Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies
Abstract
:1. Introduction
2. VOCs and Anti-SARS-CoV-2 Therapeutic Antibodies
3. Efficacy of the Therapeutic Antibodies against the VOCs
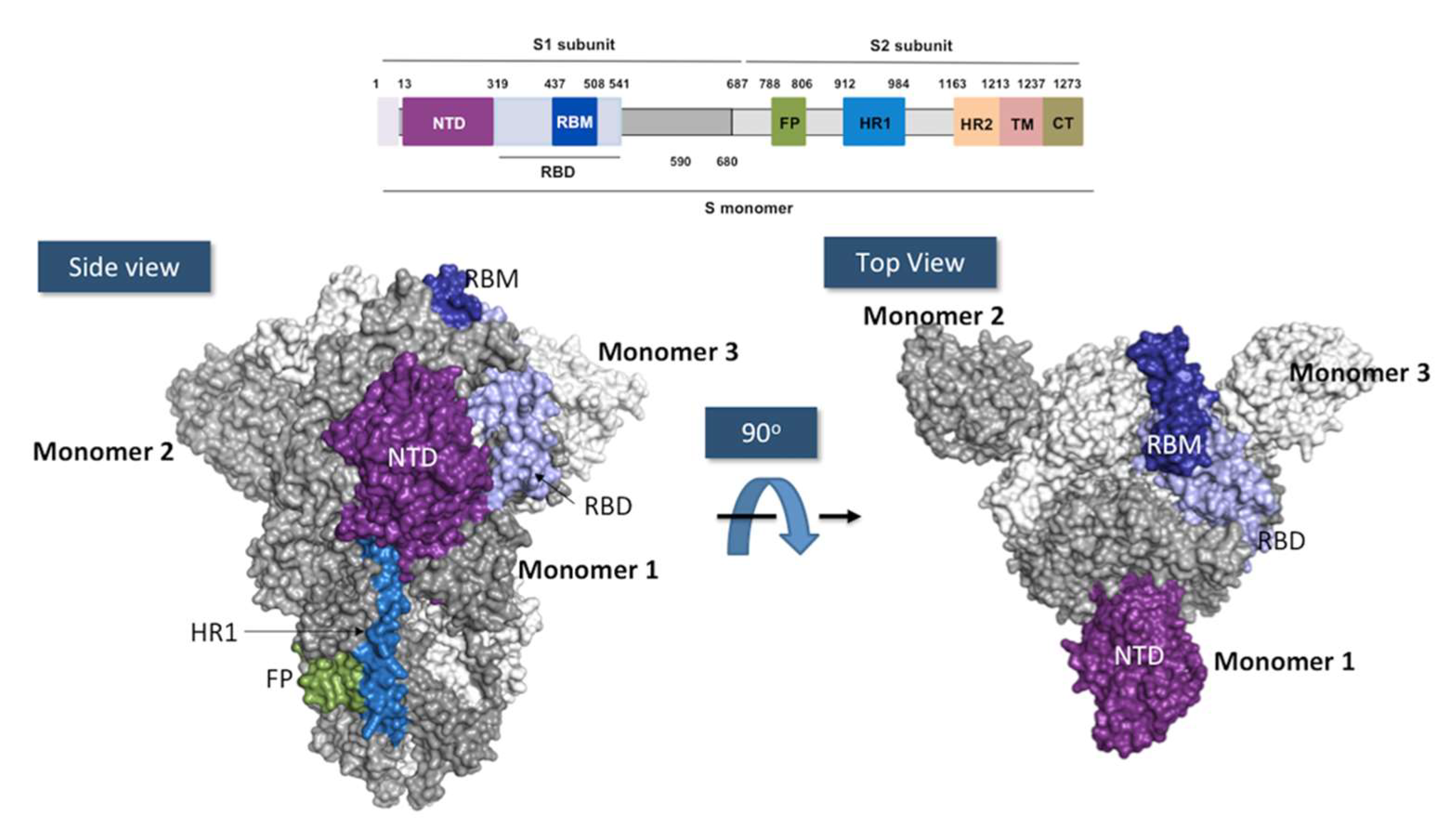
4. SARS-CoV-2 and Mechanism of Infection
RBD Structure and Interaction with hACE-2
5. Sources of the Therapeutic Antibodies
6. Gene Usage and LCDR3/HCDR3 Key Features
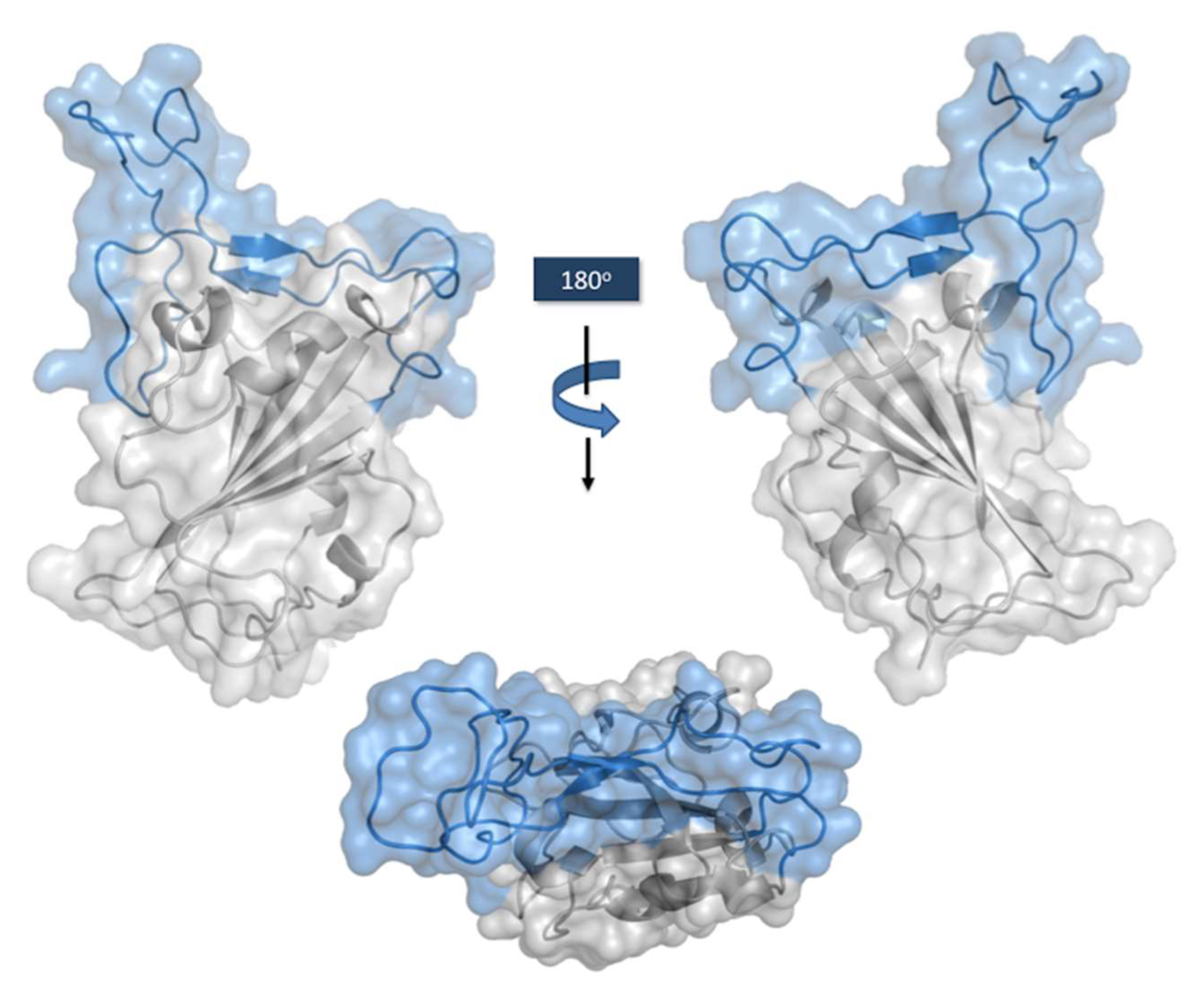
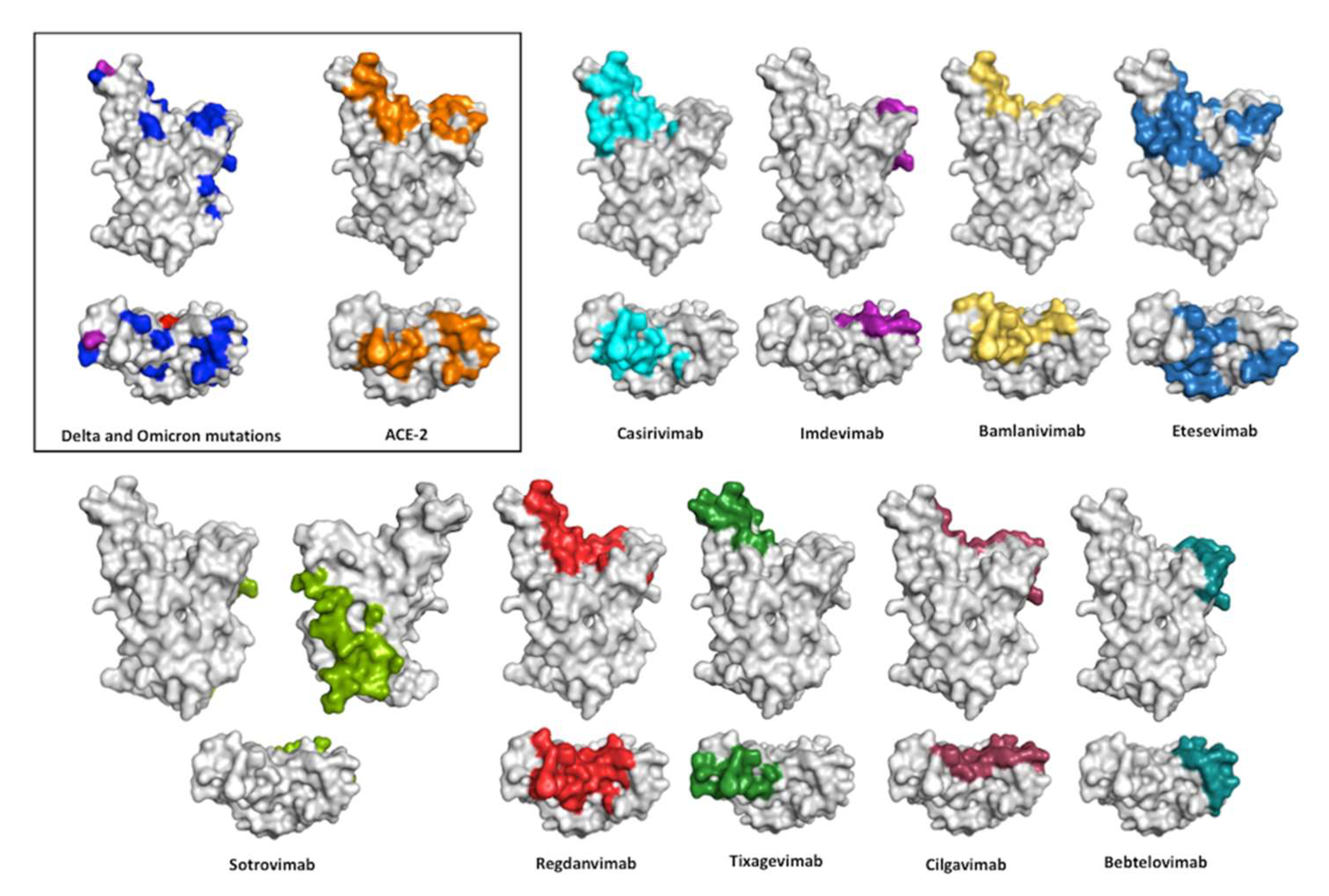
7. Interaction of the Therapeutic Antibodies with the RBD
8. Isotypes and Fc Engineering
9. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The-nCoV Outbreak Joint Field Epidemiology Investigation Team; Li, Q. An Outbreak of NCIP (2019-nCoV) Infection in China —Wuhan, Hubei Province, 2019–2020. China CDC Wkly. 2020, 2, 79–80. [Google Scholar] [CrossRef]
- Tan, W.; Zhao, X.; Ma, X.; Wang, W.; Niu, P.; Xu, W.; Gao, G.F.; Wu, G. A Novel Coronavirus Genome Identified in a Cluster of Pneumonia Cases—Wuhan, China 2019–2020. China CDC Wkly. 2020, 2, 61–62. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Virtual Press Conference on COVID-19–11 March 2020; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Mullard, A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Discov. 2021, 20, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Moekotte, A.L.; Huson, M.A.; van der Ende, A.J.; Agnandji, S.T.; Huizenga, E.; Goorhuis, A.; Grobusch, M.P. Monoclonal antibodies for the treatment of Ebola virus disease. Expert Opin. Investig. Drugs 2016, 25, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Renn, A.; Fu, Y.; Hu, X.; Hall, M.D.; Simeonov, A. Fruitful Neutralizing Antibody Pipeline Brings Hope To Defeat SARS-Cov-2. Trends Pharmacol. Sci. 2020, 41, 815–829. [Google Scholar] [CrossRef]
- Corti, D.; Purcell, L.A.; Snell, G.; Veesler, D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 2021, 184, 3086–3108. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Emergency Use Authorization (EUA) for Casirivimab and Imdevimab; Center for Drug Evaluation and Research (CDER) Review; FDA: Silver Spring, MD, USA, 2020. [Google Scholar]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2020. [Google Scholar]
- European Medicines Agency. Ronapreve; Assessment Report; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- European Medicines Agency. COVID-19: EMA Recommends Authorisation of Two Monoclonal Antibody Medicines; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- U.S. Food and Drug Administration. Emergency Use Authorization (EUA) for Bamlanivimab 700 mg and Etesevimab 1400 mg IV Administered Together; Center for Drug Evaluation and Research (CDER) Review; FDA: Silver Spring, MD, USA, 2020. [Google Scholar]
- European Medicines Agency. Bamlanivimab and Etesevimab for COVID-19: Withdrawal from the Rolling Review Process. Available online: https://www.ema.europa.eu/en/medicines/human/withdrawn-applications/bamlanivimab-etesevimab-covid-19 (accessed on 5 July 2022).
- U.S. Food and Drug Administration. Emergency Use Authorization (EUA) for Sotrovimab 1500 mg; Center for Drug Evaluation and Research (CDER) Review; FDA: Silver Spring, MD, USA, 2020. [Google Scholar]
- European Medicines Agency. COVID-19: EMA recommends authorisation of antibody medicine Xevudy; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- European Medicines Agency. Assessment Report Procedure under Article 5(3) of Regulation (EC) No 726/2004, Celltrion Use of Regdanvimab for the Treatment of COVID-19; Center for Drug Evaluation and Research (CDER) Review; EMA: Amsterdam, The Netherlands, 2020. [Google Scholar]
- U.S. Food and Drug Administration. Emergency Use Authorization (EUA) for EVUSHELD (Tixagevimab 150 mg and Cilgavimab 150 mg Injection Co-Packaged for Intramuscular Use); Center for Drug Evaluation and Research (CDER) Review; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- European Medicines Agency. EMA Recommends Authorisation of COVID-19 Medicine Evusheld; European Medicines Agency: Amsterdam, The Netherlands, 24 March 2022. [Google Scholar]
- U.S. Food and Drug Administration. Emergency Use Authorization (EUA) for Bebtelovimab (LY-CoV1404); Center for Drug Evaluation and Research (CDER) Review; FDA: Silver Spring, MD, USA, 2022. [Google Scholar]
- GISAID. Available online: https://www.gisaid.org/ (accessed on 14 July 2022).
- World Health Organization. Guidance for Surveillance of SARS-CoV-2 Variants: Interim Guidance; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 8 June 2022).
- Mohapatra, R.K.; Kandi, V.; Sarangi, A.K.; Verma, S.; Tuli, H.S.; Chakraborty, S.; Chakraborty, C.; Dhama, K. The recently emerged BA.4 and BA.5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic—Correspondence. Int. J. Surg. 2022, 103, 106698. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Errico, J.M.; Halfmann, P.J.; Zost, S.J.; Crowe, J.E.; Purcell, L.A.; Kawaoka, Y.; Corti, D.; Fremont, D.H.; Diamond, M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022, 28, 490–495. [Google Scholar] [CrossRef]
- Yang, T.J.; Yu, P.Y.; Chang, Y.C.; Liang, K.H.; Tso, H.C.; Ho, M.R.; Chen, W.Y.; Lin, H.T.; Wu, H.C.; Hsu, S.D. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat. Struct. Mol. Biol. 2021, 28, 731–739. [Google Scholar] [CrossRef]
- Radvak, P.; Kwon, H.J.; Kosikova, M.; Ortega-Rodriguez, U.; Xiang, R.; Phue, J.N.; Shen, R.F.; Rozzelle, J.; Kapoor, N.; Rabara, T.; et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat. Commun. 2021, 12, 6559. [Google Scholar] [CrossRef] [PubMed]
- Gräf, T.; Bello, G.; Venas, T.M.M.; Pereira, E.C.; Paixão, A.C.D.; Appolinario, L.R.; Lopes, R.S.; Mendonça, A.C.D.F.; da Rocha, A.S.B.; Motta, F.C.; et al. Identification of a novel SARS-CoV-2 P.1 sub-lineage in Brazil provides new insights about the mechanisms of emergence of variants of concern. Virus Evol. 2021, 7, veab091. [Google Scholar] [CrossRef] [PubMed]
- Shiehzadegan, S.; Alaghemand, N.; Fox, M.; Venketaraman, V. Analysis of the Delta Variant B.1.617.2 COVID-19. Clin. Pract. 2021, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Covariants. Available online: https://covariants.org/variants/21K.Omicron (accessed on 8 June 2022).
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.E.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e2413. [Google Scholar] [CrossRef]
- Food and Drug Administration. Coronavirus (COVID-19) Update: July 30, 2021; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- Food and Drug Administration. FDA Authorizes Bamlanivimab and Etesevimab Monoclonal Antibody Therapy for Post-Exposure Prophylaxis (Prevention) for COVID-19; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes Additional Monoclonal Antibody for Treatment of COVID-19; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- European Medicines Agency. Regkirona; Assessment Report; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes New Long-Acting Monoclonal Antibodies for Pre-exposure Prevention of COVID-19 in Certain Individuals; FDA: Silver Spring, MD, USA, 2021. [Google Scholar]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes New Monoclonal Antibody for Treatment of COVID-19 That Retains Activity against Omicron Variant; FDA: Silver Spring, MD, USA, 2022. [Google Scholar]
- Food and Drug Administration. Bamlanivimab and Etesevimab Authorized States, Territories, and U.S. Jurisdictions. 2021. Available online: https://www.fda.gov/media/151719/download (accessed on 8 June 2022).
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Limits Use of Certain Monoclonal Antibodies to Treat COVID-19 Due to the Omicron Variant; FDA: Silver Spring, MD, USA, 2022. [Google Scholar]
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 14 July 2022).
- Wang, L.; Zhou, T.; Zhang, Y.; Yang, E.S.; Schramm, C.A.; Shi, W.; Pegu, A.; Oloniniyi, O.K.; Henry, A.R.; Darko, S.; et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 2021, 373. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health. Available online: https://www.covid19treatmentguidelines.nih.gov/tables/variants-and-susceptibility-to-mabs/ (accessed on 5 July 2022).
- Chen, R.E.; Winkler, E.S.; Case, J.B.; Aziati, I.D.; Bricker, T.L.; Joshi, A.; Darling, T.L.; Ying, B.; Errico, J.M.; Shrihari, S.; et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature 2021, 596, 103–108. [Google Scholar] [CrossRef]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; di Iulio, J.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef]
- Takashita, E.; Kinoshita, N.; Yamayoshi, S.; Sakai-Tagawa, Y.; Fujisaki, S.; Ito, M.; Iwatsuki-Horimoto, K.; Halfmann, P.; Watanabe, S.; Maeda, K.; et al. Efficacy of Antiviral Agents against the SARS-CoV-2 Omicron Subvariant BA.2. N. Engl. J. Med. 2022, 386, 1475–1477. [Google Scholar] [CrossRef]
- Food and Drug Administration. Fact Sheet for Healthcare Providers Emergency Use Authorization (EUA) of Sotrovimab; FDA: Silver Spring, MD, USA, 2022. [Google Scholar]
- Ryu, D.K.; Kang, B.; Noh, H.; Woo, S.J.; Lee, M.H.; Nuijten, P.M.; Kim, J.I.; Seo, J.M.; Kim, C.; Kim, M.; et al. The in vitro and in vivo efficacy of CT-P59 against Gamma, Delta and its associated variants of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021, 578, 91–96. [Google Scholar] [CrossRef]
- Food and Drug Administration. Fact Sheet for Healthcare Providers Emergency Use Authorization of EVUSHELD—(Tixagevimab Co-Packaged with Cilgavimab); FDA: Silver Spring, MD, USA, 2022. [Google Scholar]
- Food and Drug Administration. Fact Sheet for Healthcare Providers: Emergency Use Authorization for Bebtelovimab; FDA: Silver Spring, MD, USA, 2022. [Google Scholar]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus Infections—More Than Just the Common Cold. JAMA 2020, 323, 707–708. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.L.; Lavillette, D.; Denolly, S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021, 296, 100111. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Ye, F.; Guo, Y.; Xia, L.; Zhong, X.; Chi, X.; Zhou, Q. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res. 2021, 31, 717–719. [Google Scholar] [CrossRef]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640.e610. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Tuccori, M.; Convertino, I.; Ferraro, S.; Valdiserra, G.; Cappello, E.; Fini, E.; Focosi, D. An overview of the preclinical discovery and development of bamlanivimab for the treatment of novel coronavirus infection (COVID-19): Reasons for limited clinical use and lessons for the future. Expert Opin. Drug Discov. 2021, 16, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E.; Brown-Augsburger, P.L.; Corbett, K.S.; Westendorf, K.; Davies, J.; Cujec, T.P.; Wiethoff, C.M.; Blackbourne, J.L.; Heinz, B.A.; Foster, D.; et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021, 13, eabf1906. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Kim, C.; Ryu, D.K.; Lee, J.; Kim, Y.I.; Seo, J.M.; Kim, Y.G.; Jeong, J.H.; Kim, M.; Kim, J.I.; Kim, P.; et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021, 12, 288. [Google Scholar] [CrossRef]
- Zost, S.J.; Gilchuk, P.; Chen, R.E.; Case, J.B.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; Sutton, R.E.; Suryadevara, N.; Chen, E.C.; et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020, 26, 1422–1427. [Google Scholar] [CrossRef]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Westendorf, K.; Žentelis, S.; Wang, L.; Foster, D.; Vaillancourt, P.; Wiggin, M.; Lovett, E.; van der Lee, R.; Hendle, J.; Pustilnik, A.; et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022, 39, 110812. [Google Scholar] [CrossRef]
- Popov, A.V.; Zou, X.; Xian, J.; Nicholson, I.C.; Brüggemann, M. A human immunoglobulin lambda locus is similarly well expressed in mice and humans. J. Exp. Med. 1999, 189, 1611–1620. [Google Scholar] [CrossRef]
- Almagro, J.C.; Pedraza-Escalona, M.; Arrieta, H.I.; Pérez-Tapia, S.M. Phage Display Libraries for Antibody Therapeutic Discovery and Development. Antibodies 2019, 8, 44. [Google Scholar] [CrossRef]
- Wu, Y.C.; Kipling, D.; Leong, H.S.; Martin, V.; Ademokun, A.A.; Dunn-Walters, D.K. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 2010, 116, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, I.M.; Walter, G.; Marks, J.D.; Llewelyn, M.B.; Winter, G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J. Mol. Biol. 1992, 227, 776–798. [Google Scholar] [CrossRef]
- Glanville, J.; Zhai, W.; Berka, J.; Telman, D.; Huerta, G.; Mehta, G.R.; Ni, I.; Mei, L.; Sundar, P.D.; Day, G.M.; et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc. Natl. Acad. Sci. USA 2009, 106, 20216–20221. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Ishii, K.; Bourvagnet, P.; Kuma, K.I.; Hayashida, H.; Miyata, T.; Honjo, T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 1998, 188, 2151–2162. [Google Scholar] [CrossRef]
- Raybould, M.I.J.; Marks, C.; Krawczyk, K.; Taddese, B.; Nowak, J.; Lewis, A.P.; Bujotzek, A.; Shi, J.; Deane, C.M. Five computational developability guidelines for therapeutic antibody profiling. Proc. Natl. Acad. Sci. USA 2019, 116, 4025–4030. [Google Scholar] [CrossRef]
- Dong, J.; Zost, S.J.; Greaney, A.J.; Starr, T.N.; Dingens, A.S.; Chen, E.C.; Chen, R.E.; Case, J.B.; Sutton, R.E.; Gilchuk, P.; et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat. Microbiol. 2021, 6, 1233–1244. [Google Scholar] [CrossRef]
- Almagro, J.C.; Raghunathan, G.; Beil, E.; Janecki, D.J.; Chen, Q.; Dinh, T.; LaCombe, A.; Connor, J.; Ware, M.; Kim, P.H.; et al. Characterization of a high-affinity human antibody with a disulfide bridge in the third complementarity-determining region of the heavy chain. J. Mol. Recognit. 2012, 25, 125–135. [Google Scholar] [CrossRef]
- Gilliland, G.L.; Luo, J.; Vafa, O.; Almagro, J.C. Leveraging SBDD in protein therapeutic development: Antibody engineering. Methods Mol. Biol. 2012, 841, 321–349. [Google Scholar] [CrossRef]
- Tomlinson, I.M.; Cox, J.P.; Gherardi, E.; Lesk, A.M.; Chothia, C. The structural repertoire of the human V kappa domain. EMBO J. 1995, 14, 4628–4638. [Google Scholar] [CrossRef]
- Finlay, W.J.; Almagro, J.C. Natural and man-made V-gene repertoires for antibody discovery. Front. Immunol. 2012, 3, 342. [Google Scholar] [CrossRef]
- Yan, Q.; He, P.; Huang, X.; Luo, K.; Zhang, Y.; Yi, H.; Wang, Q.; Li, F.; Hou, R.; Fan, X.; et al. Germline IGHV3-53-encoded RBD-targeting neutralizing antibodies are commonly present in the antibody repertoires of COVID-19 patients. Emerg. Microbes Infect. 2021, 10, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Mariano, G.; Farthing, R.J.; Lale-Farjat, S.L.M.; Bergeron, J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020, 7, 605236. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.B.; Tao, S.C. Epitope Analysis of Anti-SARS-CoV-2 Neutralizing Antibodies. Curr. Med. Sci. 2021, 41, 1065–1074. [Google Scholar] [CrossRef]
- Barnes, C.O.; West, A.P., Jr.; Huey-Tubman, K.E.; Hoffmann, M.A.G.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842.e816. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- Wu, W.L.; Chiang, C.Y.; Lai, S.C.; Yu, C.Y.; Huang, Y.L.; Liao, H.C.; Liao, C.L.; Chen, H.W.; Liu, S.J. Monoclonal antibody targeting the conserved region of the SARS-CoV-2 spike protein to overcome viral variants. JCI Insight 2022, 7, e157597. [Google Scholar] [CrossRef]
- Ku, Z.; Xie, X.; Davidson, E.; Ye, X.; Su, H.; Menachery, V.D.; Li, Y.; Yuan, Z.; Zhang, X.; Muruato, A.E.; et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021, 12, 469. [Google Scholar] [CrossRef]
- Du, L.; Yang, Y.; Zhang, X. Neutralizing antibodies for the prevention and treatment of COVID-19. Cell Mol. Immunol. 2021, 18, 2293–2306. [Google Scholar] [CrossRef]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331.e2315. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Nelson-Sathi, S.; Umasankar, P.K.; Sreekumar, E.; Nair, R.R.; Joseph, I.; Nori, S.R.C.; Philip, J.S.; Prasad, R.; Navyasree, K.V.; Ramesh, S.; et al. Mutational landscape and in silico structure models of SARS-CoV-2 spike receptor binding domain reveal key molecular determinants for virus-host interaction. BMC Mol. Cell Biol. 2022, 23, 2. [Google Scholar] [CrossRef]
- Camacho-Sandoval, R.; Nieto-Patlán, A.; Carballo-Uicab, G.; Montes-Luna, A.; Jiménez-Martínez, M.C.; Vallejo-Castillo, L.; González-González, E.; Arrieta-Oliva, H.I.; Gómez-Castellano, K.; Guzmán-Bringas, O.U.; et al. Development and Evaluation of a Set of Spike and Receptor Binding Domain-Based Enzyme-Linked Immunosorbent Assays for SARS-CoV-2 Serological Testing. Diagnostics 2021, 11, 1506. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Choi, A.; Izikson, R.; Cox, M.M.; Palese, P.; Krammer, F. Age Dependence and Isotype Specificity of Influenza Virus Hemagglutinin Stalk-Reactive Antibodies in Humans. mBio 2016, 7, e01996-15. [Google Scholar] [CrossRef]
- Chen, M.; Sällberg, M.; Sönnerborg, A.; Weiland, O.; Mattsson, L.; Jin, L.; Birkett, A.; Peterson, D.; Milich, D.R. Limited humoral immunity in hepatitis C virus infection. Gastroenterology 1999, 116, 135–143. [Google Scholar] [CrossRef]
- Gregorek, H.; Madaliński, K.; Woynarowski, M.; Mikolajewicz, J.; Syczewska, M.; Socha, J. IgG subclass distribution of hepatitis B surface antigen antibodies induced in children with chronic hepatitis B infection after interferon-alpha therapy. J. Infect. Dis. 2000, 181, 2059–2062. [Google Scholar] [CrossRef]
- Gregorek, H.; Dzierzanowska-Fangrat, K.; Woynarowski, M.; Jóźwiak, P.; Witkowska-Vogtt, E.; Socha, J.; Syczewska, M.; Madaliński, K. Persistence of HBV-DNA in children with chronic hepatitis B who seroconverted to anti-HBs antibodies after interferon-alpha therapy: Correlation with specific IgG subclass responses to HBsAg. J. Hepatol. 2005, 42, 486–490. [Google Scholar] [CrossRef]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [Green Version]
- Almagro, J.C.; Daniels-Wells, T.R.; Perez-Tapia, S.M.; Penichet, M.L. Progress and Challenges in the Design and Clinical Development of Antibodies for Cancer Therapy. Front. Immunol. 2017, 8, 1751. [Google Scholar] [CrossRef] [PubMed]
- Zalevsky, J.; Chamberlain, A.K.; Horton, H.M.; Karki, S.; Leung, I.W.; Sproule, T.J.; Lazar, G.A.; Roopenian, D.C.; Desjarlais, J.R. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 2010, 28, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; McTamney, P.M.; Arends, R.H.; Abram, M.E.; Aksyuk, A.A.; Diallo, S.; Flores, D.J.; Kelly, E.J.; Ren, K.; Roque, R.; et al. The SARS-CoV-2 monoclonal antibody combination, AZD7442, is protective in nonhuman primates and has an extended half-life in humans. Sci. Transl. Med. 2022, 14, eabl8124. [Google Scholar] [CrossRef]
- Winkler, E.S.; Gilchuk, P.; Yu, J.; Bailey, A.L.; Chen, R.E.; Chong, Z.; Zost, S.J.; Jang, H.; Huang, Y.; Allen, J.D.; et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell 2021, 184, 1804–1820.e1816. [Google Scholar] [CrossRef]
- Chan, C.E.Z.; Seah, S.G.K.; Chye, D.H.; Massey, S.; Torres, M.; Lim, A.P.C.; Wong, S.K.K.; Neo, J.J.Y.; Wong, P.S.; Lim, J.H.; et al. The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. PLoS ONE 2021, 16, e0253487. [Google Scholar] [CrossRef]
- Lee, J.H.; Toy, L.; Kos, J.T.; Safonova, Y.; Schief, W.R.; Havenar-Daughton, C.; Watson, C.T.; Crotty, S. Vaccine genetics of IGHV1-2 VRC01-class broadly neutralizing antibody precursor naïve human B cells. NPJ Vaccines 2021, 6, 113. [Google Scholar] [CrossRef] [PubMed]
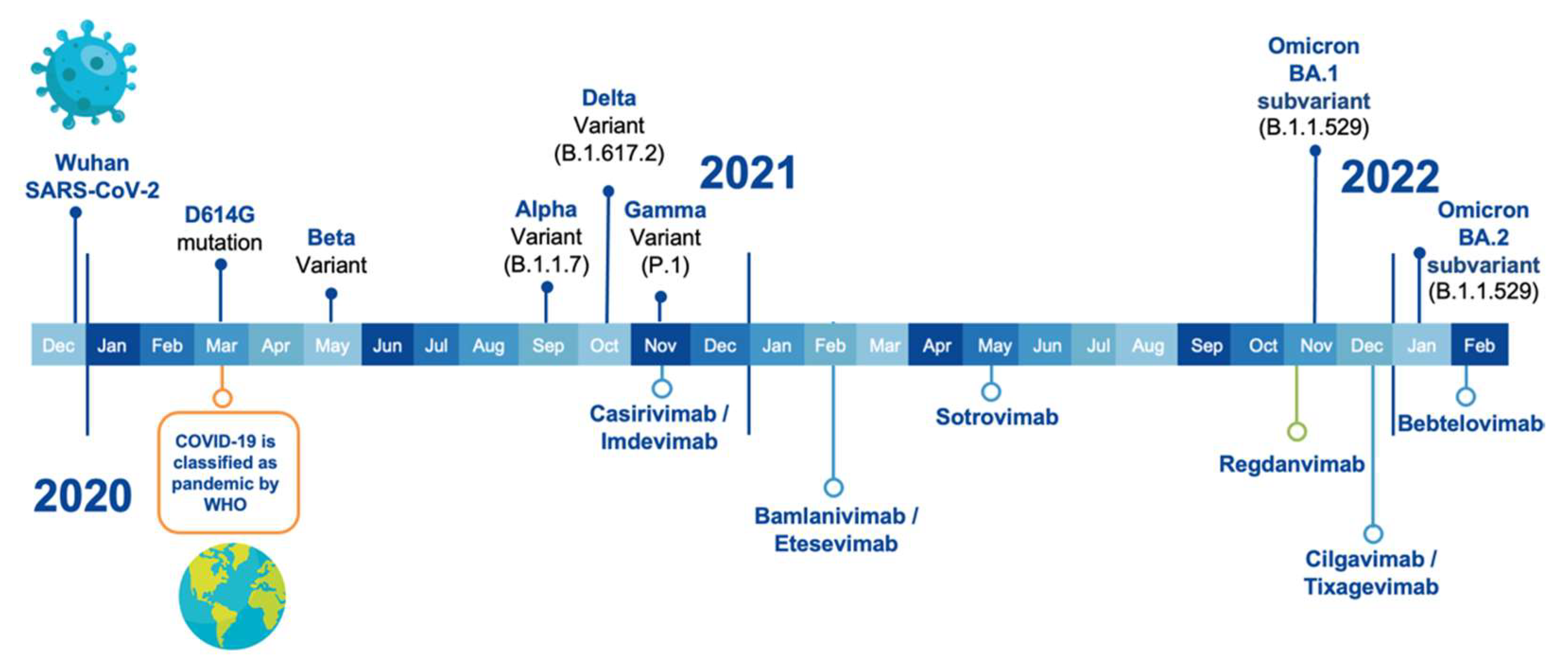
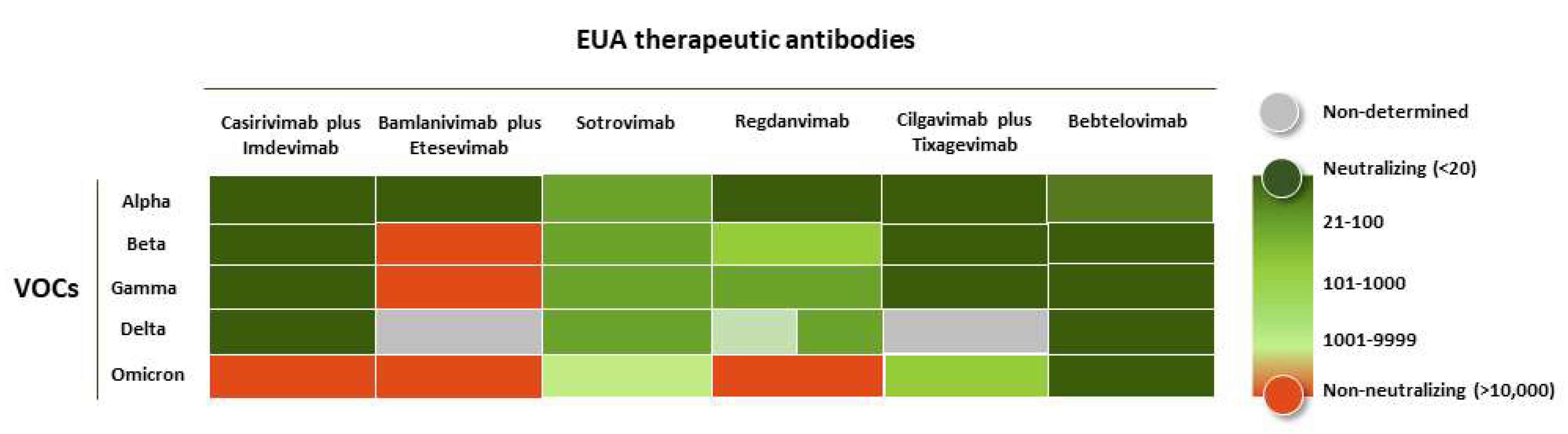

| INN (a) | Other Names | Commercial Name | Company | EUA | |
|---|---|---|---|---|---|
| FDA | EMA | ||||
| Casirivimab | REGN 10933 | REGEN-COV, Ronapreve | Regeneron Pharmaceuticals | 21 November 2020 [9,10] | 11 November 2021 [11,12] |
| Imdevimab | REGN 10987 | ||||
| Bamlanivimab | BAM, LY3819253, LY-CoV555 | N/A (b) | Eli Lilly and Company | 9 February 2021 [13] | EMA withdrew the application on 29 October 2021 [14] |
| Etesevimab | CB6, ETE, LY3832479, LY-CoV016 | ||||
| Sotrovimab | S309, VIR-7831 GSK 4182136 | Xevudy | GlaxoSmithKline (GSK) | 26 May 2021 [15] | 16 December 2021 [16] |
| Regdanvimab | CT-P59 | Regkirona | Celltrion | N/A | 11 November 2021 [12,17] |
| Cilgavimab | COV2-2130, AZD1061 | Evusheld | Astra Zeneca | 8 December 2021 [18] | 24 March 2022 [19] |
| Tixagevimab | COV2-2196, AZD8895 | ||||
| Bebtelovimab | LY-CoV1404 | N/A | Eli Lilly and Company | 11 February 2022 [20] | N/A |
| Isotype | IGHV | IGHD | IGHJ | IGK/LV | IGK/LJ | HCDR3 | Fc | |
|---|---|---|---|---|---|---|---|---|
| Casirivimab | IgG1k | 3-11 * 01 | 1-14 * 02 | 4 * 02 | 1-33 * 01 | 1 * 01 | 11 | None |
| Imdevimab | IgG1λ | 3-33 * 03 | 2-8 * 02 | 1 * 01 | 2-14 * 01 | 3 * 02 | 11 | None |
| Bamlanivimab | IgG1k | 1-69 * 09 | 3-16 * 01 /6 * 0104 | 6-01 | 1-39 * 01 | 2 * 2 | 16 | None |
| Etesevimab | IgG1k | 3-66 * 01 | 2-8 * 02 /4 * 01 | 4-01 | 1-39 * 01 | 2 * 01 | 11 | LALA |
| Sotrovimab | IgG1k | 1-18 * 01 | 3-16 * 01 | 1 * 01 | 3-20 * 01 | 1 * 01 | 18 | LS |
| Regdanvimab | IgG1λ | 2-70 * 12 | 1-14 * 01 | 6 * 02 | 1-51 * 01 | 3 * 02 | 18 | None |
| Cilgavimab | IgG1k | 3-15 * 01 | 3-22 * 01 | 4 * 02 | 4-1 * 02 | 1 * 01 | 20 | YTE and TM |
| Tixagevimab | IgG1k | 1-5 * 03 | 6-13 * 01 | 3 * 02 | 3-20 * 01 | 1 * 01 | 14 | |
| Bebtelovimab | IgG1λ | 2-5 * 02 | 3-3 * 02 | 1 * 01 | 2-14 * 01 | 3 * 02 | 9 | None |
| Class | KD | Blocking Assay (IC50) | Neutralization Assay (EC50) | Dose (mg) | |
|---|---|---|---|---|---|
| Casirivimab [9,63] | I | 0.046 | 0.056 | 0.04 | 600 |
| Imdevimab [9,63] | III/IV | 0.047 | 0.165 | 0.04 | 600 |
| Bamlanivimab [13] | II | 0.071 | 0.170 | 0.14 | 700 |
| Etesevimab [13] | I | 6.450 | 0.320 | 0.97 | 1400 |
| Sotrovimab [15] | IV | 0.210 | 33.600 | 0.67 | 500 |
| Regdanvimab [37] | I | 0.065 | - | 0.05 | 2400 (a) |
| Cilgavimab [18] | I | 2.150 | 0.531 | 0.012 | 150 |
| Tixagevimab [18] | III | 2.180 | 0.318 | 0.06 | 150 |
| Bebtelovimab [20] | III | 0.075 | 0.380 | 0.04 | 175 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almagro, J.C.; Mellado-Sánchez, G.; Pedraza-Escalona, M.; Pérez-Tapia, S.M. Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies. Int. J. Mol. Sci. 2022, 23, 9763. https://doi.org/10.3390/ijms23179763
Almagro JC, Mellado-Sánchez G, Pedraza-Escalona M, Pérez-Tapia SM. Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies. International Journal of Molecular Sciences. 2022; 23(17):9763. https://doi.org/10.3390/ijms23179763
Chicago/Turabian StyleAlmagro, Juan C., Gabriela Mellado-Sánchez, Martha Pedraza-Escalona, and Sonia M. Pérez-Tapia. 2022. "Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies" International Journal of Molecular Sciences 23, no. 17: 9763. https://doi.org/10.3390/ijms23179763
APA StyleAlmagro, J. C., Mellado-Sánchez, G., Pedraza-Escalona, M., & Pérez-Tapia, S. M. (2022). Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies. International Journal of Molecular Sciences, 23(17), 9763. https://doi.org/10.3390/ijms23179763






