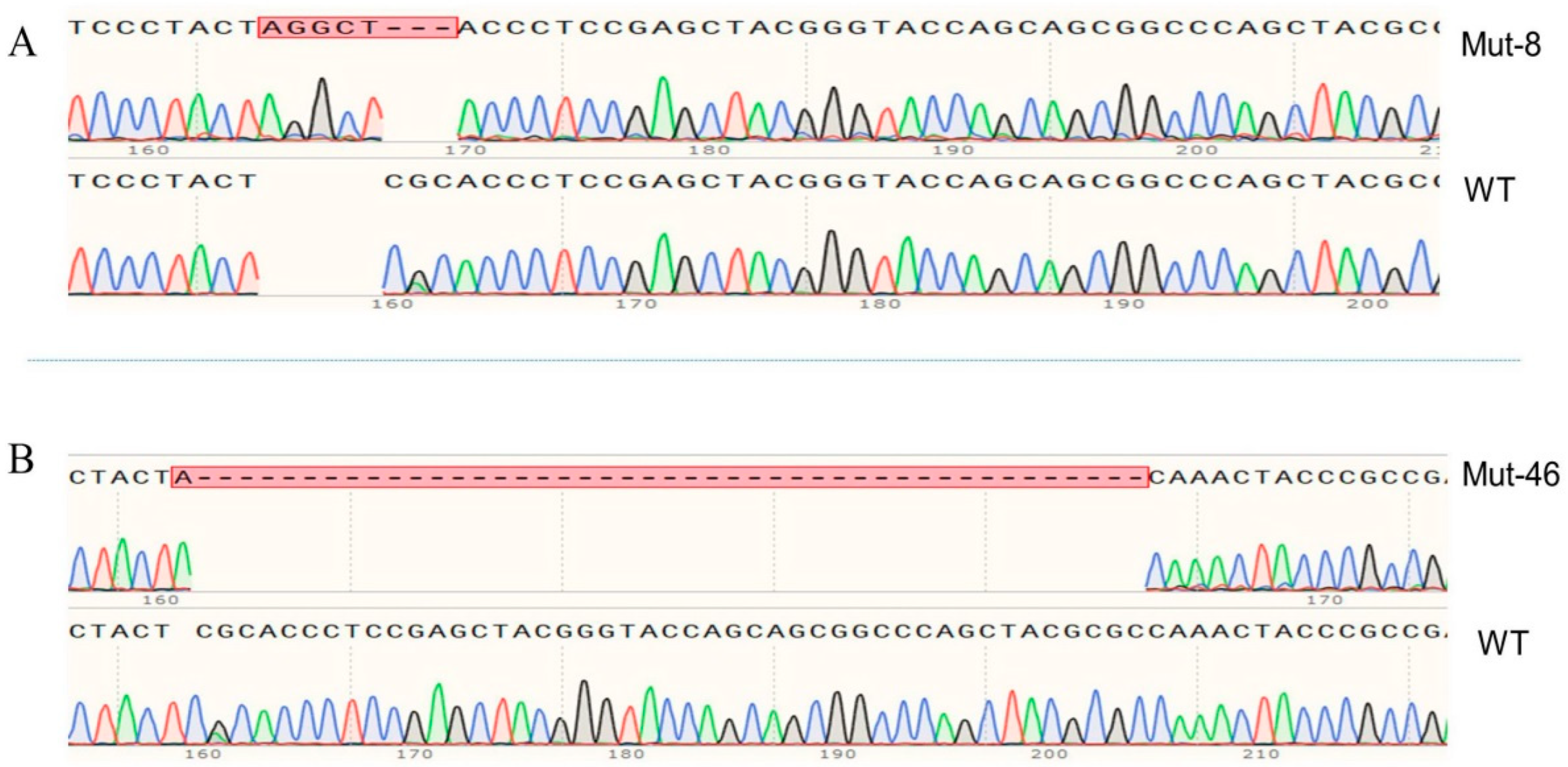
2.3. The mutation of PxVMP26 Produced by CRISPR/Cas9
2.4. Establishment of the Homozygous PxVMP26 Mutant Lines
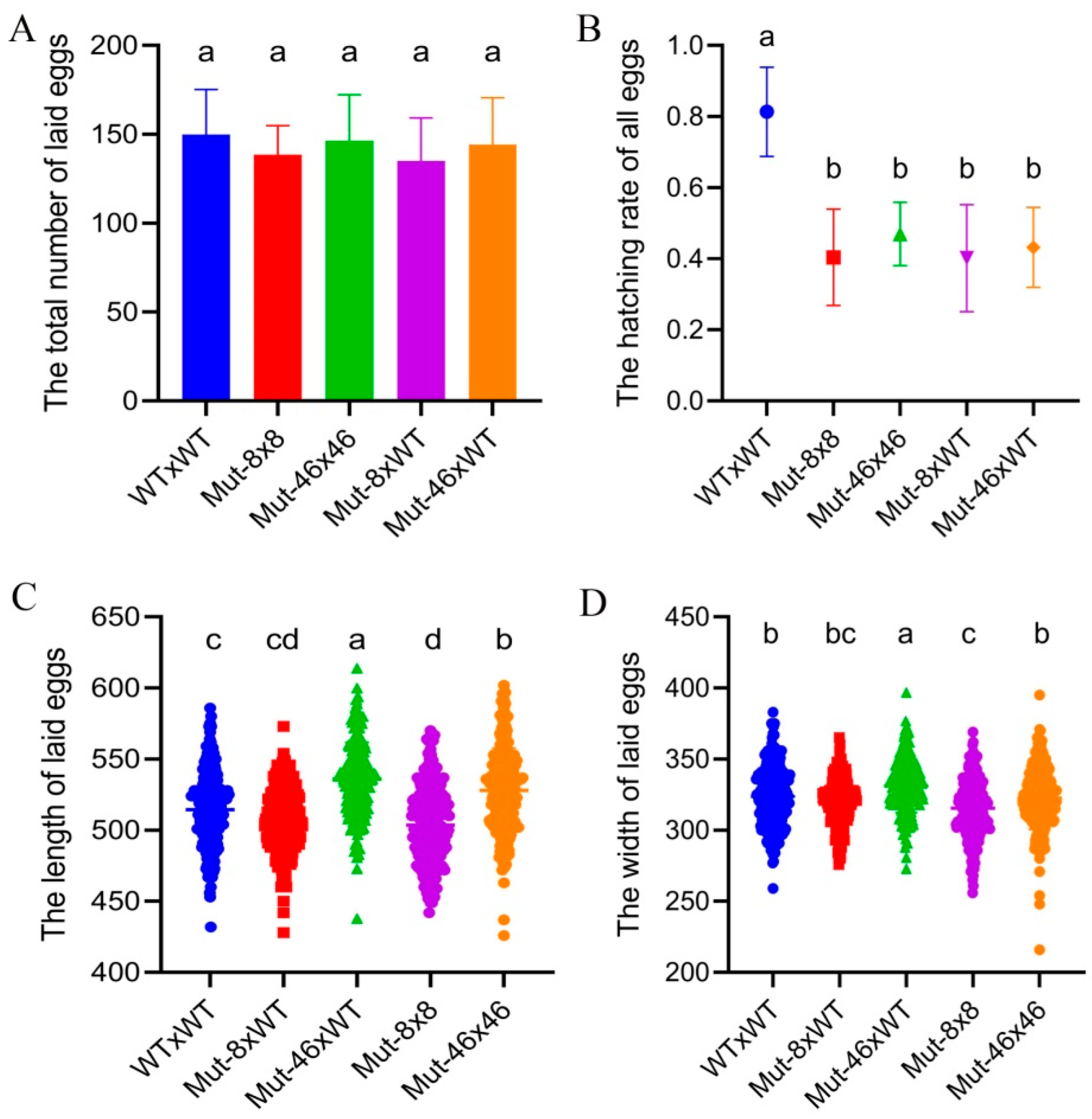
2.5. Effects of PxVMP26 Knockout on the Fertility of P. xylostella
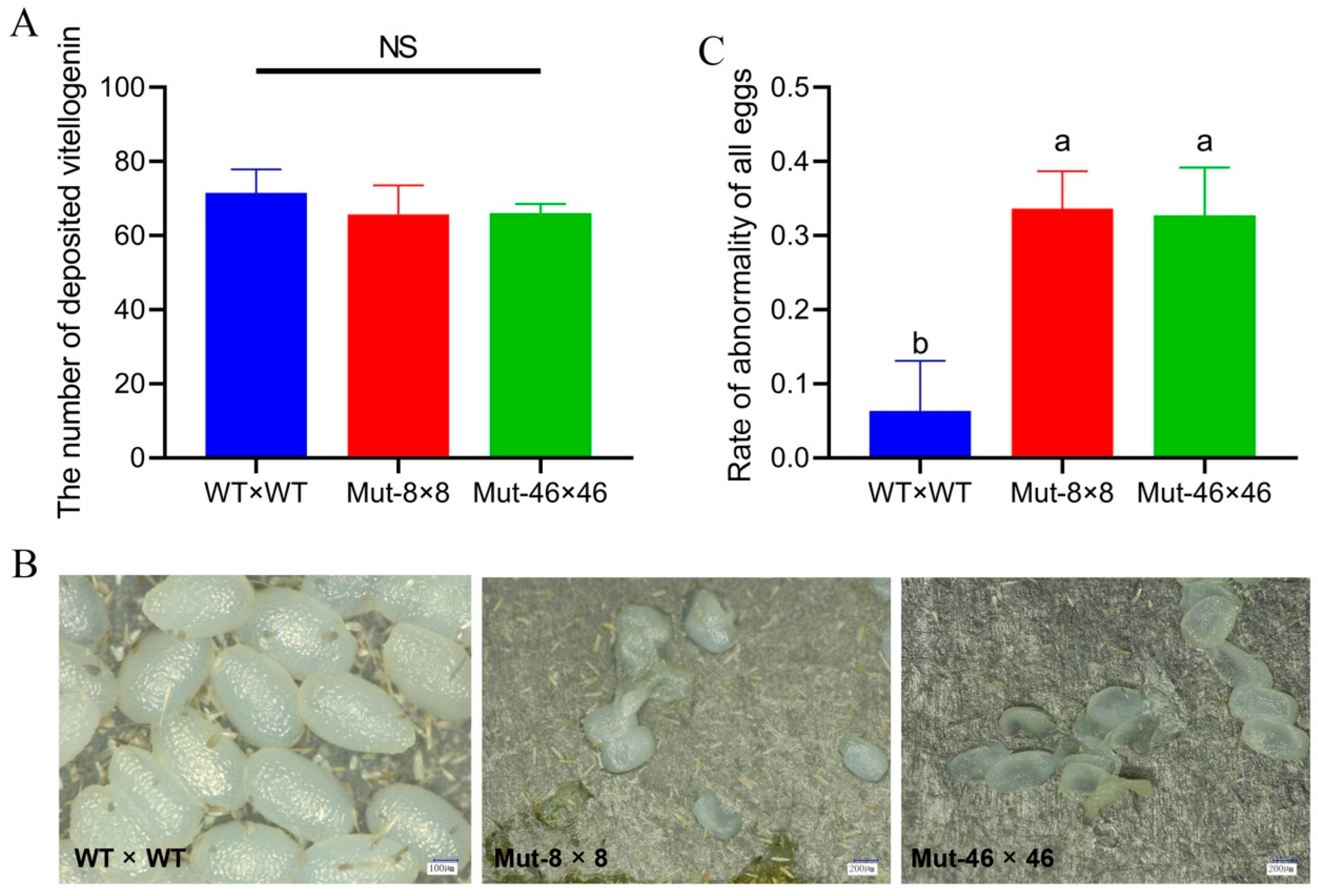
2.6. Effects of PxVMP26 Knockout on Vitelline Deposition and Water Retention of P. xylostella Eggs
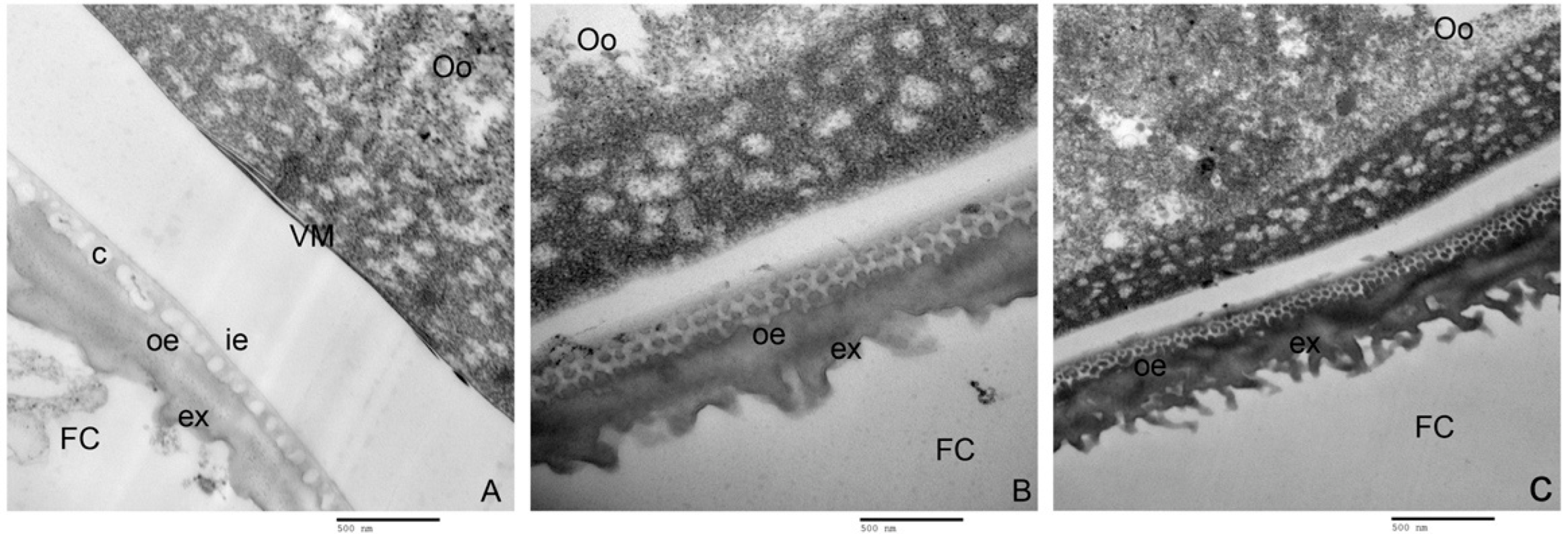
2.7. Effects of PxVMP26 Knockout on the Formation of Vitelline Membrane
3. Discussion
4. Materials and Methods
4.1. Insect Colony
4.2. Total RNA Extraction and cDNA Synthesis
4.3. PxVMP26 Cloning
4.4. Sequence Comparison and Phylogenetic Analysis of PxVMP26
4.5. Expression Patterns of PxVMP26
4.5.1. Sample Preparation
4.5.2. qRT-PCR
4.5.3. Protein Preparation and Western Blot
4.6. Immunofluorescence Analysis
4.7. sgRNA Design and Synthesis
4.8. sgRNA/Cas9 Protein Microinjection
4.9. Genetic Crosses and PCR-Based Genotyping
4.10. Phenotypic Observation and Bioassays
4.11. Microstructure Observation of Vitelline Membrane
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woods, H.A.; Bonnecaze, R.T.; Zrubek, B. Oxygen and water flux across eggshells of Manduca sexta. J. Exp. Biol. 2005, 208, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Margaritis, L.H. Structure and physiology of the eggshell. In Comprehensive Insect Physiology, Biochemistry and Pharmacology; Gilbert, L.I., Kercut, G.A., Eds.; Pergamon: Oxford, UK, 1985; Volume 1, pp. 153–230. [Google Scholar]
- Irles, P.; Piulachs, M.D. Citrus, a key insect eggshell protein. Insect Biochem. Mol. Biol. 2011, 41, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Woods, H.A. Water loss and gas exchange by eggs of Manduca sexta: Trading off costs and benefits. J. Insect Physiol. 2010, 56, 480–487. [Google Scholar] [CrossRef]
- Lou, Y.H.; Pan, P.L.; Ye, Y.X.; Cheng, C.; Xu, H.J.; Zhang, C.X. Identification and functional analysis of a novel chorion protein essential for egg maturation in the brown planthopper. Insect Mol. Biol. 2018, 27, 393–403. [Google Scholar] [CrossRef]
- Furriols, M.; Casanova, J. Germline and somatic vitelline proteins colocalize in aggregates in the follicular epithelium of Drosophila ovaries. Fly 2014, 8, 113–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velentzas, A.D.; Velentzas, P.D.; Katarachia, S.A.; Anagnostopoulos, A.K.; Sagioglou, N.E.; Thanou, E.V.; Tsioka, M.M.; Mpakou, V.E.; Kollia, Z.; Gavriil, V.E.; et al. The indispensable contribution of s38 protein to ovarian-eggshell morphogenesis in Drosophila melanogaster. Sci. Rep. 2018, 8, 16103. [Google Scholar] [CrossRef]
- Zrubek, B.; Woods, H.A. Insect eggs exert rapid control over an oxygen–water tradeoff. Proc. Biol. Sci. 2006, 273, 831–834. [Google Scholar] [CrossRef]
- Isoe, J.; Koch, L.E.; Isoe, Y.E.; Rascon, A.A., Jr.; Brown, H.E.; Massani, B.B.; Miesfeld, R.L. Identification and characterization of a mosquito-specific eggshell organizing factor in Aedes aegypti mosquitoes. PLoS Biol. 2019, 17, e3000068. [Google Scholar] [CrossRef]
- Mineo, A.; Furriols, M.; Casanova, J. Accumulation of the Drosophila Torso-like protein at the blastoderm plasma membrane suggests that it translocates from the eggshell. Development 2015, 142, 1299–1304. [Google Scholar] [CrossRef]
- Taylor, S.E.; Tuffery, J.; Bakopoulos, D.; Lequeux, S.; Warr, C.G.; Johnson, T.K.; Dearden, P.K. The torso-like gene functions to maintain the structure of the vitelline membrane in Nasonia vitripennis, implying its co-option into Drosophila axis formation. Biol. Open 2019, 8, bio046284. [Google Scholar] [CrossRef]
- Dos Santos, G.; Schroeder, A.J.; Goodman, J.L.; Strelets, V.B.; Crosby, M.A.; Thurmond, J.; Emmert, D.B.; Gelbart, W.M.; FlyBase, C. FlyBase: Introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015, 43, D690–D697. [Google Scholar] [CrossRef] [PubMed]
- Papantonis, A.; Swevers, L.; Iatrou, K. Chorion genes: A landscape of their evolution, structure, and regulation. Annu. Rev. Entomol. 2015, 60, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Marinotti, O.; Ngo, T.; Kojin, B.B.; Chou, S.P.; Nguyen, B.; Juhn, J.; Carballar-Lejarazú, R.; Marinotti, P.N.; Jiang, X.; Walter, M.F. Integrated proteomic and transcriptomic analysis of the Aedes aegypti eggshell. BMC Dev. Biol. 2014, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Amenya, D.A.; Chou, W.; Li, J.; Yan, G.; Gershon, P.D.; James, A.A.; Marinotti, O. Proteomics reveals novel components of the Anopheles gambiae eggshell. J. Insect Physiol. 2010, 56, 1414–1419. [Google Scholar] [CrossRef]
- Gonçalves, V.R.; Jr Sobrinho, I.S.; Jr Malagó, W.; Henrique-Silva, F.; de Brito, R.A. Transcriptome analysis of female reproductive tissues of Anastrepha obliqua and molecular evolution of eggshell proteins in the fraterculus group. Insect Mol. Biol. 2013, 22, 551–561. [Google Scholar] [CrossRef]
- Wei, D.; Li, R.; Zhang, M.Y.; Liu, Y.W.; Zhang, Z.; Smagghe, G.; Wang, J.J. Comparative proteomic profiling reveals molecular characteristics associated with oogenesis and oocyte maturation during ovarian development of Bactrocera dorsalis (Hendel). Int. J. Mol. Sci. 2017, 18, 1379. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, Y.X.; Liu, Y.W.; Li, W.J.; Chen, Z.X.; Smagghe, G.; Wang, J.J. Gene expression profiling of ovary identified eggshell proteins regulated by 20-hydroxyecdysone in Bactrocera dorsalis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 206–216. [Google Scholar] [CrossRef]
- Li, W.; Song, Y.; Xu, H.J.; Wei, D.; Wang, J. Vitelline membrane protein gene ZcVMP26Ab and its role in preventing water loss in Zeugodacus cucurbitae (Coquillett) embryos. Entomol. Gen. 2021, 41, 279–288. [Google Scholar] [CrossRef]
- Manogaran, A.; Waring, G.L. The N-terminal prodomain of sV23 is essential for the assembly of a functional vitelline membrane network in Drosophila. Dev. Biol. 2004, 270, 261–271. [Google Scholar] [CrossRef][Green Version]
- Fakhouri, M.; Elalayli, M.; Sherling, D.; Hall, J.D.; Miller, E.; Sun, X.; Wells, L.; LeMosy, E.K. Minor proteins and enzymes of the Drosophila eggshell matrix. Dev. Biol. 2006, 293, 127–141. [Google Scholar] [CrossRef]
- Wu, T.; Manogaran, A.L.; Beauchamp, J.M.; Waring, G.L. Drosophila vitelline membrane assembly: A critical role for an evolutionarily conserved cysteine in the “VM domain” of sV23. Dev. Biol. 2010, 347, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Elalayli, M.; Hall, J.D.; Fakhouri, M.; Neiswender, H.; Ellison, T.T.; Han, Z.; Roon, P.; LeMosy, E.K. Palisade is required in the Drosophila ovary for assembly and function of the protective vitelline membrane. Dev. Biol. 2008, 319, 359–369. [Google Scholar] [CrossRef][Green Version]
- Zhang, Z.; Stevens, L.M.; Stein, D. Sulfation of eggshell components by Pipe defines dorsal-ventral polarity in the Drosophila embryo. Curr. Biol. 2009, 19, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Kendirgi, F.; Swevers, L.; Iatrou, K. An ovarian follicular epithelium protein of the silkworm (Bombyx mori) that associates with the vitelline membrane and contributes to the structural integrity of the follicle. FEEBS Lett. 2002, 524, 59–68. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, Q.; Li, S.; He, N. Silkworm egg proteins at the germ-band formation stage and a functional analysis of BmEP80 protein. Insect Biochem. Mol. Biol. 2011, 41, 572–581. [Google Scholar] [CrossRef]
- Sdralia, N.; Swevers, L.; Iatrou, K. BmVMP90, a large vitelline membrane protein of the domesticated silkmoth Bombyx mori, is an essential component of the developing ovarian follicle. Insect Biochem. Mol. Biol. 2012, 42, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Gao, P.; Zhao, Q.; Tang, S.; Shen, X.; Zhang, G.; Qiu, Z.; Xia, D.; Huang, Y.; Xu, Y. Mutation of a vitelline membrane protein, BmEP80, is responsible for the silkworm “Ming” lethal egg mutant. Gene 2013, 515, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Xia, D.; Qiu, Z.; Gao, P.; Tang, S.; Shen, X.; Zhu, F.; Zhao, Q. Expression of a vitelline membrane protein, BmVMP23, is repressed by bmo-miR-1a-3p in silkworm, Bombyx mori. FEEBS Lett. 2013, 587, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef]
- You, M.; Yue, Z.; He, W.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef]
- Peng, L.; Wang, L.; Yang, Y.F.; Zou, M.M.; He, W.Y.; Wang, Y.; Wang, Q.; Vasseur, L.; You, M.S. Transcriptome profiling of the Plutella xylostella (Lepidoptera: Plutellidae) ovary reveals genes involved in oogenesis. Gene 2017, 637, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Gribble, M.; Baker, D.; Marois, E.; Russell, S.; et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef]
- Reid, W.; O’Brochta, D.A. Applications of genome editing in insects. Curr. Opin. Insect Sci. 2016, 13, 43–54. [Google Scholar] [CrossRef]
- Taning, C.N.T.; Van Eynde, B.; Yu, N.; Ma, S.; Smagghe, G. CRISPR/Cas9 in insects: Applications, best practices and biosafety concerns. J. Insect Physiol. 2017, 98, 245–257. [Google Scholar] [CrossRef]
- Peng, L.; Wang, Q.; Zou, M.M.; Qin, Y.D.; Vasseur, L.; Chu, L.N.; Zhai, Y.L.; Dong, S.J.; Liu, L.L.; He, W.Y.; et al. CRISPR/Cas9-mediated vitellogenin receptor knockout leads to functional deficiency in the reproductive development of Plutella xylostella. Front. Physiol. 2020, 10, 1585. [Google Scholar] [CrossRef]
- Zou, M.M.; Wang, Q.; Chu, L.N.; Vasseur, L.; Zhai, Y.L.; Qin, Y.D.; He, W.Y.; Yang, G.; Zhou, Y.Y.; Peng, L.; et al. CRISPR/Cas9-induced vitellogenin knockout lead to incomplete embryonic development in Plutella xylostella. Insect Biochem. Mol. Biol. 2020, 123, 103406. [Google Scholar] [CrossRef]
- Griffith, C.M.; Lai-Fook, J. Structure and formation of the chorion in the butterfly. Calpodes. Tissue Cell 1986, 18, 589–601. [Google Scholar] [CrossRef]
- Xu, Y.M. Studies on Inner Eggshell Proteins and a Lethal Egg Mutant (l-em) of the Silkworm, Bombyx mori. Ph.D. Thesis, Southwest University, Chongqing, China, 2013. [Google Scholar]
- Zhan, S.; Merlin, C.; Boore, J.L.; Reppert, S.M. The monarch butterfly genome yields insights into long-distance migration. Cell 2011, 147, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Iatrou, K. The ecdysone regulatory cascade and ovarian development in lepidopteran insects: Insights from the silkmoth paradigm. Insect Biochem. Mol. Biol. 2003, 33, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Cavaliere, V.; Bernardi, F.; Romani, P.; Duchi, S.; Gargiulo, G. Building up the Drosophila eggshell: First of all the eggshell genes must be transcribed. Dev. Dyn. 2008, 237, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zou, Z.; Zha, X.; Xiang, Z.; He, N. A syntenic coding region for vitelline membrane proteins in four lepidopteran insects. Insect Biochem. Mol. Biol. 2012, 1, 155–160. [Google Scholar]
- Mineo, A.; Furriols, M.; Casanova, J. Transfer of dorsoventral and terminal information from the ovary to the embryo by a common group of eggshell proteins in Drosophila. Genetics 2017, 205, 1529–1536. [Google Scholar] [CrossRef]
- Wei, D.; Xu, H.Q.; Chen, D.; Zhang, S.Y.; Li, W.J.; Smagghe, G.; Wang, J.J. Genome-wide gene expression profiling of the melon fly, Zeugodacus cucurbitae, during thirteen life stages. Sci. Data 2020, 7, 45. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Y.; Zeng, B.; Wang, Y.; James, A.A.; Gurr, G.M.; Yang, G.; Lin, X.; Huang, Y.; You, M. CRISPR/Cas9 mediated knockout of the abdominal-A homeotic gene in the global pest, diamondback moth (Plutella xylostella). Insect Biochem. Mol. Biol. 2016, 75, 98–106. [Google Scholar] [CrossRef]
- Zhang, L.; Reed, R.D. A practical guide to CRISPR/Cas9 genome editing in Lepidoptera. In Diversity and Evolution of Butterfly wing Patterns; Sekimura, T., Nijhout, H.F., Eds.; Springer: Singapore, 2017; pp. 155–172. [Google Scholar]
- Chen, W.; Dong, Y.; Saqib, H.S.A.; Vasseur, L.; Zhou, W.; Zheng, L.; Lai, Y.; Ma, X.; Lin, L.; Xu, X.; et al. Functions of duplicated glucosinolate sulfatases in the development and host adaptation of Plutella xylostella. Insect Biochem. Mol. Biol. 2020, 119, 103316. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Bennett, E.J. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014, 204, 467–476. [Google Scholar] [CrossRef]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef]
- Yamashita, O.; Irie, K. Larval hatching from vitellogenin-deficient eggs developed in male hosts of the silkworm. Nature 1980, 283, 385–386. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Scali, M.; Vignani, R.; Spadafora, A.; Sensi, E.; Mazzuca, S.; Cresti, M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 2003, 24, 2369–2375. [Google Scholar] [CrossRef] [PubMed]
| Purpose | Primer Name | Primer Sequence 5′-3′ | Position |
|---|---|---|---|
| Common PCR | C-VMP26 F | ATGGTGCCCCTGGCGGAG | 1–18 |
| C-VMP26 R | TCAGTAGCCATATCCAGGCCTTGG | 829–852 | |
| qRT-PCR | Q-RPL32 F | CAATCAGGCCAATTTACCGC | 5–24 |
| Q-RPL32 R | CTGCGTTTACGCCAGTTACG | 94–113 | |
| Q-RPL8 F | CGGTCGTGCCTACCACAAATACA | 560–582 | |
| Q-RPL8 R | CGTGAGGATGCTCCACAGGGT | 628–648 | |
| Q-EF-1α F | GCCTCCCTACAGCGAATC | 477–494 | |
| Q-EF-1α R | CGTGAGGATGCTCCACAGGGT | 620–637 | |
| Q-VMP26 F | CGATGCAGCCGCTGGAAGA | 104–122 | |
| Q- VMP26 R | GTAGTCGTAGTCCCTCGCAG | 266–285 | |
| sgRNA synthesis | sgRNA-F1 * | TAATACGACTCACTATA TAGCCCGCGTATCCCGGGCT GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCC | - |
| sgRNA-F2 * | TAATACGACTCACTATA GTAGCTCGGAGGGTGCGAGT GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCC | ||
| sgRNA-ComR # | AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAA | ||
| Genotyping | VMP26-F | ACATGGACAACGTGCTACCG | 131–150 |
| VMP26-R | TTTGAGTTCTACGTCGCCCG | 611–630 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, Y.-L.; Dong, S.-J.; Zou, M.-M.; Qin, Y.-D.; Liu, L.-L.; Cao, M.-H.; Huang, M.-Q.; Vasseur, L.; You, M.-S.; Peng, L. Vitelline Membrane Protein 26 Mutagenesis, Using CRISPR/Cas9, Results in Egg Collapse in Plutella xylostella. Int. J. Mol. Sci. 2022, 23, 9538. https://doi.org/10.3390/ijms23179538
Zhai Y-L, Dong S-J, Zou M-M, Qin Y-D, Liu L-L, Cao M-H, Huang M-Q, Vasseur L, You M-S, Peng L. Vitelline Membrane Protein 26 Mutagenesis, Using CRISPR/Cas9, Results in Egg Collapse in Plutella xylostella. International Journal of Molecular Sciences. 2022; 23(17):9538. https://doi.org/10.3390/ijms23179538
Chicago/Turabian StyleZhai, Yi-Long, Shi-Jie Dong, Ming-Min Zou, Yu-Dong Qin, Li-Li Liu, Min-Hui Cao, Meng-Qi Huang, Liette Vasseur, Min-Sheng You, and Lu Peng. 2022. "Vitelline Membrane Protein 26 Mutagenesis, Using CRISPR/Cas9, Results in Egg Collapse in Plutella xylostella" International Journal of Molecular Sciences 23, no. 17: 9538. https://doi.org/10.3390/ijms23179538
APA StyleZhai, Y.-L., Dong, S.-J., Zou, M.-M., Qin, Y.-D., Liu, L.-L., Cao, M.-H., Huang, M.-Q., Vasseur, L., You, M.-S., & Peng, L. (2022). Vitelline Membrane Protein 26 Mutagenesis, Using CRISPR/Cas9, Results in Egg Collapse in Plutella xylostella. International Journal of Molecular Sciences, 23(17), 9538. https://doi.org/10.3390/ijms23179538





