Complete Genome Sequence Analysis of Bacillus subtilis Bbv57, a Promising Biocontrol Agent against Phytopathogens
Abstract
:1. Introduction
2. Results
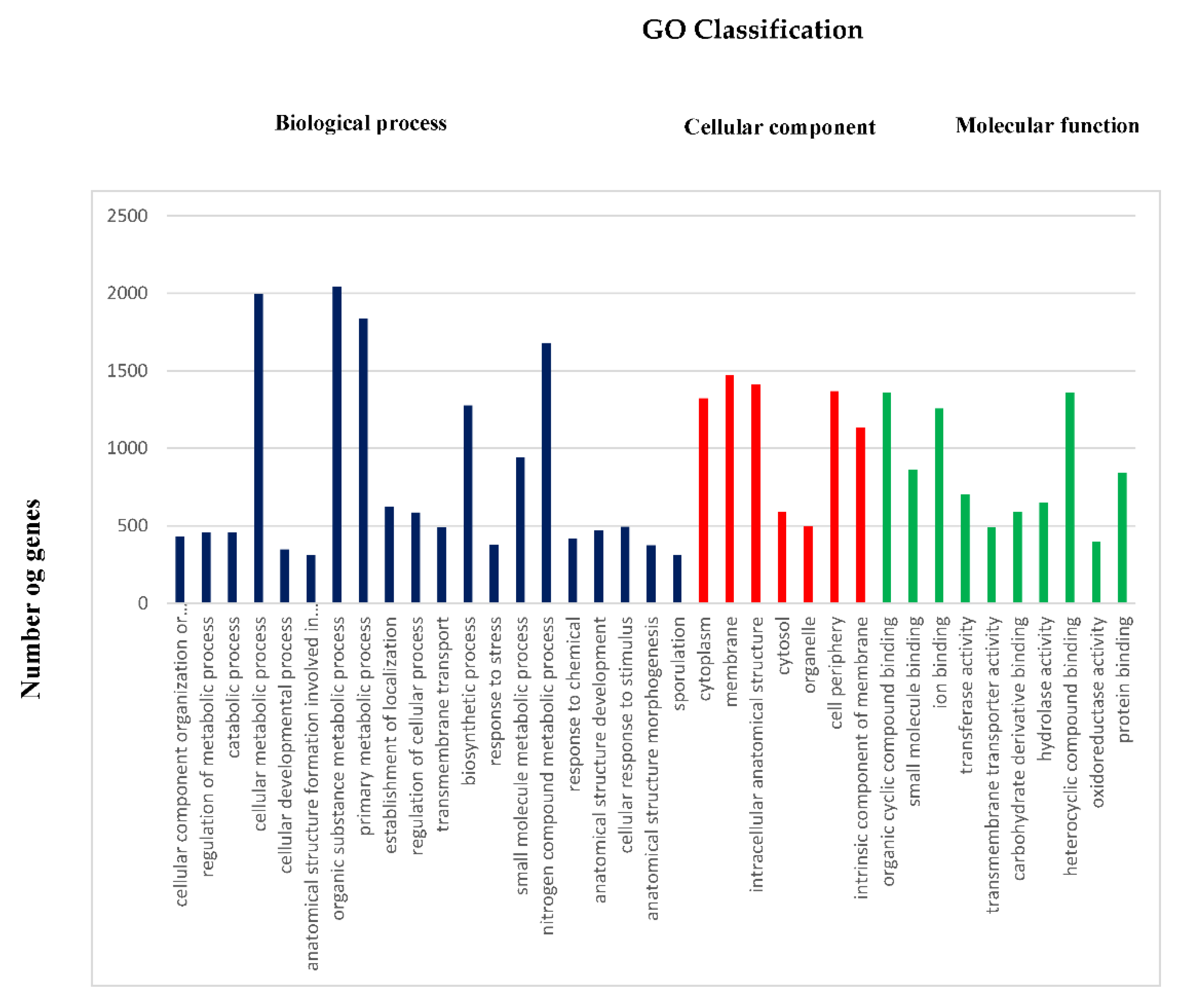
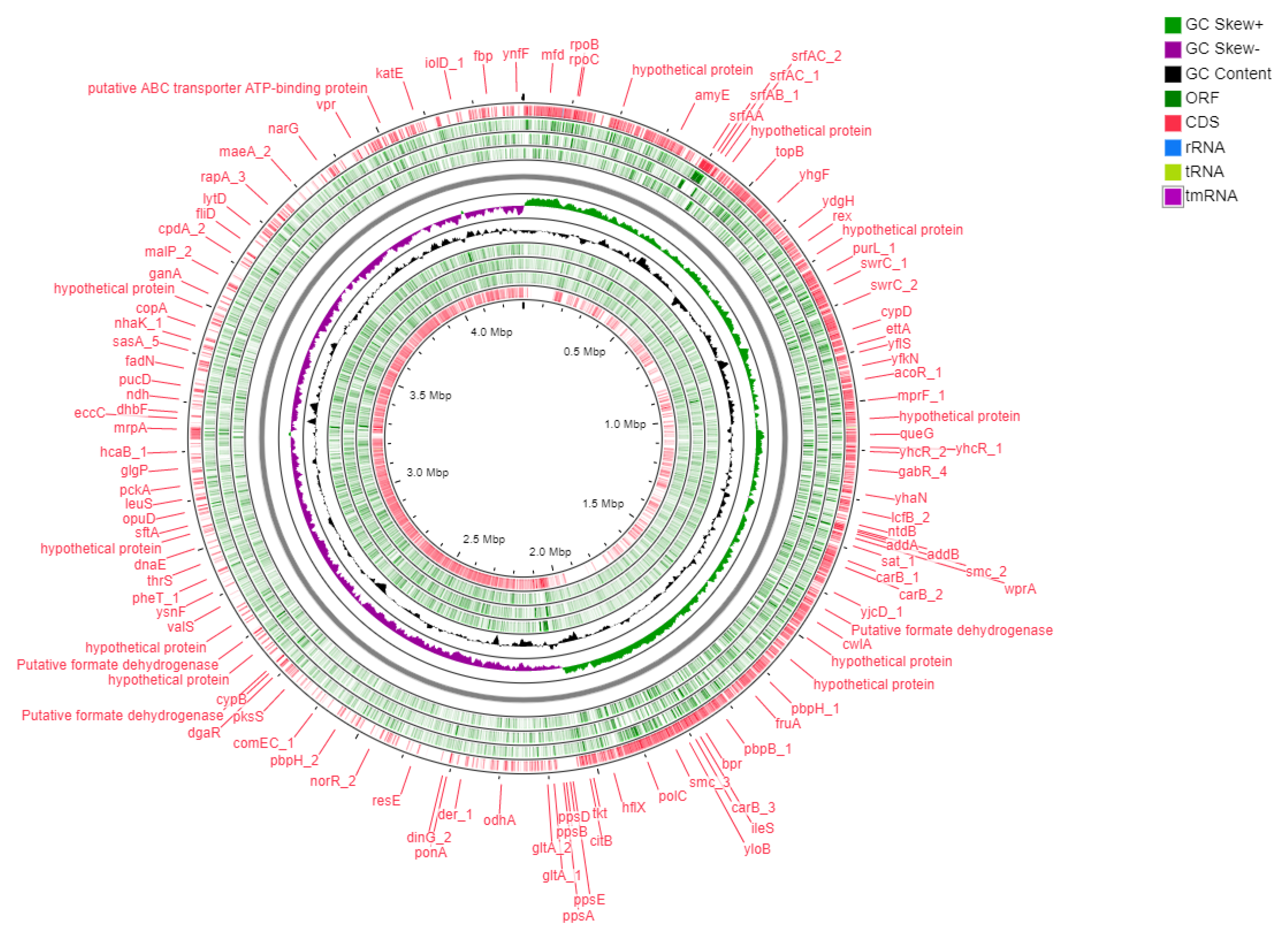
2.1. Genomic Features of Bacillus subtilis Bbv57
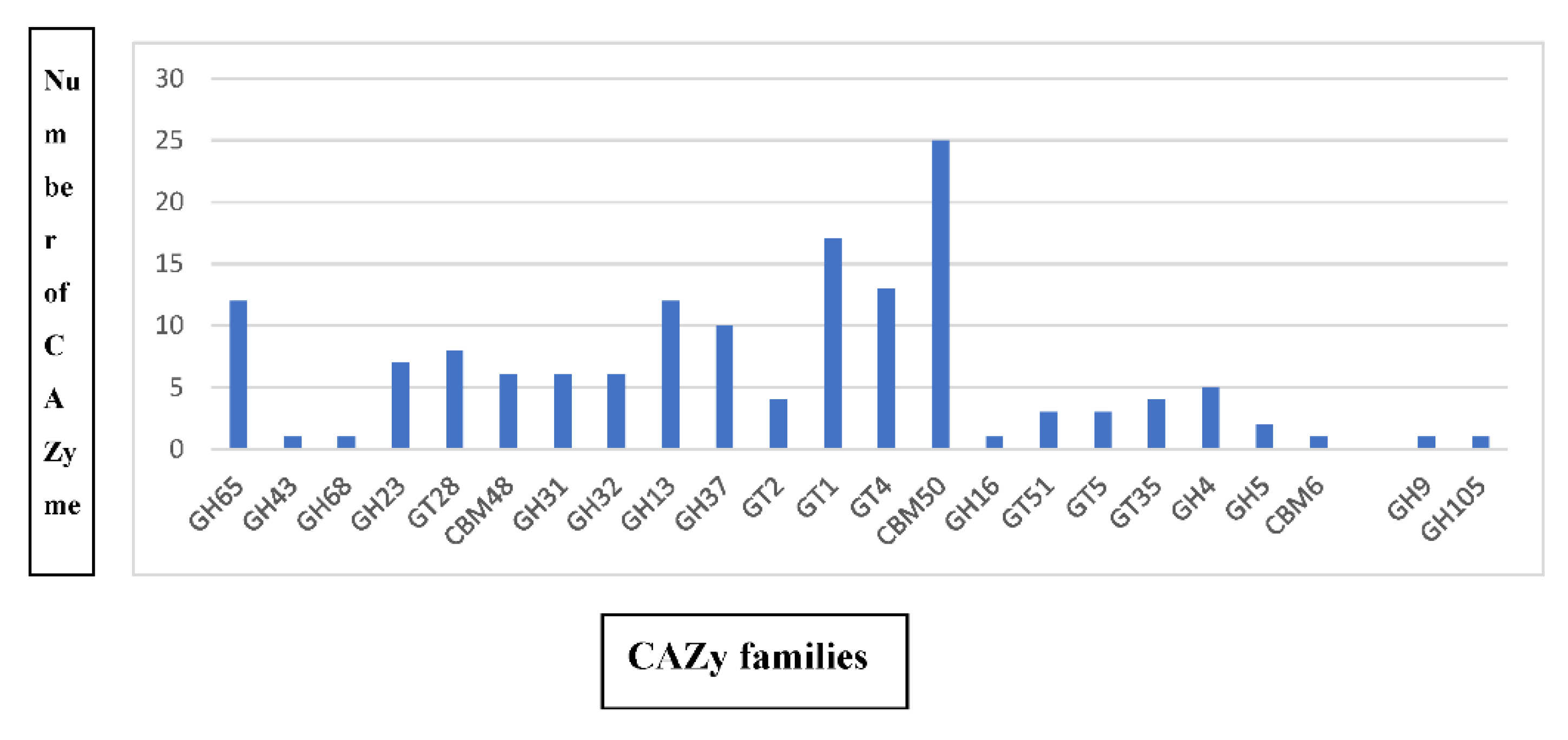
2.3. Bbv57 Harbors Novel Genes Encoding for CAZymes
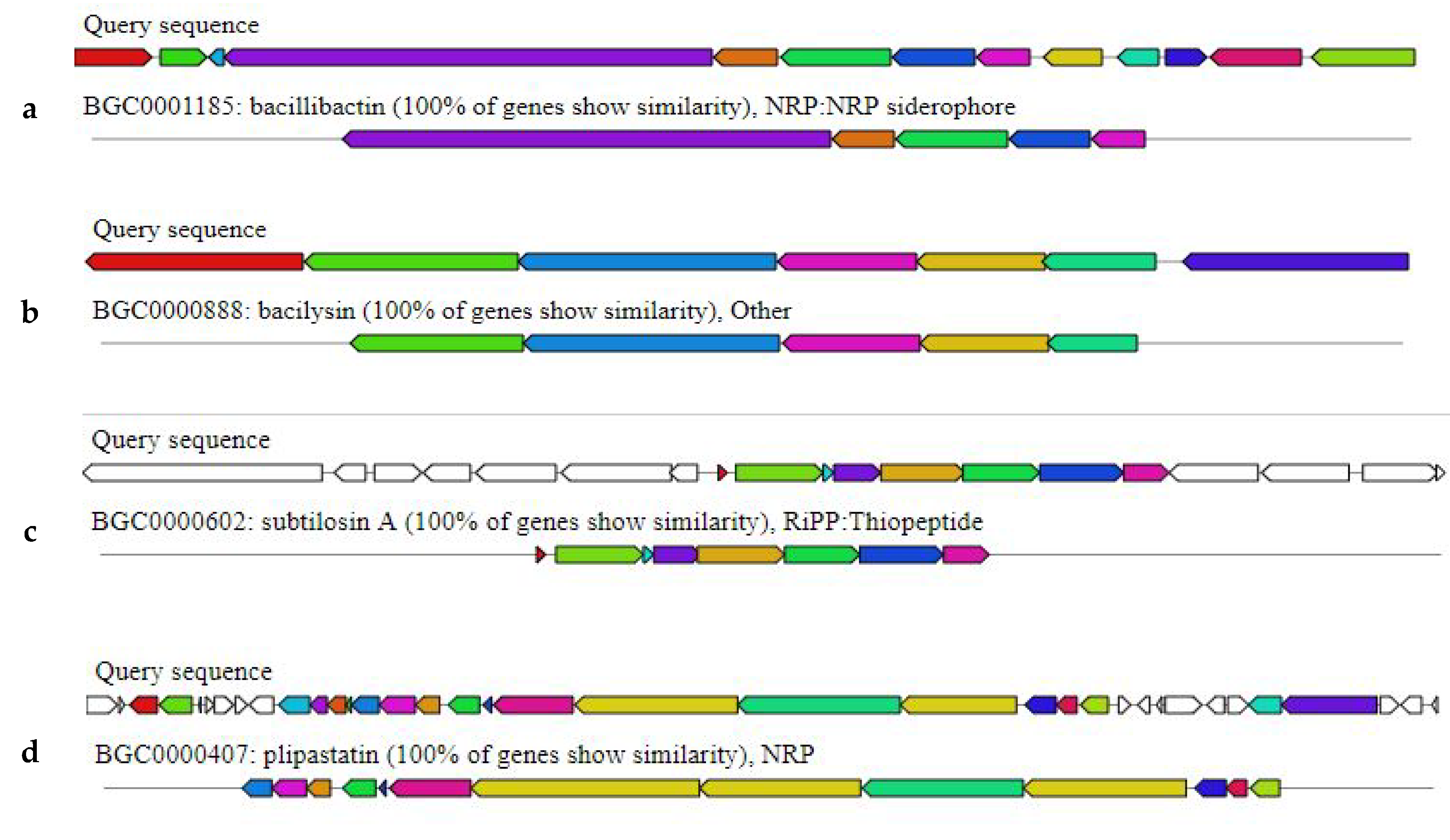
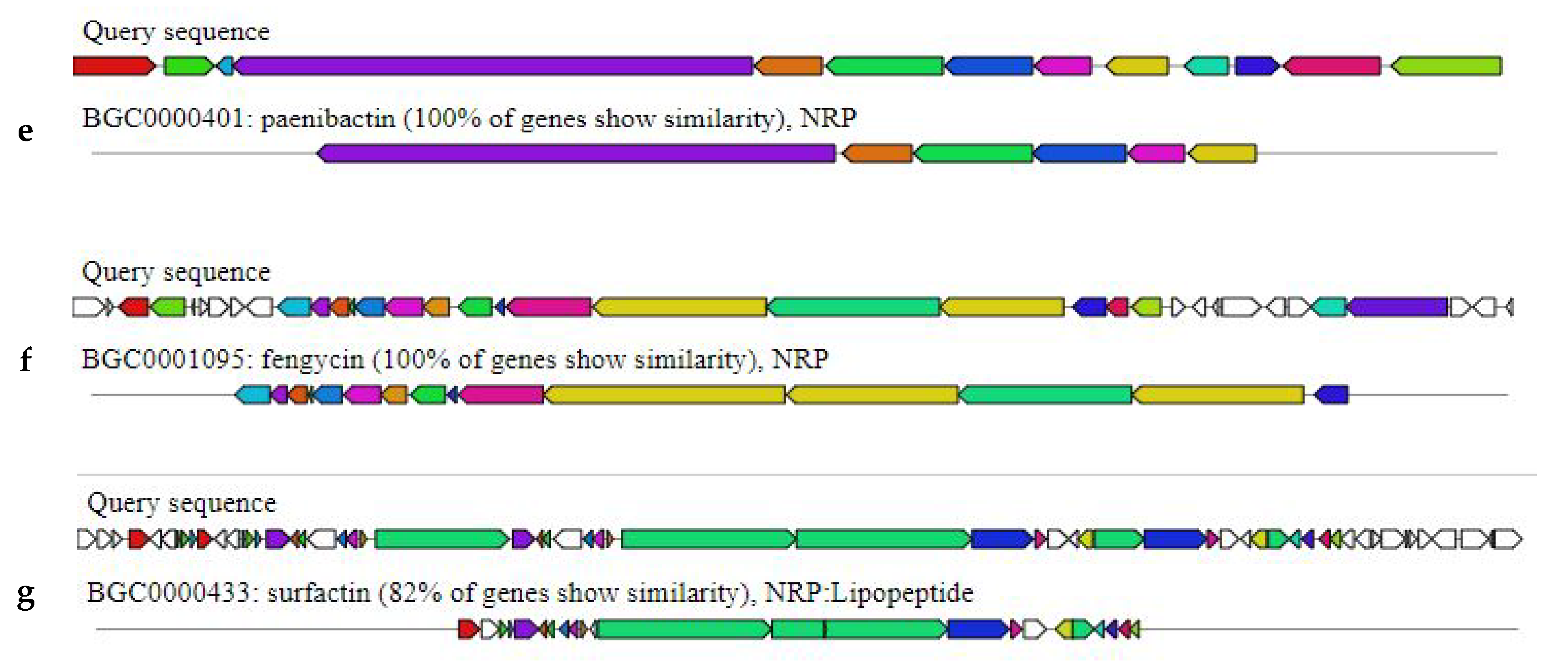
2.4. Bacillus subtilis Bbv57 Harbors Genes Encoding for Antimicrobial Secondary Metabolites
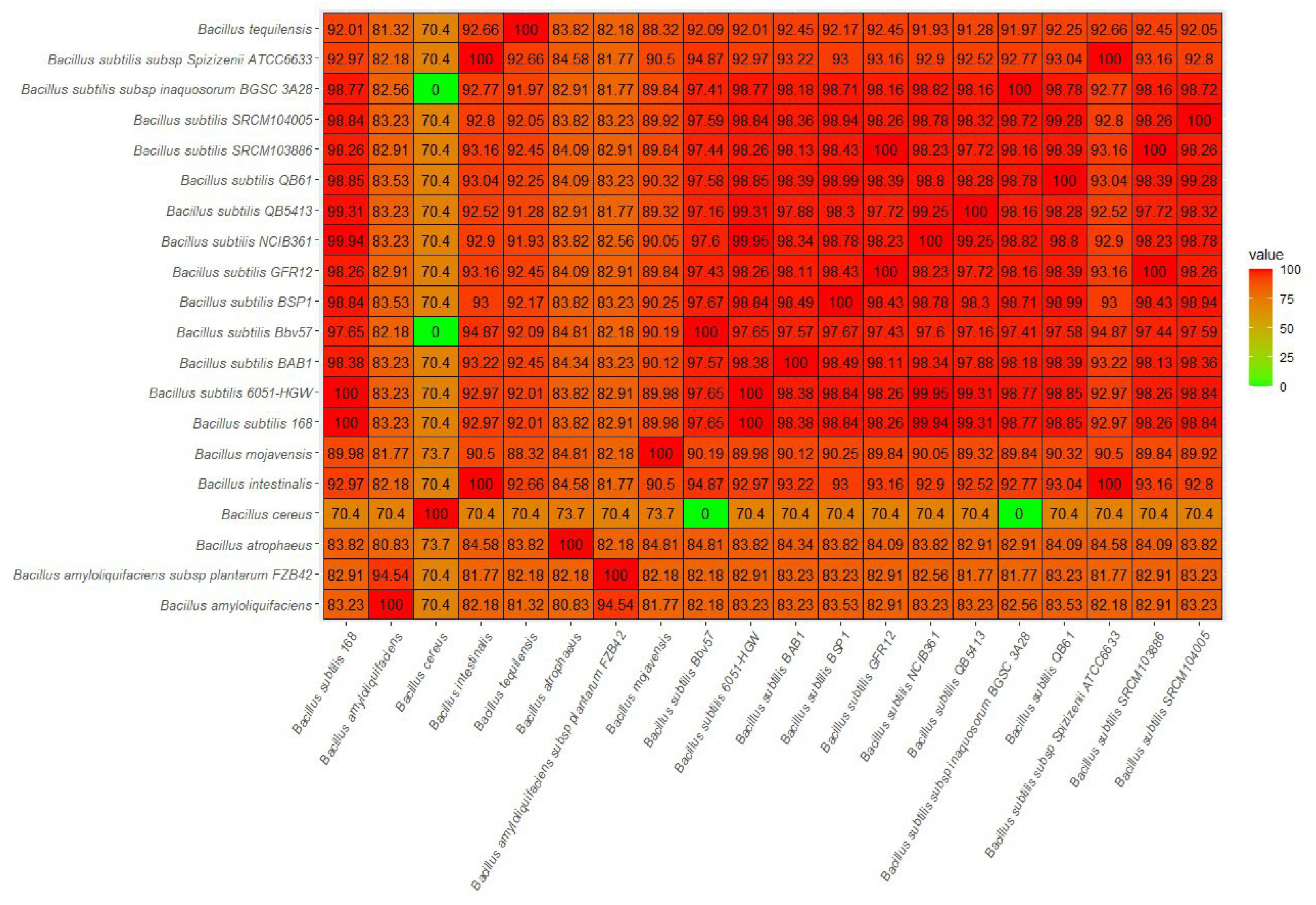
2.5. Pangenome Analysis of B. subtilis Bbv57
3. Discussion
4. Materials and Methods
4.1. Isolation and Maintenance of Bacterial Strain Bbv57
4.2. Genome Sequencing of Strain B. subtilis Bbv57
4.3. Genome Assembly and Annotation
4.4. Molecular Confirmation of Bacillus subtilis Bbv57
4.5. Prediction of Genes Encoding for CAZymes and Secondary Metabolites in Bacillus subtilis Bbv57
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peng, Y.; Li, S.J.; Yan, J.; Tang, Y.; Cheng, J.P.; Gao, A.J.; Yao, X.; Ruan, J.J.; Xu, B.L. Research Progress on Phytopathogenic Fungi and Their Role as Biocontrol Agents. Front. Microbiol. 2021, 12, 1209. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, B.M.; Carvajal-Yepes, M.; Kumar, P.L.; Kawarazuka, N.; Liu, Y.; Mulema, A.A.; McCutcheon, S.; Ibabao, X. Sustainable management of transboundary pests requires holistic and inclusive solutions. Food Secur. 2022, 1–9. [Google Scholar] [CrossRef]
- Harish, S.; Parthasarathy, S.; Durgadevi, D.; Anandhi, K.; Raguchander, T. Plant Growth-Promoting Rhizobacteria: Harnessing Its Potential for Sustainable Plant Disease Management; Springer: Singapore, 2019; pp. 151–187. [Google Scholar] [CrossRef]
- Prabhukarthikeyan, S.; Manikandan, R.; Durgadevi, D.; Keerthana, U.; Harish, S.; Karthikeyan, G.; Raguchander, T. Bio-suppression of turmeric rhizome rot disease and understanding the molecular basis of tripartite interaction among Curcuma longa, Pythium aphanidermatum and Pseudomonas fluorescens. Biol. Control 2017, 111, 23–31. [Google Scholar] [CrossRef]
- Durgadevi, D.; Harish, S.; Manikandan, R.; Prabhukarthikeyan, S.; Alice, D.; Raguchander, T. Proteomic profiling of defense/resistant genes induced during the tripartite interaction of Oryza sativa, Rhizoctonia solani AG1-1A, and Bacillus subtilis against rice sheath blight. Physiol. Mol. Plant Pathol. 2021, 115, 101669. [Google Scholar] [CrossRef]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef]
- Prabhukarthikeyan, S.; Keerthana, U.; Baite, M.S.; Panneerselvam, P.; Mitra, D.; Kumar, R.N.; Parameswaran, C.; Cayalvizhi, B.; Kumar, A.M.; Harish, S.; et al. Bacillus rhizobacteria: A versatile biostimulant for sustainable agriculture. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2021; pp. 33–44. [Google Scholar] [CrossRef]
- Harish, S.; Kavino, M.; Kumar, N.; Balasubramanian, P.; Samiyappan, R. Induction of defense-related proteins by mixtures of plant growth promoting endophytic bacteria against Banana bunchy top virus. Biol. Control 2009, 51, 16–25. [Google Scholar] [CrossRef]
- Ramyabharathi, S.; Meena, K.S.; Rajendran, L.; Raguchander, T.; Jonathan, E.I. Potential of a rhizobacterium Bacillus subtilis (Bbv 57) on Fusarium oxysporum f. sp. gerberae and Meloidogyne incognita infecting Gerbera grown in protected cultivation. Eur. J. Plant Pathol. 2020, 158, 615–632. [Google Scholar] [CrossRef]
- Ramyabharathi, S.A.M.; Raguchander, T.; Jonathan, E.I. Isolation and characterization of novel Bacillus strain from cut flowers in Tamil Nadu. Pestology 2014, 38, 4. [Google Scholar]
- Ramyabharathi, S.; Meena, K.S.; Rajendran, L.; Karthikeyan, G.; Jonathan, E.I.; Raguchander, T. Biocontrol of wilt-nematode complex infecting gerbera by Bacillus subtilis under protected cultivation. Egypt. J. Biol. Pest Control 2018, 28, 21. [Google Scholar] [CrossRef] [Green Version]
- Jonathan, E.I.; Raguchander, T.; Meena, K.S.; Kavitha, P.G. Plant growth promoting rhizobacteria in the management of Radopholus similis and Meloidogyne incognita in black pepper. Madras Agric. J. 2012, 99, 356–358. [Google Scholar]
- Meena, K.S.; Archana, S.; Jonathan, E.I.; Raguchander, T. Study on viability and effect of liquid formulation of Bacillus subtilis, Bbv 57 against nematode-fungus disease complex in tomato, Lycopersicon esculentum Mill. Indian J. Nematol. 2015, 45, 184–194. [Google Scholar] [CrossRef]
- Baek, I.; Lee, K.; Goodfellow, M.; Chun, J. Comparative Genomic and Phylogenomic Analyses Clarify Relationships within and Between Bacillus cereus and Bacillus thuringiensis: Proposal for the Recognition of Two Bacillus thuringiensis Genomovars. Front. Microbiol. 2019, 10, 1978. [Google Scholar] [CrossRef] [PubMed]
- Leal, C.; Fontaine, F.; Aziz, A.; Egas, C.; Clément, C.; Trotel-Aziz, P. Genome sequence analysis of the beneficial Bacillus subtilis PTA-271 isolated from a Vitis vinifera (cv. Chardonnay) rhizospheric soil: Assets for sustainable biocontrol. Environ. Microbiome 2021, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Balderas-Ruíz, K.A.; Bustos, P.; Santamaria, R.I.; González, V.; Cristiano-Fajardo, S.A.; Barrera-Ortíz, S.; Mezo-Villalobos, M.; Aranda-Ocampo, S.; Guevara-García, A.; Galindo, E.; et al. Bacillus velezensis 83 a bacterial strain from mango phyllosphere, useful for biological control and plant growth promotion. AMB Express 2020, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Theobald, S.; Vesth, T.C.; Rendsvig, J.K.; Nielsen, K.F.; Riley, R.; De Abreu, L.M.; Salamov, A.; Frisvad, J.C.; Larsen, T.O.; Andersen, M.R.; et al. Uncovering secondary metabolite evolution and biosynthesis using gene cluster networks and genetic dereplication. Sci. Rep. 2018, 8, 17957. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Ryu, C.-M.; Zhang, S. Induced Systemic Resistance and Promotion of Plant Growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef]
- Nagórska, K.; Bikowski, M.; Obuchowski, M. Multicellular behaviour and production of a wide variety of toxic substances support usage of Bacillus subtilis as a powerful biocontrol agent. Acta Biochim. Pol. 2007, 54, 495–508. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Zachow, C.; Fatehi, J.; Cardinale, M.; Tilcher, R.; Berg, G. Strain-specific colonization pattern of Rhizoctonia antagonists in the root system of sugar beet. FEMS Microbiol. Ecol. 2010, 74, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Malfanova, N.; Kamilova, F.; Validov, S.; Shcherbakov, A.; Chebotar, V.; Tikhonovich, I.; Lugtenberg, B. Characterization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microb. Biotechnol. 2011, 4, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.-H.; Gusti, A.R.; Zhang, Q.; Xu, J.-L.; Zhang, L.-H. Identification of Quorum-Quenching N-Acyl Homoserine Lactonases from Bacillus Species. Appl. Environ. Microbiol. 2002, 68, 1754–1759. [Google Scholar] [CrossRef] [PubMed]
- Samaras, A.; Roumeliotis, E.; Ntasiou, P.; Karaoglanidis, G. Bacillus subtilis MBI600 Promotes Growth of Tomato Plants and Induces Systemic Resistance Contributing to the Control of Soilborne Pathogens. Plants 2021, 10, 1113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, D.; Kendall, J.R.A.; Borriss, R.; Druzhinina, I.S.; Kubicek, C.P.; Shen, Q.; Zhang, R. Comparative Genomic Analysis of Bacillus amyloliquefaciens and Bacillus subtilis Reveals Evolutional Traits for Adaptation to Plant-Associated Habitats. Front. Microbiol. 2016, 7, 2039. [Google Scholar] [CrossRef]
- Guo, Q.; Li, S.; Lu, X.; Zhang, X.; Wang, P.; Ma, P. Complete Genome Sequence of Bacillus subtilis BAB-1, a Biocontrol Agent for Suppression of Tomato Gray Mold. Genome Announc. 2014, 2, e00744-14. [Google Scholar] [CrossRef]
- Guo, S.; Li, X.; He, P.; Ho, H.; Wu, Y.; He, Y. Whole-genome sequencing of Bacillus subtilis XF-1 reveals mechanisms for biological control and multiple beneficial properties in plants. J. Ind. Microbiol. Biotechnol. 2015, 42, 925–937. [Google Scholar] [CrossRef]
- Kim, P.I.; Ryu, J.; Kim, Y.H.; Chi, Y.-T. Production of Biosurfactant Lipopeptides Iturin A, Fengycin and Surfactin A from Bacillus subtilis CMB32 for Control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 2010, 20, 138–145. [Google Scholar] [CrossRef]
- Chung, S.; Kong, H.; Buyer, J.S.; Lakshman, D.K.; Lydon, J.; Kim, S.-D.; Roberts, D.P. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl. Microbiol. Biotechnol. 2008, 80, 115–123. [Google Scholar] [CrossRef]
- Romero, D.; De Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.-W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The Iturin and Fengycin Families of Lipopeptides Are Key Factors in Antagonism of Bacillus subtilis Toward Podosphaera fusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef] [Green Version]
- Toure, Y.; Ongena, M.; Jacques, P.; Guiro, A.; Thonart, P. Role of lipopeptides produced by Bacillus subtilis GA1 in the reduction of grey mould disease caused by Botrytis cinerea on apple. J. Appl. Microbiol. 2004, 96, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Roongsawang, N.; Thaniyavarn, J.; Thaniyavarn, S.; Kameyama, T.; Haruki, M.; Imanaka, T.; Morikawa, M.; Kanaya, S. Isolation and characterization of a halotolerant Bacillus subtilis BBK-1 which produces three kinds of lipopeptides: Bacillomycin L, plipastatin, and surfactin. Extremophiles 2002, 6, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.F. A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bibsonomy.org/bibtex/f230a919c34360709aa298734d63dca3 (accessed on 15 July 2022).
- Joshi, N.; Sickle, F. A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. 2011. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Alonge, M.; Soyk, S.; Ramakrishnan, S.; Wang, X.; Goodwin, S.; Sedlazeck, F.J.; Lippman, Z.B.; Schatz, M.C. RaGOO: Fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 2019, 20, 224. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef]
- Götz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Feature | Value |
|---|---|
| Genome size (bp) | 4,30,2465 |
| G + C content | 44.5% |
| Total number of genes | 4363 |
| Total size of protein-coding genes | 3,735,486 |
| Protein-coding genes | 4281 |
| Average CDs size (bp) | 872.57 |
| rRNA number | 5 |
| tRNA number | 76 |
| tmRNA number | 1 |
| Pseudogenes (total) | 27 |
| COG Code | Number | Proportion | Description |
|---|---|---|---|
| J | 210 | 4.91 | Translation, ribosomal structure, and biogenesis |
| A | 1 | 0.02 | RNA processing and modification |
| K | 352 | 8.22 | Transcription |
| L | 157 | 3.67 | Replication, recombination, and repair |
| B | 1 | 0.02 | Chromatin structure and dynamics |
| D | 47 | 1.10 | Cell cycle control, cell division, and chromosome partitioning |
| Y | 0 | 0 | Nuclear structure |
| V | 66 | 1.54 | Defense mechanism |
| T | 189 | 4.41 | Signal transduction mechanisms |
| M | 234 | 5.47 | Cell wall/membrane/envelope biogenesis |
| N | 68 | 1.59 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 56 | 1.31 | Intracellular trafficking, secretion, and vesicular transport |
| O | 106 | 2.48 | Post-translational modification, protein turnover, and chaperons |
| C | 229 | 5.35 | Energy production and conversion |
| G | 335 | 7.83 | Carbohydrate transport and metabolism |
| E | 406 | 9.48 | Amino acid transport and metabolism |
| F | 121 | 2.83 | Nucleotide transport and metabolism |
| H | 136 | 3.18 | Coenzyme transport and metabolism |
| I | 120 | 2.80 | Lipid transport and metabolism |
| P | 285 | 6.66 | Inorganic ion transport and metabolism |
| Q | 92 | 2.15 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 0 | 0 | General function prediction only |
| S | 1086 | 25.37 | Function unknown |
| - | 117 | 2.73 | Not in COGs |
| Sl. No. | Isolate | Predicted Functions | Reference |
|---|---|---|---|
| 1. | B. subtilis EBPBS4 | Iturin, surfactin, bacillomycin D, fengycin, ericinmycosubtilin, subtilosin, and mersacidin | [5] |
| 2. | Bacillus subtilis MBI600 | Fengycin, surfactin, bacillaene, bacillibactin, subtilosin A, basilysin, carbohydrate transport and metabolism, aminoacid transport and metabolism, nitrate transporter, magnesium transporter, and potassium uptake | [26] |
| 3. | Bacillus subtilis PTA-271 | Catecholicsiderophore, surfactin, fengycin, acetoin, 2,3-butanediol, and N-acyl-L-homoserine lactone | [15] |
| 4. | Bacillus subtilis | Carbohydrate transport and metabolism, amino acid transport and metabolism, endo-1, 4-ß-glucanase, endo- ß -1,3-,4glucanase, xylose isomerase, and pectatelyase | [27] |
| 5. | Bacillus subtilis BAB-1 | Non-ribosomal peptide synthetase (NRPS) antibiotics, polyketide synthase (PKS) antibiotics, lantibiotics, surfactin, fengycin, and bacillibactin | [28] |
| 6. | Bacillus subtilis XF-1 | Antimicrobial lipopeptides (surfactin and fengycin), polyketides (macrolactin and bacillaene), bacillibactin, bacilysin, and chitosanase | [29] |
| 7. | Bacillus subtilis CMB32 | Antifungal lipopeptides | [30] |
| 8. | B. subtilis isolate ME488 | Possessing secondary metabolites ituC, ituD, bacA, bacD, mrsA, and mrsM | [31] |
| 9. | Bacillus subtilis | Iturin and fengycin | [32] |
| 10. | Bacillus subtilis GA1 | Lipopeptides | [33] |
| 11. | Bacillus subtilis BBK1 | Bacillomycin L, plipastatin, and surfactin | [34] |
| Property | Term |
|---|---|
| Sequencing finishing quality | High quality draft |
| Libraries used | Illumina paired-end library (2 × 150 bp insert size) |
| Sequencing platform | IlluminaHiseq |
| Assemblers | SPAdes |
| Gene-calling method | Prodigal |
| BioProject | PRJNA794929 |
| BioSample | SAMN24663524 |
| Source material identifier | Bacillus subtilis |
| Project relevance | Biocontrol |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiruvengadam, R.; Gandhi, K.; Vaithiyanathan, S.; Sankarasubramanian, H.; Loganathan, K.; Lingan, R.; Rajagopalan, V.R.; Muthurajan, R.; Ebenezer Iyadurai, J.; Kuppusami, P. Complete Genome Sequence Analysis of Bacillus subtilis Bbv57, a Promising Biocontrol Agent against Phytopathogens. Int. J. Mol. Sci. 2022, 23, 9732. https://doi.org/10.3390/ijms23179732
Thiruvengadam R, Gandhi K, Vaithiyanathan S, Sankarasubramanian H, Loganathan K, Lingan R, Rajagopalan VR, Muthurajan R, Ebenezer Iyadurai J, Kuppusami P. Complete Genome Sequence Analysis of Bacillus subtilis Bbv57, a Promising Biocontrol Agent against Phytopathogens. International Journal of Molecular Sciences. 2022; 23(17):9732. https://doi.org/10.3390/ijms23179732
Chicago/Turabian StyleThiruvengadam, Raguchander, Karthikeyan Gandhi, Sendhilvel Vaithiyanathan, Harish Sankarasubramanian, Karthiba Loganathan, Rajendran Lingan, Veera Ranjani Rajagopalan, Raveendran Muthurajan, Jonathan Ebenezer Iyadurai, and Prabakar Kuppusami. 2022. "Complete Genome Sequence Analysis of Bacillus subtilis Bbv57, a Promising Biocontrol Agent against Phytopathogens" International Journal of Molecular Sciences 23, no. 17: 9732. https://doi.org/10.3390/ijms23179732





