TRIM28 Is a Novel Regulator of CD133 Expression Associated with Cancer Stem Cell Phenotype
Abstract
:1. Introduction
2. Results
2.1. The Majority of Analyzed Cancer Cell Lines Do Not Contain CD133-Positive Cells
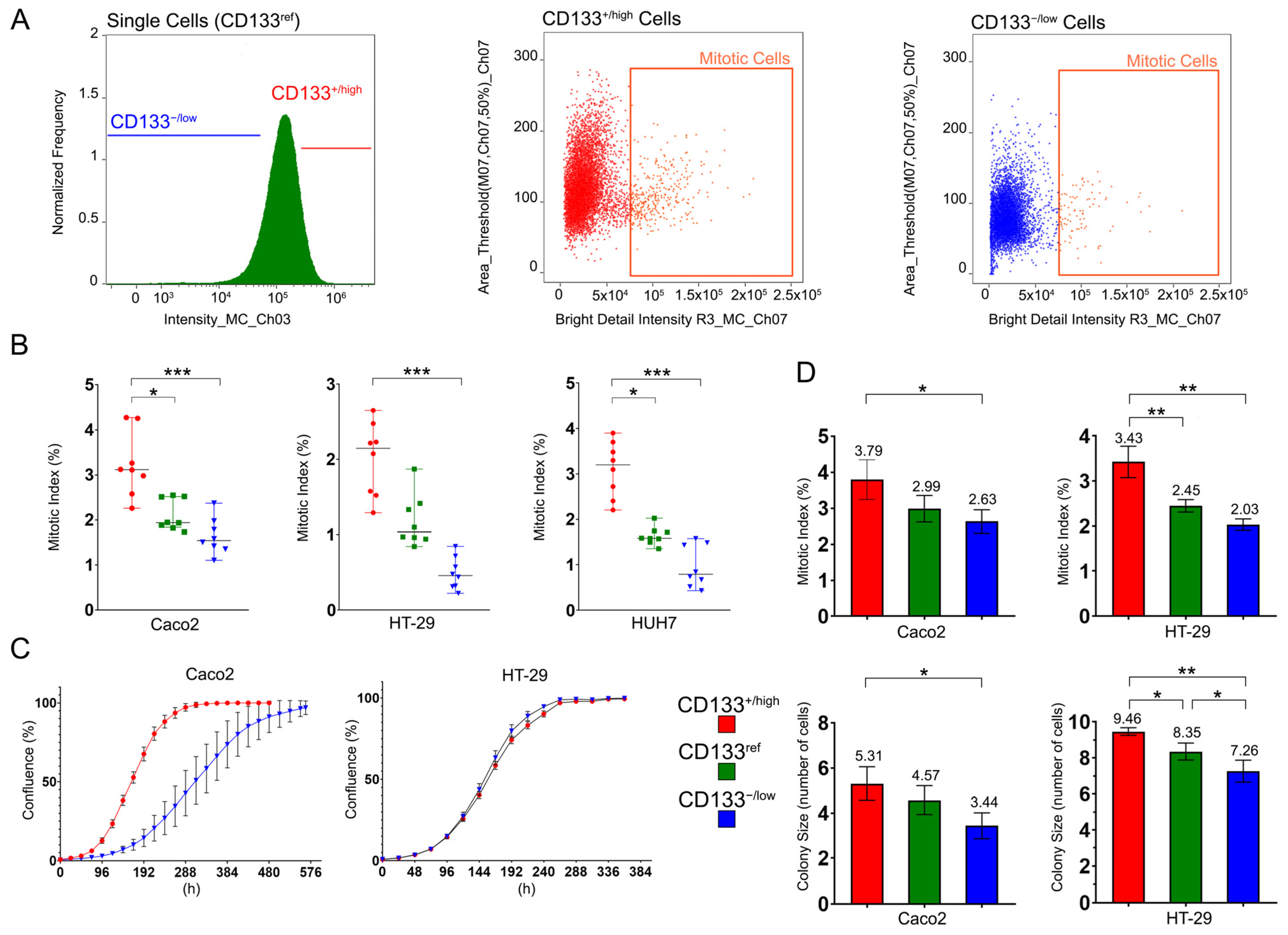
2.2. CD133+/high Cells Have Higher Proliferative Capacity Than CD133−/low and CD133ref Cells
2.3. Transcripts and Proteins Differentially Expressed in the CD133+/high and CD133−/low Cell Populations
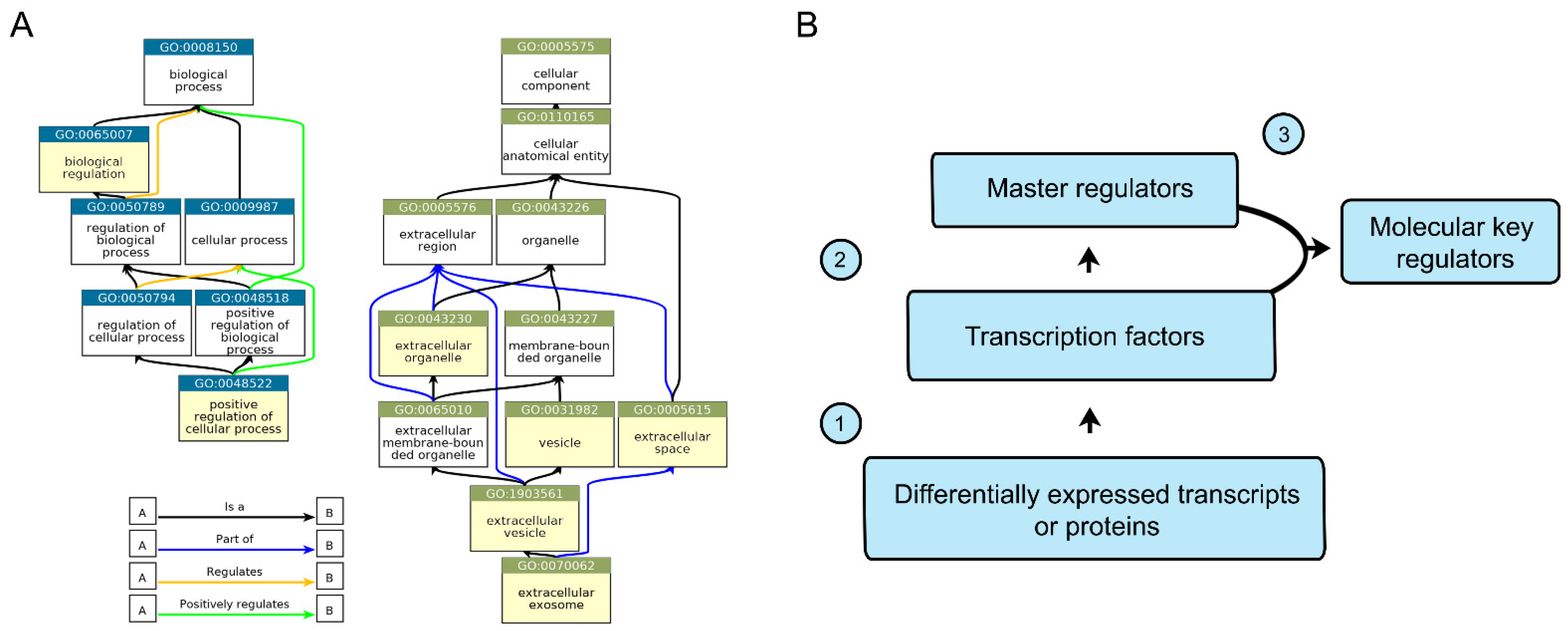
2.4. In Silico Analysis of CD133-Associated Key Molecular Regulators
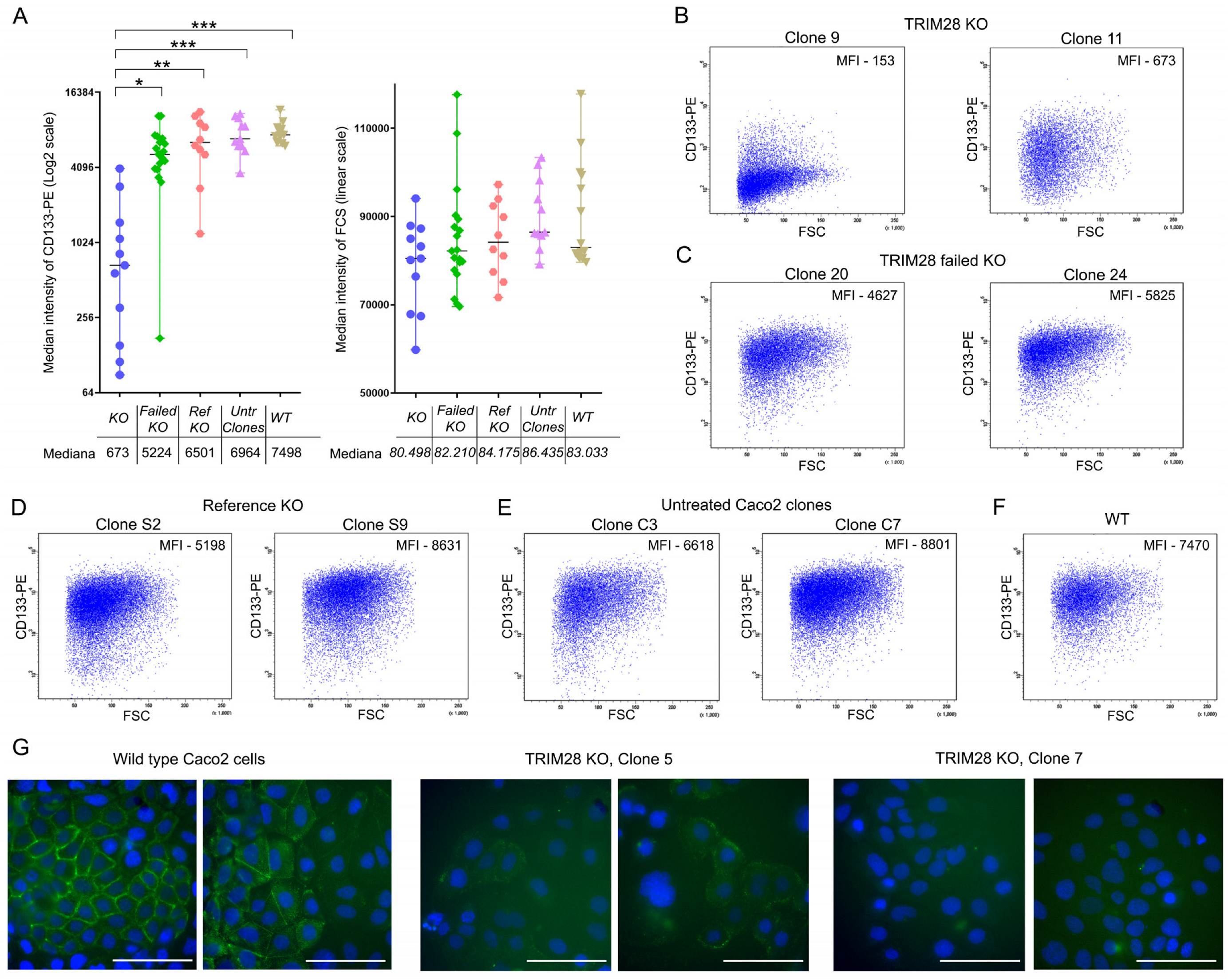
2.5. Full TRIM28 Knockout, But Not Its Knockdown Downregulates CD133 Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. FACS (Fluorescence-Activated Cell Sorting)
4.3. Imaging Flow Cytometry
4.4. Fluorescence Microscopy
4.5. Cell Growth Analysis
4.6. Short-Time Clonogenic Assay and Mitotic Index (MI) Calculation
4.7. Whole Transcriptome Analysis
4.8. Label-Free Mass-Spectrometry
4.9. In Silico Analysis
4.10. Lentivirus Transduction Knockdown
4.11. CRISPR/CAS9-Mediated Knockout
4.12. SDS-PAGE and Western Blot Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, J.; Ding, D. The Prognostic Role of the Cancer Stem Cell Marker CD44 in Ovarian Cancer: A Meta-Analysis. Cancer Cell Int. 2017, 17, 8. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Yan, T.; Di, G.; Shen, Z.; Shao, Z.; Lu, J. The Prognostic Role of Cancer Stem Cells in Breast Cancer: A Meta-Analysis of Published Literatures. Breast Cancer Res. Treat. 2010, 122, 795–801. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kaidina, A.M.; Chiang, J.H.; Yarygin, K.N.; Lupatov, A.Y. Cancer Stem Cell Molecular Markers Verified in Vivo. Biochem. Mosc. Suppl. B Biomed. Chem. 2017, 11, 43–54. [Google Scholar] [CrossRef]
- Suvorov, R.E.; Kim, Y.S.; Gisina, A.M.; Chiang, J.H.; Yarygin, K.N.; Lupatov, A.Y. Surface Molecular Markers of Cancer Stem Cells: Computation Analysis of Full-Text Scientific Articles. Bull. Exp. Biol. Med. 2018, 166, 135–140. [Google Scholar] [CrossRef]
- Yin, A.H.; Miraglia, S.; Zanjani, E.D.; Almeida-Porada, G.; Ogawa, M.; Leary, A.G.; Olweus, J.; Kearney, J.; Buck, D.W.; Johnson, R.H. AC133, a Novel Marker for Human Hematopoietic Stem and Progenitor Cells Myelomonocytic, and Megakaryocytic Repopulation in Vivo While the CD34 Dim Population Contains Lineage-Committed. Blood 1997, 90, 5002–5012. [Google Scholar] [CrossRef] [PubMed]
- Ward, H.H.; Romero, E.; Welford, A.; Pickett, G.; Bacallao, R.; Gattone, V.H.; Ness, S.A.; Wandinger-Ness, A.; Roitbak, T. Adult Human CD133/1+ Kidney Cells Isolated from Papilla Integrate into Developing Kidney Tubules. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 1344–1357. [Google Scholar] [CrossRef] [PubMed]
- Richardson, G.D.; Robson, C.N.; Lang, S.H.; Neal, D.E.; Maitland, N.J.; Collins, A.T. CD133, a Novel Marker for Human Prostatic Epithelial Stem Cells. J. Cell Sci. 2004, 117, 3539–3545. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, S.; Eckert, K.; He, D.; Sutton, R.; Doshe, M.; Jain, G.; Tushinski, R.; Reitsma, M.; Harris, B.; Tsukamoto, A.; et al. Engraftment of Sorted/Expanded Human Central Nervous System Stem Cells from Fetal Brain. J. Neurosci. Res. 2002, 69, 976–986. [Google Scholar] [CrossRef]
- Rountree, C.B.; Barsky, L.; Ge, S.; Zhu, J.; Senadheera, S.; Crooks, G.M. A CD133-Expressing Murine Liver Oval Cell Population with Bilineage Potential. Stem Cells 2007, 25, 2419–2429. [Google Scholar] [CrossRef]
- Li, Y.; Yunshan, G.; Bo, M.; Yu, Z.; Tao, W.; Gengfang, L.; Dexian, F.; Shiqian, C.; Jianli, J.; Tang, J.; et al. CD133 Overexpression Correlates with Clinicopathological Features of Gastric Cancer Patients and Its Impact on Survival: A Systematic Review and Meta-Analysis. Oncotarget 2015, 6, 42015–42027. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhao, H.; Gu, J.; Zheng, L. Prognostic Value of Cancer Stem Cell Marker CD133 Expression in Pancreatic Ductal Adenocarcinoma (PDAC): A Systematic Review and Meta-Analysis. Int. J. Clin. Exp. Pathol. 2015, 8, 12084–12092. [Google Scholar] [PubMed]
- Huang, R.; Mo, D.; Wu, J.; Ai, H.; Lu, Y. CD133 Expression Correlates with Clinicopathologic Features and Poor Prognosis of Colorectal Cancer Patients: An Updated Meta-Analysis of 37 Studies. Medicine 2018, 97, e10446. [Google Scholar] [CrossRef] [PubMed]
- Gokulan, R.C.; Devaraj, H. Stem Cell Markers CXCR-4 and CD133 Predict Aggressive Phenotype and Their Double Positivity Indicates Poor Prognosis of Oral Squamous Cell Carcinoma. Cancers 2021, 13, 5895. [Google Scholar] [CrossRef]
- Weigmann, A.; Corbeil, D.; Hellwig, A.; Huttner, W.B. Prominin, a Novel Microvilli-Specific Polytopic Membrane Protein of the Apical Surface of Epithelial Cells, Is Targeted to Plasmalemmal Protrusions of Non-Epithelial Cells. Proc. Natl. Acad. Sci. USA 1997, 94, 12425–12430. [Google Scholar] [CrossRef]
- Thamm, K.; Šimaitė, D.; Karbanová, J.; Bermúdez, V.; Reichert, D.; Morgenstern, A.; Bornhäuser, M.; Huttner, W.B.; Wilsch-Bräuninger, M.; Corbeil, D. Prominin-1 (CD133) Modulates the Architecture and Dynamics of Microvilli. Traffic 2019, 20, 39–60. [Google Scholar] [CrossRef]
- Zacchigna, S.; Oh, H.; Wilsch-Brauninger, M.; Missol-Kolka, E.; Jaszai, J.; Jansen, S.; Tanimoto, N.; Tonagel, F.; Seeliger, M.; Huttner, W.B.; et al. Loss of the Cholesterol-Binding Protein Prominin-1/CD133 Causes Disk Dysmorphogenesis and Photoreceptor Degeneration. J. Neurosci. 2009, 29, 2297–2308. [Google Scholar] [CrossRef] [PubMed]
- Bauer, N.; Wilsch-Bräuninger, M.; Karbanová, J.; Fonseca, A.V.; Strauss, D.; Freund, D.; Thiele, C.; Huttner, W.B.; Bornhäuser, M.; Corbeil, D. Haematopoietic Stem Cell Differentiation Promotes the Release of Prominin-1/CD133-Containing Membrane Vesicles-a Role of the Endocytic-Exocytic Pathway. EMBO Mol. Med. 2011, 3, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, D.; Marzesco, A.M.; Wilsch-Bräuninger, M.; Huttner, W.B. The Intriguing Links between Prominin-1 (CD133), Cholesterol-Based Membrane Microdomains, Remodeling of Apical Plasma Membrane Protrusions, Extracellular Membrane Particles, and (Neuro)Epithelial Cell Differentiation. FEBS Lett. 2010, 584, 1659–1664. [Google Scholar] [CrossRef]
- Marzesco, A.M.; Janich, P.; Wilsch-Bräuninger, M.; Dubreuil, V.; Langenfeld, K.; Corbeil, D.; Huttner, W.B. Release of Extracellular Membrane Particles Carrying the Stem Cell Marker Prominin-1 (CD133) from Neural Progenitors and Other Epithelial Cells. J. Cell Sci. 2005, 118, 2849–2858. [Google Scholar] [CrossRef]
- Mak, A.B.; Nixon, A.M.L.; Kittanakom, S.; Stewart, J.M.; Chen, G.I.; Curak, J.; Gingras, A.C.; Mazitschek, R.; Neel, B.G.; Stagljar, I.; et al. Regulation of CD133 by HDAC6 Promotes β-Catenin Signaling to Suppress Cancer Cell Differentiation. Cell Rep. 2012, 2, 951–963. [Google Scholar] [CrossRef] [Green Version]
- Gong, A.; Huang, S. FoxM1 and Wnt/β-Catenin Signaling in Glioma Stem Cells. Cancer Res. 2012, 72, 5658–5662. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jiang, Y.; Liu, Y.; Zou, F.; Liu, Y.C.; Wang, S.; Xu, N.; Xu, W.; Cui, C.; Xing, Y.; et al. Activation of PI3K/Akt Pathway by CD133-P85 Interaction Promotes Tumorigenic Capacity of Glioma Stem Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 6829–6834. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Y.; Xing, Y.; Cao, B.; Yang, F.; Yang, T.; Ai, Z.; Wei, Y.; Jiang, J. The Interaction between Cancer Stem Cell Marker CD133 and Src Protein Promotes Focal Adhesion Kinase (FAK) Phosphorylation and Cell Migration. J. Biol. Chem. 2016, 291, 15540–15550. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, O.; Shimizu, K.; Semba, S.; Chiba, S.; Ku, Y.; Yokozaki, H.; Hori, Y. Hypoxia Induces Tumor Aggressiveness and the Expansion of CD133-Positive Cells in a Hypoxia-Inducible Factor-1α-Dependent Manner in Pancreatic Cancer Cells. Pathobiology 2011, 78, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Griguer, C.E.; Oliva, C.R.; Gobin, E.; Marcorelles, P.; Benos, D.J.; Lancaster, J.R.; Gillespie, G.Y. CD133 Is a Marker of Bioenergetic Stress in Human Glioma. PLoS ONE 2008, 3, e3655. [Google Scholar] [CrossRef]
- Iida, H.; Suzuki, M.; Goitsuka, R.; Ueno, H. Hypoxia Induces CD133 Expression in Human Lung Cancer Cells by Up-Regulation of OCT3/4 and SOX2. Int. J. Oncol. 2012, 40, 71–79. [Google Scholar] [CrossRef]
- You, H.; Ding, W.; Rountree, C.B. Epigenetic Regulation of Cancer Stem Cell Marker CD133 by Transforming Growth Factor-β. Hepatology 2010, 51, 1635–1644. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, N.; Luo, Y.; Peng, Z.; Zhang, T.; Li, S. MicroRNA-150 Inhibits Human CD133-Positive Liver Cancer Stem Cells through Negative Regulation of the Transcription Factor c-Myb. Int. J. Oncol. 2012, 40, 747–756. [Google Scholar] [CrossRef]
- Ma, S.; Tang, K.H.; Chan, Y.P.; Lee, T.K.; Kwan, P.S.; Castilho, A.; Ng, I.; Man, K.; Wong, N.; To, K.F.; et al. MiR-130b Promotes CD133+ Liver Tumor-Initiating Cell Growth and Self-Renewal via Tumor Protein 53-Induced Nuclear Protein 1. Cell Stem Cell 2010, 7, 694–707. [Google Scholar] [CrossRef]
- Gisina, A.; Novikova, S.; Kim, Y.; Sidorov, D.; Bykasov, S.; Volchenko, N.; Kaprin, A.; Zgoda, V.; Yarygin, K.; Lupatov, A. CEACAM5 Overexpression Is a Reliable Characteristic of CD133-Positive Colorectal Cancer Stem Cells. Cancer Biomark. 2021, 32, 85–98. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, Q.; Liu, Y.; Sun, L.; Han, P.; Wang, R.; Zhao, J.; Hu, S.; Zhao, X. Human CD133-Positive Hematopoietic Progenitor Cells Enhance the Malignancy of Breast Cancer Cells. BMC Cancer 2020, 20, 1158. [Google Scholar] [CrossRef] [PubMed]
- Kohga, K.; Tatsumi, T.; Takehara, T.; Tsunematsu, H.; Shimizu, S.; Yamamoto, M.; Sasakawa, A.; Miyagi, T.; Hayashi, N. Expression of CD133 Confers Malignant Potential by Regulating Metalloproteinases in Human Hepatocellular Carcinoma. J. Hepatol. 2010, 52, 872–879. [Google Scholar] [CrossRef]
- Ghazalpour, A.; Bennett, B.; Petyuk, V.A.; Orozco, L.; Hagopian, R.; Mungrue, I.N.; Farber, C.R.; Sinsheimer, J.; Kang, H.M.; Furlotte, N.; et al. Comparative Analysis of Proteome and Transcriptome Variation in Mouse. PLoS Genet. 2011, 7, e1001393. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.S. Genome-Wide Correlation between MRNA and Protein in a Single Cell. Angew. Chem. Int. Ed. 2011, 50, 583–585. [Google Scholar] [CrossRef] [PubMed]
- Poverennaya, E.V.; Ilgisonis, E.V.; Ponomarenko, E.A.; Kopylov, A.T.; Zgoda, V.G.; Radko, S.P.; Lisitsa, A.V.; Archakov, A.I. Why Are the Correlations between MRNA and Protein Levels so Low among the 275 Predicted Protein-Coding Genes on Human Chromosome 18? J. Proteome Res. 2017, 16, 4311–4318. [Google Scholar] [CrossRef]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde Dehydrogenase 1 Is a Marker for Normal and MalignantHuman Colonic Stem Cells (SC) and Tracks SC Overpopulationduring Colon Tumorigenesis. Cancer Res. 2010, 69, 3382–3389. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chan, K.W.; Lee, T.K.-W.; Tang, K.H.; Wo, J.Y.-H.; Zheng, B.-J.; Guan, X.-Y. Aldehyde Dehydrogenase Discriminates the CD133 Liver Cancer Stem Cell Populations. Mol. Cancer Res. 2008, 6, 1146–1153. [Google Scholar] [CrossRef]
- Dubreuil, V.; Marzesco, A.M.; Corbeil, D.; Huttner, W.B.; Wilsch-Bräuninger, M. Midbody and Primary Cilium of Neural Progenitors Release Extracellular Membrane Particles Enriched in the Stem Cell Marker Prominin-1. J. Cell Biol. 2007, 176, 483–495. [Google Scholar] [CrossRef]
- Bobinger, T.; Roeder, S.S.; Spruegel, M.I.; Froehlich, K.; Beuscher, V.D.; Hoelter, P.; Lücking, H.; Corbeil, D.; Huttner, H.B. Variation of Membrane Particle–Bound CD133 in Cerebrospinal Fluid of Patients with Subarachnoid and Intracerebral Hemorrhage. J. Neurosurg. 2021, 134, 600–607. [Google Scholar] [CrossRef]
- Bunch, H.; Lawney, B.P.; Lin, Y.F.; Asaithamby, A.; Murshid, A.; Wang, Y.E.; Chen, B.P.C.; Calderwood, S.K. Transcriptional Elongation Requires DNA Break-Induced Signalling. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Goodarzi, A.A.; Kurka, T.; Jeggo, P.A. KAP-1 Phosphorylation Regulates CHD3 Nucleosome Remodeling during the DNA Double-Strand Break Response. Nat. Struct. Mol. Biol. 2011, 18, 831–839. [Google Scholar] [CrossRef]
- Chen, L.; Muñoz-Antonia, T.; Cress, W.D. Trim28 Contributes to EMT via Regulation of E-Cadherin and N-Cadherin in Lung Cancer Cell Lines. PLoS ONE 2014, 9, e101040. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ivanov, A.; Chen, L.; Fredericks, W.J.; Seto, E.; Rauscher, F.J.; Chen, J. MDM2 Interaction with Nuclear Corepressor KAP1 Contributes to P53 Inactivation. EMBO J. 2005, 24, 3279–3290. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Kim, J.; Xu, Q.; Hu, G.; Kim, J.; Xu, Q.; Leng, Y.; Orkin, S.H.; Elledge, S.J. A Genome-Wide RNAi Screen Identifies a New Transcriptional Module Required for Self-Renewal. Genes Dev. 2009, 23, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Schwab, L.P.; Peacock, D.L.; Majumdar, D.; Ingels, J.F.; Jensen, L.C.; Smith, K.D.; Cushing, R.C.; Seagroves, T.N. Hypoxia-Inducible Factor 1α Promotes Primary Tumor Growth and Tumor-Initiating Cell Activity in Breast Cancer. Breast Cancer Res. 2012, 14, R6. [Google Scholar] [CrossRef] [PubMed]
- Soeda, A.; Park, M.; Lee, D.; Mintz, A.; Androutsellis-Theotokis, A.; McKay, R.D.; Engh, J.; Iwama, T.; Kunisada, T.; Kassam, A.B.; et al. Hypoxia Promotes Expansion of the CD133-Positive Glioma Stem Cells through Activation of HIF-1α. Oncogene 2009, 28, 3949–3959. [Google Scholar] [CrossRef]
- Won, C.; Kim, B.H.; Yi, E.H.; Choi, K.J.; Kim, E.K.; Jeong, J.M.; Lee, J.H.; Jang, J.J.; Yoon, J.H.; Jeong, W.I.; et al. Signal Transducer and Activator of Transcription 3-Mediated CD133 up-Regulation Contributes to Promotion of Hepatocellular Carcinoma. Hepatology 2015, 62, 1160–1173. [Google Scholar] [CrossRef]
- Lehnus, K.S.; Donovan, L.K.; Huang, X.; Zhao, N.; Warr, T.J.; Pilkington, G.J.; An, Q. CD133 Glycosylation Is Enhanced by Hypoxiain Cultured Glioma Stem Cells. Int. J. Oncol. 2013, 42, 1011–1017. [Google Scholar] [CrossRef]
- Park, E.K.; Lee, J.C.; Park, J.W.; Bang, S.Y.; Yi, S.A.; Kim, B.K.; Park, J.H.; Kwon, S.H.; You, J.S.; Nam, S.W.; et al. Transcriptional Repression of Cancer Stem Cell Marker CD133 by Tumor Suppressor P53. Cell Death Dis. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Lo Nigro, A.; Roelandt, P.; Catherine, M. Verfaillie Expression and Function of Pluripotency Genes in Adult Stem Cells. In Adult Stem Cells; Springer: Berlin/Heidelberg, Germany, 2011; pp. 95–112. ISBN 978-1-61779-001-0. [Google Scholar]
- Jeter, C.R.; Liu, B.; Liu, X.; Chen, X.; Liu, C.; Calhoun-davis, T.; Repass, J.; Zaehres, H.; Shen, J.J.; Tang, D.G.; et al. NANOG Promotes Cancer Stem Cell Characteristics and Prostate Cancer Resistance to Androgen Deprivation Collene. Oncogene 2012, 30, 3833–3845. [Google Scholar] [CrossRef] [Green Version]
- Van Niekerk, G.; Davids, L.M.; Hattingh, S.M.; Engelbrecht, A.M. Cancer Stem Cells: A Product of Clonal Evolution? Int. J. Cancer 2017, 140, 993–999. [Google Scholar] [CrossRef]
- Karagonlar, Z.F.; Akbari, S.; Karabicici, M.; Sahin, E.; Avci, S.T.; Ersoy, N.; Ates, K.E.; Balli, T.; Karacicek, B.; Kaplan, K.N.; et al. A Novel Function for KLF4 in Modulating the De-Differentiation of EpCAM-/CD133- NonStem Cells into EpCAM+/CD133+ Liver Cancer Stem Cells in HCC Cell Line HuH7. Cells 2020, 9, 1198. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Aigner, M.; Nakata, S.; Engel, F.; Schlotter, M.; Kloor, M.; Brand, K.; Schmitt, S.; Steinert, G.; Rahbari, N.; et al. A Gene Signature Distinguishing CD133hi from CD133− Colorectal Cancer Cells: Essential Role for EGR1 and Downstream Factors. Pathology 2011, 43, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Puca, F.; Colamaio, M.; Federico, A.; Gemei, M.; Tosti, N.; Bastos, A.U.; del Vecchio, L.; Pece, S.; Battista, S.; Fusco, A. HMGA1 Silencing Restores Normal Stem Cell Characteristics in Colon Cancer Stem Cells by Increasing P53 Levels. Oncotarget 2014, 5, 3234–3245. [Google Scholar] [CrossRef] [PubMed]
- Koşar, Z.; Erbaş, A. Can the Concentration of a Transcription Factor Affect Gene Expression? Front. Soft Matter 2022, 2, 1–6. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Chen, L.; Yabuuchi, A.; Eminli, S.; Takeuchi, A.; Lu, C.W.; Hochedlinger, K.; Daley, G.Q. Cross-Regulation of the Nanog and Cdx2 Promoters. Cell Res. 2009, 19, 1052–1061. [Google Scholar] [CrossRef]
- De Souza, A.T.; Dai, X.; Spencer, A.G.; Reppen, T.; Menzie, A.; Roesch, P.L.; He, Y.; Caguyong, M.J.; Bloomer, S.; Herweijer, H.; et al. Transcriptional and Phenotypic Comparisons of Ppara Knockout and SiRNA Knockdown Mice. Nucleic Acids Res. 2006, 34, 4486–4494. [Google Scholar] [CrossRef]
- Mazurek, S.; Oleksiewicz, U.; Czerwińska, P.; Wróblewska, J.; Klimczak, M.; Wiznerowicz, M. Disruption of RING and PHD Domains of TRIM28 Evokes Differentiation in Human IPSCs. Cells 2021, 10, 1933. [Google Scholar] [CrossRef]
- Jovčevska, I.; Zupanec, N.; Kočevar, N.; Cesselli, D.; Podergajs, N.; Stokin, C.L.; Myers, M.P.; Muyldermans, S.; Ghassabeh, G.H.; Motaln, H.; et al. TRIM28 and β-Actin Identified via Nanobody-Based Reverse Proteomics Approach as Possible Human Glioblastoma Biomarkers. PLoS ONE 2014, 9, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Porčnik, A.; Novak, M.; Breznik, B.; Majc, B.; Hrastar, B.; Šamec, N.; Zottel, A.; Jovčevska, I.; Vittori, M.; Rotter, A.; et al. Trim28 Selective Nanobody Reduces Glioblastoma Stem Cell Invasion. Molecules 2021, 26, 5141. [Google Scholar] [CrossRef] [PubMed]
- Czerwińska, P.; Shah, P.K.; Tomczak, K.; Mazurek, S.; Sozańska, B.; Biecek, P.; Wiznerowicz, M. TRIM28 Multi-Domain Protein Regulates Cancer Stem Cell Population in Breast Tumor Development. Oncotarget 2016, 8, 863–882. [Google Scholar] [CrossRef]
- Li, J.; Xi, Y.; Li, W.; McCarthy, R.L.; Stratton, S.A.; Zou, W.; Li, W.; Dent, S.Y.; Jain, A.K.; Barton, M.C. TRIM28 Interacts with EZH2 and SWI/SNF to Activate Genes That Promote Mammosphere Formation. Oncogene 2017, 36, 2991–3001. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.A. Melanoma Stem Cell-like Phenotype and Significant Suppression of Immune Response within a Tumor Are Regulated by Trim28 Protein. Cancers 2020, 12, 2998. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.X.; Cai, J.J.; Chen, L.C.; Yue, Q.; Gong, Y.; Yao, Y.; Mao, Y. TRIM28 as an Independent Prognostic Marker Plays Critical Roles in Glioma Progression. J. Neurooncol. 2016, 126, 19–26. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, J.; Li, Q.; Ma, H.; Xu, Z.; Gao, Y. KAP1 Is Overexpressed in Hepatocellular Carcinoma and Its Clinical Significance. Int. J. Clin. Oncol. 2016, 21, 927–933. [Google Scholar] [CrossRef]
- Cui, Y.; Yang, S.; Fu, X.; Feng, J.; Xu, S.; Ying, G. High Levels of KAP1 Expression Are Associated with Aggressive Clinical Features in Ovarian Cancer. Int. J. Mol. Sci. 2015, 16, 363–377. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, E.; Li, C.; Huang, L.; Xiao, L.; Cheng, L.; Huang, X.; Song, Y.; Xu, D. TRIM28, a New Molecular Marker Predicting Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Cancer Epidemiol. 2013, 37, 71–78. [Google Scholar] [CrossRef]
- Czerwińska, P.; Mazurek, S.; Wiznerowicz, M. The Complexity of TRIM28 Contribution to Cancer. J. Biomed. Sci. 2017, 24, 63. [Google Scholar] [CrossRef]
- Poverennaya, E.V.; Kopylov, A.T.; Ponomarenko, E.A.; Ilgisonis, E.V.; Zgoda, V.G.; Tikhonova, O.V.; Novikova, S.E.; Farafonova, T.E.; Kiseleva, Y.Y.; Radko, S.P.; et al. State of the Art of Chromosome 18-Centric HPP in 2016: Transcriptome and Proteome Profiling of Liver Tissue and HepG2 Cells. J. Proteome Res. 2016, 15, 4030–4038. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially Modified Protein Abundance Index (EmPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Kholodenko, I.; Kalinovsky, D.; Svirshchevskaya, E.; Doronin, I.; Konovalova, M.; Kibardin, A.; Shamanskaya, T.; Larin, S.; Deyev, S.; Kholodenko, R. Multimerization through Pegylation Improves Pharmacokinetic Properties of ScFv Fragments of GD2-Specific Antibodies. Molecules 2019, 24, 3835. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Disease | Cell Line | CD133-Positive Population (%) |
|---|---|---|
| Non-small cell lung carcinoma | A549 | 0 |
| H23 | 0 | |
| H358 | 0 | |
| H460 | 0 | |
| H1299 | 0 | |
| Colorectal carcinoma | Caco2 | 99.7 ± 0.3 |
| HT-29 | 69.9 ± 21.8 | |
| SW480 | 0 | |
| HCT116 | 0 | |
| SW-837 | 0 | |
| Glioblastoma | LN-229 | 0 |
| T98G | 0 | |
| U-87 MG | 0 | |
| Hepatocarcinoma | HUH7 | 93.4 ± 4.4 |
| Kidney carcinoma | A704 | 0 |
| Pancreas carcinoma | PANC-1 | 0 |
| Thyroid carcinoma | FTC-133 | 5.7 ± 4.1 |
| Breast carcinoma | MDA-MB-231 | 0 |
| Neuroblastoma | IMR-32 | 0 |
| Fibrosarcoma | HT-1080 | 0 |
| Osteosarcoma | U-2 OS | 0 |
| Key Regulators | TFs Rank | MRs Rank | Total Rank |
|---|---|---|---|
| TRIM28 | 5 | 5 | 10 |
| RELA | 5 | 3 | 8 |
| MYB | 4 | 4 | 8 |
| CREB1 | 4 | 2 | 6 |
| REST | 4 | 2 | 6 |
| TP53 | 4 | 2 | 6 |
| CEBPA | 3 | 3 | 6 |
| GABPB1 | 3 | 2 | 5 |
| NANOG | 2 | 2 | 4 |
| E2F1 | 2 | 2 | 4 |
| E2F3 | 2 | 2 | 4 |
| E2F4 | 2 | 2 | 4 |
| E2F7 | 2 | 2 | 4 |
| EGR1 | 2 | 2 | 4 |
| HIF1A | 2 | 2 | 4 |
| HMGA1 | 2 | 2 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.S.; Potashnikova, D.M.; Gisina, A.M.; Kholodenko, I.V.; Kopylov, A.T.; Tikhonova, O.V.; Kurbatov, L.K.; Saidova, A.A.; Tvorogova, A.V.; Kholodenko, R.V.; et al. TRIM28 Is a Novel Regulator of CD133 Expression Associated with Cancer Stem Cell Phenotype. Int. J. Mol. Sci. 2022, 23, 9874. https://doi.org/10.3390/ijms23179874
Kim YS, Potashnikova DM, Gisina AM, Kholodenko IV, Kopylov AT, Tikhonova OV, Kurbatov LK, Saidova AA, Tvorogova AV, Kholodenko RV, et al. TRIM28 Is a Novel Regulator of CD133 Expression Associated with Cancer Stem Cell Phenotype. International Journal of Molecular Sciences. 2022; 23(17):9874. https://doi.org/10.3390/ijms23179874
Chicago/Turabian StyleKim, Yan S., Daria M. Potashnikova, Alisa M. Gisina, Irina V. Kholodenko, Arthur T. Kopylov, Olga V. Tikhonova, Leonid K. Kurbatov, Aleena A. Saidova, Anna V. Tvorogova, Roman V. Kholodenko, and et al. 2022. "TRIM28 Is a Novel Regulator of CD133 Expression Associated with Cancer Stem Cell Phenotype" International Journal of Molecular Sciences 23, no. 17: 9874. https://doi.org/10.3390/ijms23179874
APA StyleKim, Y. S., Potashnikova, D. M., Gisina, A. M., Kholodenko, I. V., Kopylov, A. T., Tikhonova, O. V., Kurbatov, L. K., Saidova, A. A., Tvorogova, A. V., Kholodenko, R. V., Belousov, P. V., Vorobjev, I. A., Zgoda, V. G., Yarygin, K. N., & Lupatov, A. Y. (2022). TRIM28 Is a Novel Regulator of CD133 Expression Associated with Cancer Stem Cell Phenotype. International Journal of Molecular Sciences, 23(17), 9874. https://doi.org/10.3390/ijms23179874







