Intrarectal Capsazepine Administration Modulates Colonic Mucosal Health in Mice
Abstract
1. Introduction
2. Results
2.1. Rectal Administration of CPZ Inhibits TRPA1 and TRPV1 Mediated Nociceptive Responses and Modulates the Expression of Peptidergic Genes in DRGs
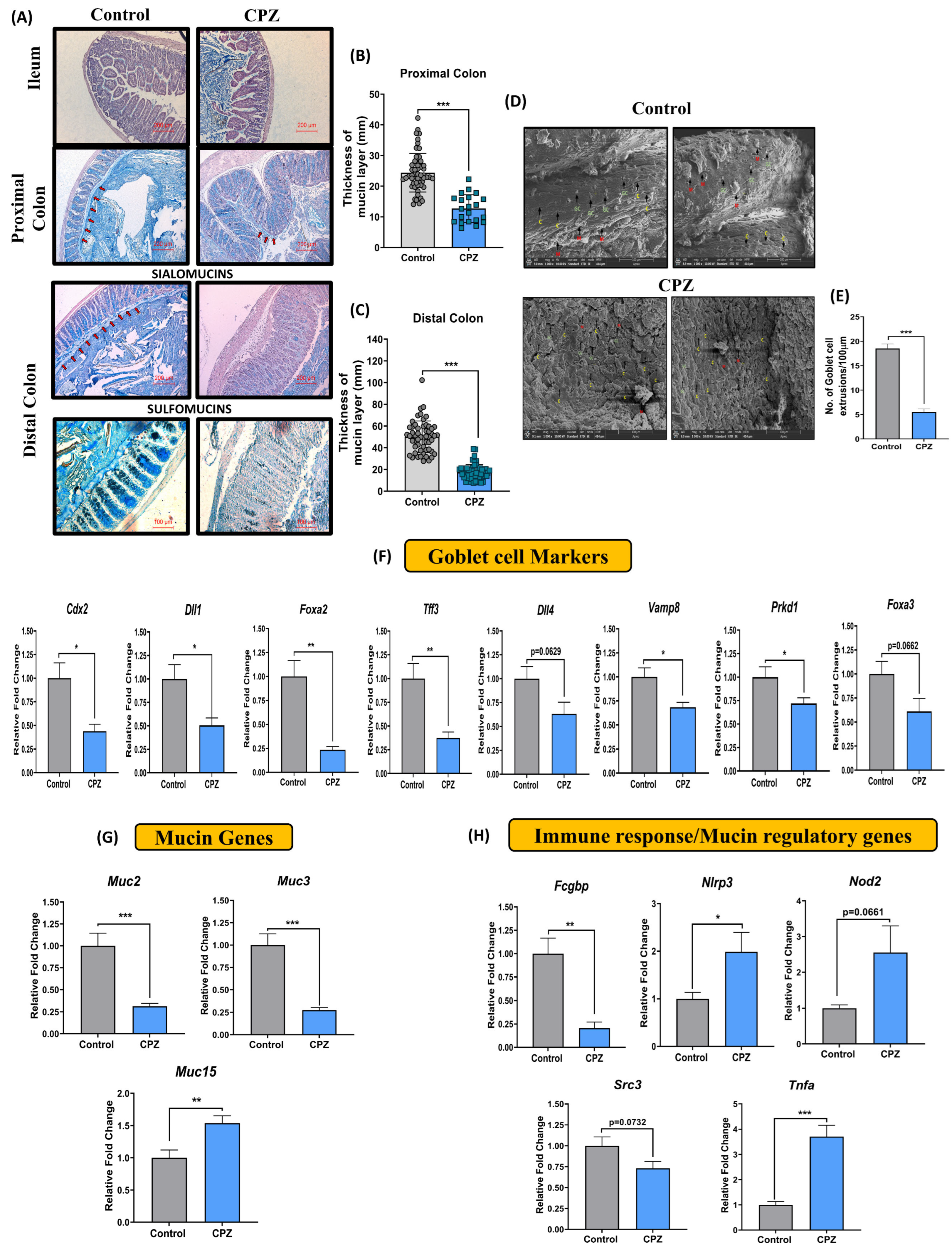
2.2. Intrarectal CPZ Administration Negatively Regulates Mucus Homeostasis
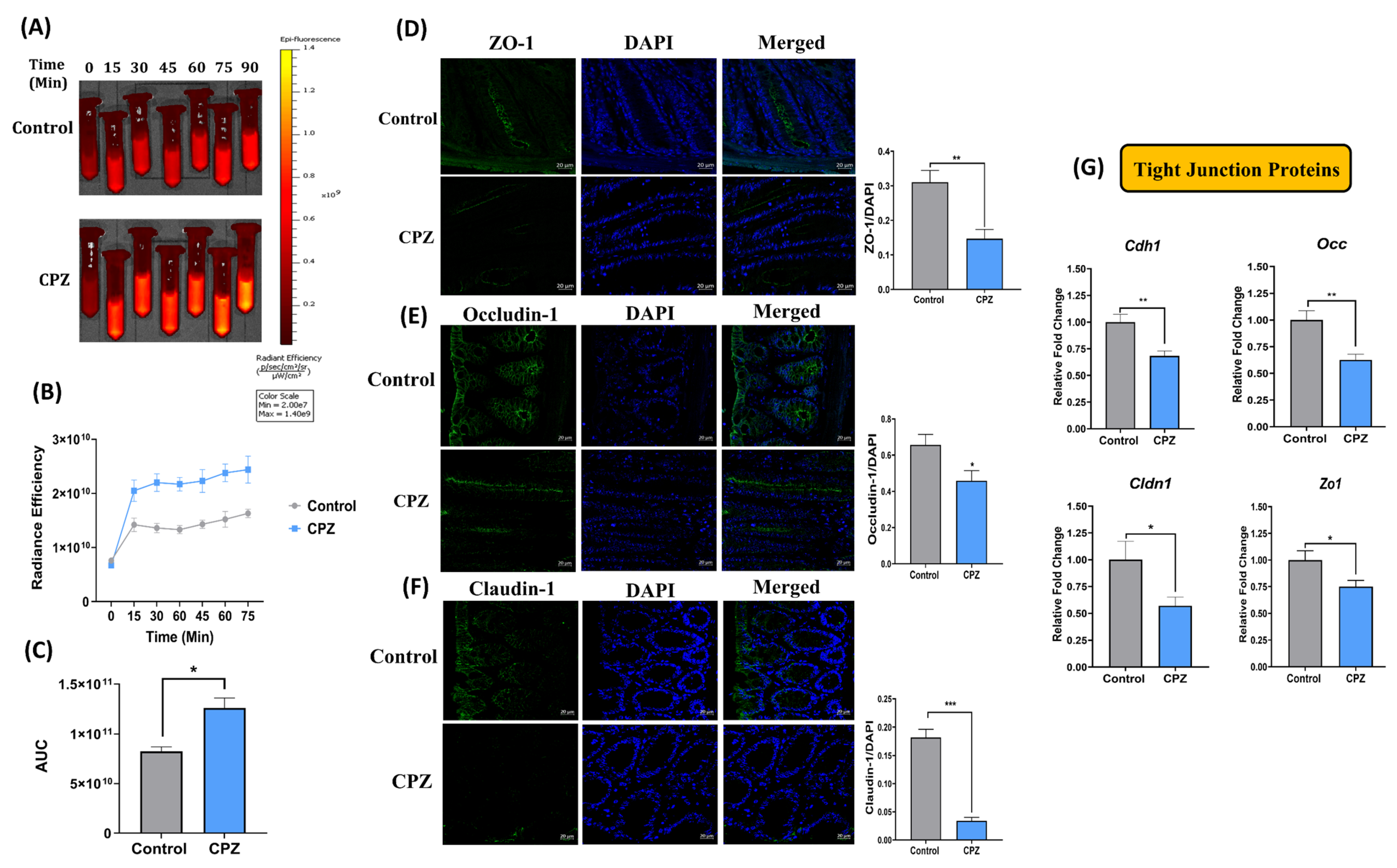
2.3. Rectal Administration of CPZ Increases Intestinal Permeability and Compromises Gut Barrier Function
2.4. Rectal CPZ Treatment Alters Mucus Associated Cecal Metabolites and Mucin Glycosylation Enzymes
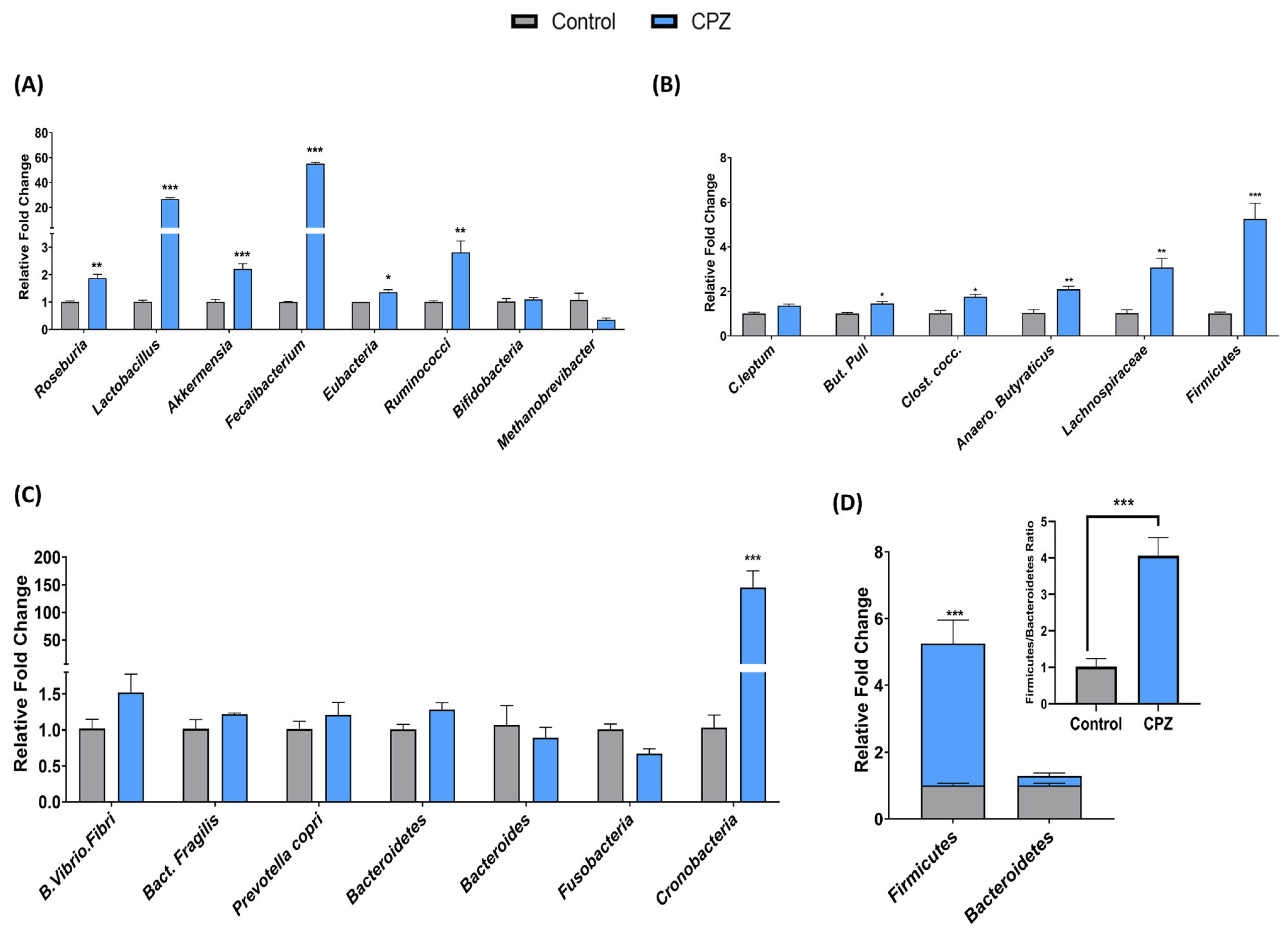
2.5. Intrarectal Administration of CPZ Induced Gut Microbiota Changes
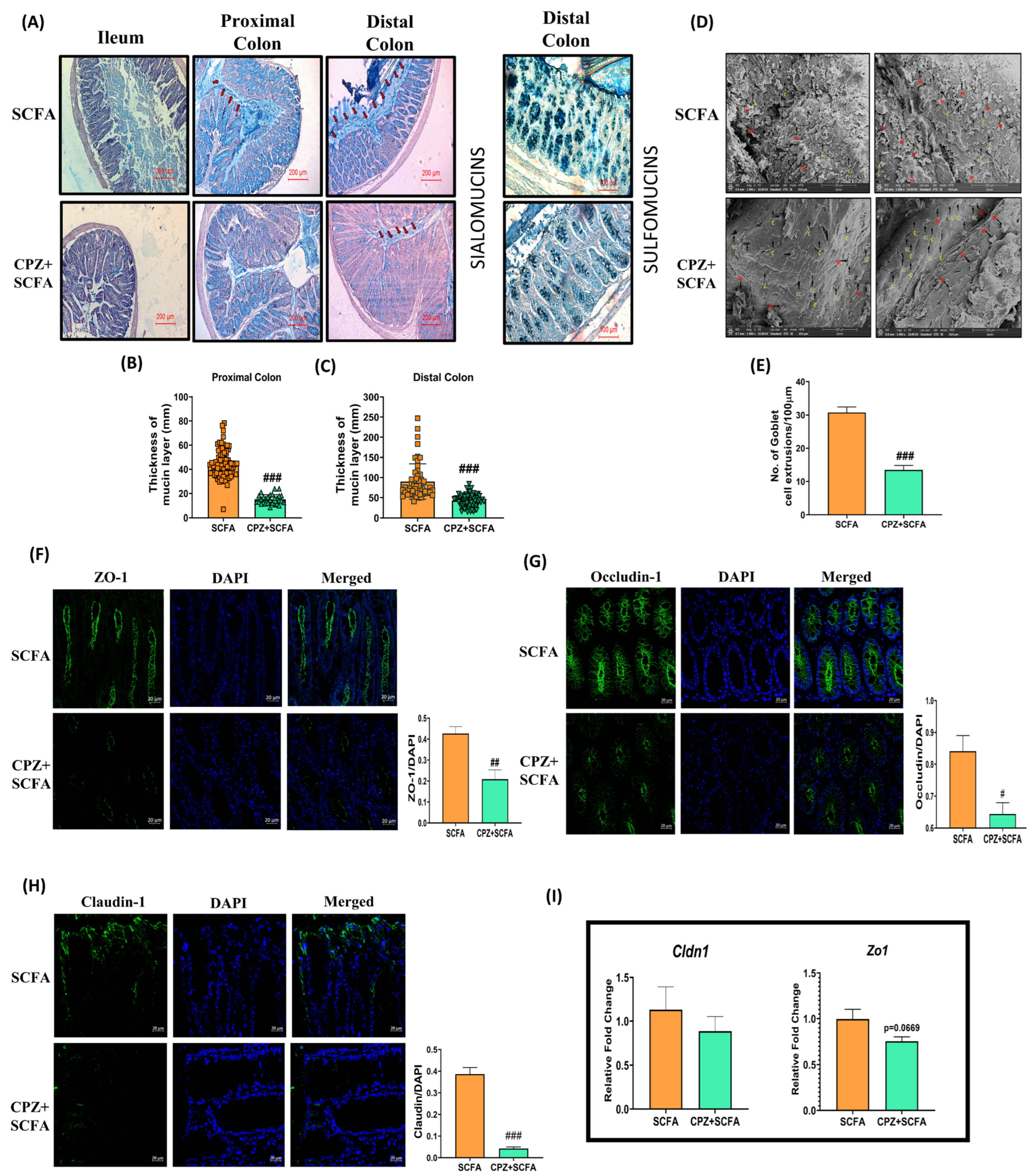
2.6. Rectal Co-Administration of SCFA Mix Fails to Prevent CPZ—Induced Compromise in Colonic Mucin Homeostasis
2.7. Principal Component and Correlation Analysis
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Intrarectal Administration of CPZ, TRPA1 and TRPV1-Mediated Nociceptive Behavioral Confirmatory Tests and SCFA Mix Treatment
4.3. In-Vitro Gut Permeability Assay Using FITC Loaded Solid-Lipid Nanoparticle
4.3.1. Formulation of FITC Loaded Solid-Lipid Nanoparticle
4.3.2. In-Vitro Gut Permeability Assay
4.4. Gene Expression Analysis in DRGs and NanoString nCounter Multiplex Gene Expression Assay
4.5. Histology and Immunohistochemistry Analysis
4.6. Scanning Electron Microscopy
4.7. Analysis of Microbial Abundance in Cecal Content
4.8. Untargeted Metabolome Profiling of Cecal Content
4.9. In Vitro Mouse Fecal Batch Fermentation with CPZ and Relative Bacterial Abundances
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Engel, M.A.; Leffler, A.; Niedermirtl, F.; Babes, A.; Zimmermann, K.; Filipović, M.R.; Izydorczyk, I.; Eberhardt, M.; Kichko, T.I.; Mueller–Tribbensee, S.M. TRPA1 and substance P mediate colitis in mice. Gastroenterology 2011, 141, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Holzer, P. Implications of tachykinins and calcitonin gene-related peptide in inflammatory bowel disease. Digestion 1998, 59, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Hockley, J.R.; Taylor, T.S.; Callejo, G.; Wilbrey, A.L.; Gutteridge, A.; Bach, K.; Winchester, W.J.; Bulmer, D.C.; McMurray, G.; Smith, E.S.J. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 2019, 68, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Story, G.M.; Peier, A.M.; Reeve, A.J.; Eid, S.R.; Mosbacher, J.; Hricik, T.R.; Earley, T.J.; Hergarden, A.C.; Andersson, D.A.; Hwang, S.W. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 2003, 112, 819–829. [Google Scholar] [CrossRef]
- Chen, Y.; Mu, J.; Zhu, M.; Mukherjee, A.; Zhang, H. Transient receptor potential channels and inflammatory bowel disease. Front. Immunol. 2020, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Tribbensee, S.M.; Karna, M.; Khalil, M.; Neurath, M.F.; Reeh, P.W.; Engel, M.A. Differential contribution of TRPA1, TRPV4 and TRPM8 to colonic nociception in mice. PLoS ONE 2015, 10, e0128242. [Google Scholar] [CrossRef] [PubMed]
- Laird, J.; Martinez-Caro, L.; Garcia-Nicas, E.; Cervero, F. A new model of visceral pain and referred hyperalgesia in the mouse. PAIN® 2001, 92, 335–342. [Google Scholar] [CrossRef]
- Miampamba, M.; Chéry-Croze, S.; Détolle-Sarbach, S.; Guez, D.; Chayvialle, J.-A. Antinociceptive effects of oral clonidine and S12813-4 in acute colon inflammation in rats. Eur. J. Pharmacol. 1996, 308, 251–259. [Google Scholar] [CrossRef]
- Kimball, E.; Wallace, N.; Schneider, C.; D’Andrea, M.; Hornby, P. Vanilloid receptor 1 antagonists attenuate disease severity in dextran sulphate sodium-induced colitis in mice. Neurogastroenterol. Motil. 2004, 16, 811–818. [Google Scholar] [CrossRef]
- Fujino, K.; Takami, Y.; de la Fuente, S.G.; Ludwig, K.A.; Mantyh, C.R. Inhibition of the vanilloid receptor subtype-1 attenuates TNBS-colitis. J. Gastrointest. Surg. 2004, 8, 842–848. [Google Scholar] [CrossRef]
- Jain, P.; Materazzi, S.; De Logu, F.; Degl’Innocenti, D.R.; Fusi, C.; Puma, S.L.; Marone, I.M.; Coppi, E.; Holzer, P.; Geppetti, P. Transient receptor potential ankyrin 1 contributes to somatic pain hypersensitivity in experimental colitis. Sci. Rep. 2020, 10, 8632. [Google Scholar] [CrossRef] [PubMed]
- Kistner, K.; Siklosi, N.; Babes, A.; Khalil, M.; Selescu, T.; Zimmermann, K.; Wirtz, S.; Becker, C.; Neurath, M.F.; Reeh, P.W. Systemic desensitization through TRPA1 channels by capsazepine and mustard oil-a novel strategy against inflammation and pain. Sci. Rep. 2016, 6, 28621. [Google Scholar] [CrossRef] [PubMed]
- Kihara, N.; De la Fuente, S.; Fujino, K.; Takahashi, T.; Pappas, T.; Mantyh, C. Vanilloid receptor-1 containing primary sensory neurones mediate dextran sulphate sodium induced colitis in rats. Gut 2003, 52, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Mesmin, L.; Chassaing, B.; Desvaux, M.; De Paepe, K.; Gresse, R.; Sauvaitre, T.; Forano, E.; de Wiele, T.V.; Schüller, S.; Juge, N. Experimental models to study intestinal microbes–mucus interactions in health and disease. FEMS Microbiol. Rev. 2019, 43, 457–489. [Google Scholar] [CrossRef]
- Corfield, A.P. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms 2018, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, A.; Kinoshita, M.; Kuroda, R.; Kakehi, K. Capsazepine partially inhibits neurally mediated gastric mucus secretion following activation of protease-activated receptor 2. Clin. Exp. Pharmacol. Physiol. 2002, 29, 360–361. [Google Scholar] [CrossRef]
- Manneck, D.; Manz, G.; Braun, H.-S.; Rosendahl, J.; Stumpff, F. The TRPA1 Agonist Cinnamaldehyde Induces the Secretion of HCO3− by the Porcine Colon. Int. J. Mol. Sci. 2021, 22, 5198. [Google Scholar] [CrossRef]
- Kumar, V.; Mahajan, N.; Khare, P.; Kondepudi, K.K.; Bishnoi, M. Role of TRPV1 in colonic mucin production and gut microbiota profile. Eur. J. Pharmacol. 2020, 888, 173567. [Google Scholar] [CrossRef]
- Garcia, A.; Olmo, B.; Lopez-Gonzalvez, A.; Cornejo, L.; Rupérez, F.; Barbas, C. Capillary electrophoresis for short chain organic acids in faeces: Reference values in a Mediterranean elderly population. J. Pharm. Biomed. Anal. 2008, 46, 356–361. [Google Scholar] [CrossRef]
- Cummings, J.H.; Macfarlane, G.T.; Englyst, H.N. Prebiotic digestion and fermentation. Am. J. Clin. Nutr. 2001, 73, 415s–420s. [Google Scholar] [CrossRef]
- Kumar, V.; Khare, P.; Devi, K.; Kaur, J.; Kumar, V.; Kiran Kondepudi, K.; Chopra, K.; Bishnoi, M. Short-chain fatty acids increase intracellular calcium levels and enhance gut hormone release from STC-1 cells via transient receptor potential Ankyrin1. Fundam. Clin. Pharmacol. 2021, 35, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Simpson, J.L.; Scott, R.; Boyle, M.J.; Gibson, P.G. Inflammatory subtypes in asthma: Assessment and identification using induced sputum. Respirology 2006, 11, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, L.; Koetsier, M.; Van Deventer, S.; Van Tol, E. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E1 and E2 production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-chain fatty acids manifest stimulative and protective effects on intestinal barrier function through the inhibition of NLRP3 inflammasome and autophagy. Cell. Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef]
- Huda-Faujan, N.; Abdulamir, A.; Fatimah, A.; Anas, O.M.; Shuhaimi, M.; Yazid, A.; Loong, Y. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 2010, 4, 53. [Google Scholar] [CrossRef]
- Breurer, R.; Buto, S.; Christ, M.; Bean, J.; Vernia, P.; Paoluzi, P.; DiPaolo, M.; Caprillo, R. Rectal irrigation with short chain fatty acids for distal ulcerative colitis. Dig. Dis. Sci. 1991, 36, 185–187. [Google Scholar] [CrossRef]
- Patz, J.; Jacobsohn, W.; Gottschalk-Sabag, S.; Zeides, S.; Braverman, D. Treatment of refractory distal ulcerative colitis with short chain fatty acid enemas. Am. J. Gastroenterol. 1996, 91, 731–734. [Google Scholar]
- Vernia, P.; Marcheggiano, A.; Caprilli, R.; Frieri, G.; Corrao, G.; Valpiani, D.; Paolo, M.D.; Paoluzi, P.; Torsoli, A. Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment. Pharmacol. Ther. 1995, 9, 309–313. [Google Scholar] [CrossRef]
- Kripke, S.A.; Fox, A.D.; Berman, J.M.; Settle, R.G.; Rombeau, J.L. Stimulation of intestinal mucosal growth with intracolonic infusion of short-chain fatty acids. J. Parenter. Enter. Nutr. 1989, 13, 109–116. [Google Scholar] [CrossRef]
- Vellani, V.; Kinsey, A.M.; Prandini, M.; Hechtfischer, S.C.; Reeh, P.; Magherini, P.C.; Giacomoni, C.; McNaughton, P.A. Protease activated receptors 1 and 4 sensitize TRPV1 in nociceptive neurones. Mol. Pain 2010, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, L.A.; Kichko, T.I.; Fischer, M.J.; Reeh, P.W. TRPA1-dependent calcium transients and CGRP release in DRG neurons require extracellular calcium. J. Cell Biol. 2020, 219, e201702151. [Google Scholar] [CrossRef]
- Salameh, E.; Meleine, M.; Gourcerol, G.; Do Rego, J.-C.; Do Rego, J.-L.; Legrand, R.; Breton, J.; Aziz, M.; Guérin, C.; Coëffier, M. Chronic colitis-induced visceral pain is associated with increased anxiety during quiescent phase. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G692–G700. [Google Scholar] [CrossRef]
- Kogure, Y.; Wang, S.; Tanaka, K.i.; Hao, Y.; Yamamoto, S.; Nishiyama, N.; Noguchi, K.; Dai, Y. Elevated H2O2 levels in trinitrobenzene sulfate-induced colitis rats contributes to visceral hyperalgesia through interaction with the transient receptor potential ankyrin 1 cation channel. J. Gastroenterol. Hepatol. 2016, 31, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Petersson, J.; Schreiber, O.; Hansson, G.C.; Gendler, S.J.; Velcich, A.; Lundberg, J.O.; Roos, S.; Holm, L.; Phillipson, M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G327–G333. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Jakobsson, H.E.; Holmén-Larsson, J.; Schütte, A.; Ermund, A.; Rodríguez-Piñeiro, A.M.; Arike, L.; Wising, C.; Svensson, F.; Bäckhed, F. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 2015, 18, 582–592. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, V.; Mahajan, N.; Kaur, J.; Devi, K.; Dharavath, R.N.; Singh, R.P.; Kondepudi, K.K.; Bishnoi, M. Mucin secretory action of capsaicin prevents high fat diet-induced gut barrier dysfunction in C57BL/6 mice colon. Biomed. Pharmacother. 2022, 145, 112452. [Google Scholar] [CrossRef]
- Shimotoyodome, A.; Meguro, S.; Hase, T.; Tokimitsu, I.; Sakata, T. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2000, 125, 525–531. [Google Scholar] [CrossRef]
- Jiminez, J.A.; Uwiera, T.C.; Abbott, D.W.; Uwiera, R.R.; Inglis, G.D. Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal inflammation in mice infected with Citrobacter rodentium. MSphere 2017, 2, e00243-17. [Google Scholar] [CrossRef]
- Yue, X.; Wen, S.; Long-Kun, D.; Man, Y.; Chang, S.; Min, Z.; Shuang-Yu, L.; Xin, Q.; Jie, M.; Liang, W. Three important short-chain fatty acids (SCFAs) attenuate the inflammatory response induced by 5-FU and maintain the integrity of intestinal mucosal tight junction. BMC Immunol. 2022, 23, 19. [Google Scholar] [CrossRef]
- Borisova, M.A.; Achasova, K.M.; Morozova, K.N.; Andreyeva, E.N.; Litvinova, E.A.; Ogienko, A.A.; Morozova, M.V.; Berkaeva, M.B.; Kiseleva, E.; Kozhevnikova, E.N. Mucin-2 knockout is a model of intercellular junction defects, mitochondrial damage and ATP depletion in the intestinal epithelium. Sci. Rep. 2020, 10, 21135. [Google Scholar] [CrossRef] [PubMed]
- Leon-Coria, A.; Kumar, M.; Workentine, M.; Moreau, F.; Surette, M.; Chadee, K. Muc2 mucin and nonmucin microbiota confer distinct innate host defense in disease susceptibility and colonic injury. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.W.; Krystofiak, E.S.; Leo-Macias, A.; Cui, R.; Sesso, A.; Weigert, R.; Ebrahim, S.; Kachar, B. Nanoarchitecture and dynamics of the mouse enteric glycocalyx examined by freeze-etching electron tomography and intravital microscopy. Commun. Biol. 2020, 3, 5. [Google Scholar] [CrossRef]
- Huang, J.; Che, M.-I.; Huang, Y.-T.; Shyu, M.-K.; Huang, Y.-M.; Wu, Y.-M.; Lin, W.-C.; Huang, P.-H.; Liang, J.-T.; Lee, P.-H. Overexpression of MUC15 activates extracellular signal-regulated kinase 1/2 and promotes the oncogenic potential of human colon cancer cells. Carcinogenesis 2009, 30, 1452–1458. [Google Scholar] [CrossRef][Green Version]
- Sidiq, T.; Yoshihama, S.; Downs, I.; Kobayashi, K.S. Nod2: A critical regulator of ileal microbiota and Crohn’s disease. Front. Immunol. 2016, 7, 367. [Google Scholar] [CrossRef] [PubMed]
- Scuderi, S.A.; Casili, G.; Lanza, M.; Filippone, A.; Paterniti, I.; Esposito, E.; Campolo, M. Modulation of NLRP3 Inflammasome Attenuated Inflammatory Response Associated to Diarrhea-Predominant Irritable Bowel Syndrome. Biomedicines 2020, 8, 519. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Huang, Y.; Wang, Y.; Wang, P.; Song, H.; Wang, F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS ONE 2019, 14, e0218384. [Google Scholar] [CrossRef]
- Van Hauwermeiren, F.; Vandenbroucke, R.; Grine, L.; Lodens, S.; Van Wonterghem, E.; De Rycke, R.; De Geest, N.; Hassan, B.; Libert, C. TNFR1-induced lethal inflammation is mediated by goblet and Paneth cell dysfunction. Mucosal Immunol. 2015, 8, 828–840. [Google Scholar] [CrossRef]
- Al-Sadi, R.; Guo, S.; Ye, D.; Ma, T.Y. TNF-α modulation of intestinal epithelial tight junction barrier is regulated by ERK1/2 activation of Elk-1. Am. J. Pathol. 2013, 183, 1871–1884. [Google Scholar] [CrossRef]
- Wang, H.; Kim, J.J.; Denou, E.; Gallagher, A.; Thornton, D.J.; Shajib, M.S.; Xia, L.; Schertzer, J.D.; Grencis, R.K.; Philpott, D.J. New role of nod proteins in regulation of intestinal goblet cell response in the context of innate host defense in an enteric parasite infection. Infect. Immun. 2015, 84, 275–285. [Google Scholar] [CrossRef]
- Amendola, A.; Butera, A.; Sanchez, M.; Strober, W.; Boirivant, M. Nod2 deficiency is associated with an increased mucosal immunoregulatory response to commensal microorganisms. Mucosal Immunol. 2014, 7, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Arike, L.; Hansson, G.C. The densely O-glycosylated MUC2 mucin protects the intestine and provides food for the commensal bacteria. J. Mol. Biol. 2016, 428, 3221–3229. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Basson, A.; Trotter, A.; Rodriguez-Palacios, A.; Cominelli, F. Mucosal interactions between genetics, diet, and microbiome in inflammatory bowel disease. Front. Immunol. 2016, 7, 290. [Google Scholar] [CrossRef]
- Li, J.; Hou, L.; Wang, C.; Jia, X.; Qin, X.; Wu, C. Short term intrarectal administration of sodium propionate induces antidepressant-like effects in rats exposed to chronic unpredictable mild stress. Front. Psychiatry 2018, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Gaudier, E.; Rival, M.; Buisine, M.-P.; Robineau, I.; Hoebler, C. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol. Res. 2009, 58, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.; Branch, W.; Naylor, C.; MacFarlane, G. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Chambers, E.S.; Preston, T.; Frost, G.; Morrison, D.J. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr. Nutr. Rep. 2018, 7, 198–206. [Google Scholar] [CrossRef]
- Van der Beek, C.M.; Canfora, E.E.; Lenaerts, K.; Troost, F.J.; Olde Damink, S.W.; Holst, J.J.; Masclee, A.A.; Dejong, C.H.; Blaak, E.E. Distal, not proximal, colonic acetate infusions promote fat oxidation and improve metabolic markers in overweight/obese men. Clin. Sci. 2016, 130, 2073–2082. [Google Scholar] [CrossRef]
- Ahmad, A.; Fauzia, E.; Kumar, M.; Mishra, R.K.; Kumar, A.; Khan, M.A.; Raza, S.S.; Khan, R. Gelatin-coated polycaprolactone nanoparticle-mediated naringenin delivery rescue human mesenchymal stem cells from oxygen glucose deprivation-induced inflammatory stress. ACS Biomater. Sci. Eng. 2018, 5, 683–695. [Google Scholar] [CrossRef]
- Li, B.-R.; Wu, J.; Li, H.-S.; Jiang, Z.-H.; Zhou, X.-M.; Xu, C.-H.; Ding, N.; Zha, J.-M.; He, W.-Q. In vitro and in vivo approaches to determine intestinal epithelial cell permeability. J. Vis. Exp. JoVE 2018, 14, e57032. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Analyses Multivariées de Marqueurs Génétiques: Développements Méthodologiques, Applications et Extensions. Ph.D. Thesis, Claude Bernard University Lyon 1, Villeurbanne, France, 2008. [Google Scholar]


| Sr. No. | Bacterial Primer | Forward Sequence | Reverse Sequence |
|---|---|---|---|
| 1. | Total Bacteria | ACTCCTACGGGAGGCAGCAGT | ATTACCGCGGCTGCTGGC |
| 2. | Butyrivibrio fibrisolvens | CTAACACATGCAAGTCGAACG | CCGTGTCTCAGTCCCAAT |
| 3. | Bacteroides fragilis | GGCGCACGGGTGAGTAACA | CAATATTCCTCACTGCTGC |
| 4. | Prevotella copri | CCGGACTCCTGCCCCTGCAA | GTTGCGCCAGGCACTGCGAT |
| 5. | Clostridium leptum | CCGCATAAGACCTCAGTACCGC | GGGATTTGCTTGCCTTCACAGGG |
| 6. | Clostridium coccoides | ACTCCTACGGGAGGCAGC | GCTTCTTAGTCARGTACCG |
| 7. | Lactobacillus | CACCGCTACACATGGAG | AGCAGTAGGGAATCTTCCA |
| 8. | Bacteroides | TCCTACGGGAGGCAGCAGT | CAATCGGAGTTCTTCGTG |
| 9. | Bifidobacterium | TCGCGTCYGGTGTGAAAG | CCACATCCAGCRTCCAC |
| 10. | Akkermansia muciniphila | CAGCACGTGAAGGTGGGGAC | CCTTGCGGTTGGCTTCAGAT |
| 11. | Butyricicoccus pullicaecorum | AGTACGGCCGCAAGGTTGAAA | CTGCCATTGTAGTACGTGTG |
| 12. | Anaerostipes butyraticus | CACCATGTCATTTACTCAAGAATATCAGA | TTATTTGTTAGATCTTCTCCAGATGTTAGC |
| 13. | Roseburia spp. | GCGGTRCGGCAAGTCTGA | CCTCCGACACTCTAGTMCGA |
| 14. | Fecalibacterium | GGAGGAAGAAGGTCTTCGG | AATTCCGCCTACCTCTGCACT |
| 15. | Ruminococci | GGCGGCYTRCTGGGCTTT | CCAGGTGGATWACTTATTGTGTTAA |
| 16. | Eubacteria | ACTCCTACGGGAGGCAGCAG | ATTACCGCGGCTGCTGG |
| 17. | Methanobrevibacter | CCGGGTATCTAATCCGGTTC | CTCCCAGGGTAGAGGTGAAA |
| 18. | Bacteroidetes | ACGCTAGCTACAGGCTTAACA | ACGCTACTTGGCTGGTTCA |
| 19. | Firmicutes | GCGTGAGTGAAGAAGT | CTACGCTCCCTTTACAC |
| 20. | Lachnospiraceae | CGGTACCTGACTAAGAAGC | AGTTTYATTCTTGCGAACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, V.; Kumar, V.; Devi, K.; Kumar, A.; Khan, R.; Singh, R.P.; Rajarammohan, S.; Kondepudi, K.K.; Chopra, K.; Bishnoi, M. Intrarectal Capsazepine Administration Modulates Colonic Mucosal Health in Mice. Int. J. Mol. Sci. 2022, 23, 9577. https://doi.org/10.3390/ijms23179577
Kumar V, Kumar V, Devi K, Kumar A, Khan R, Singh RP, Rajarammohan S, Kondepudi KK, Chopra K, Bishnoi M. Intrarectal Capsazepine Administration Modulates Colonic Mucosal Health in Mice. International Journal of Molecular Sciences. 2022; 23(17):9577. https://doi.org/10.3390/ijms23179577
Chicago/Turabian StyleKumar, Vibhu, Vijay Kumar, Kirti Devi, Ajay Kumar, Rehan Khan, Ravindra Pal Singh, Sivasubramanian Rajarammohan, Kanthi Kiran Kondepudi, Kanwaljit Chopra, and Mahendra Bishnoi. 2022. "Intrarectal Capsazepine Administration Modulates Colonic Mucosal Health in Mice" International Journal of Molecular Sciences 23, no. 17: 9577. https://doi.org/10.3390/ijms23179577
APA StyleKumar, V., Kumar, V., Devi, K., Kumar, A., Khan, R., Singh, R. P., Rajarammohan, S., Kondepudi, K. K., Chopra, K., & Bishnoi, M. (2022). Intrarectal Capsazepine Administration Modulates Colonic Mucosal Health in Mice. International Journal of Molecular Sciences, 23(17), 9577. https://doi.org/10.3390/ijms23179577







