Genomic Dissection and Diurnal Expression Analysis Reveal the Essential Roles of the PRR Gene Family in Geographical Adaptation of Soybean
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of the PRR Gene Family in Soybean
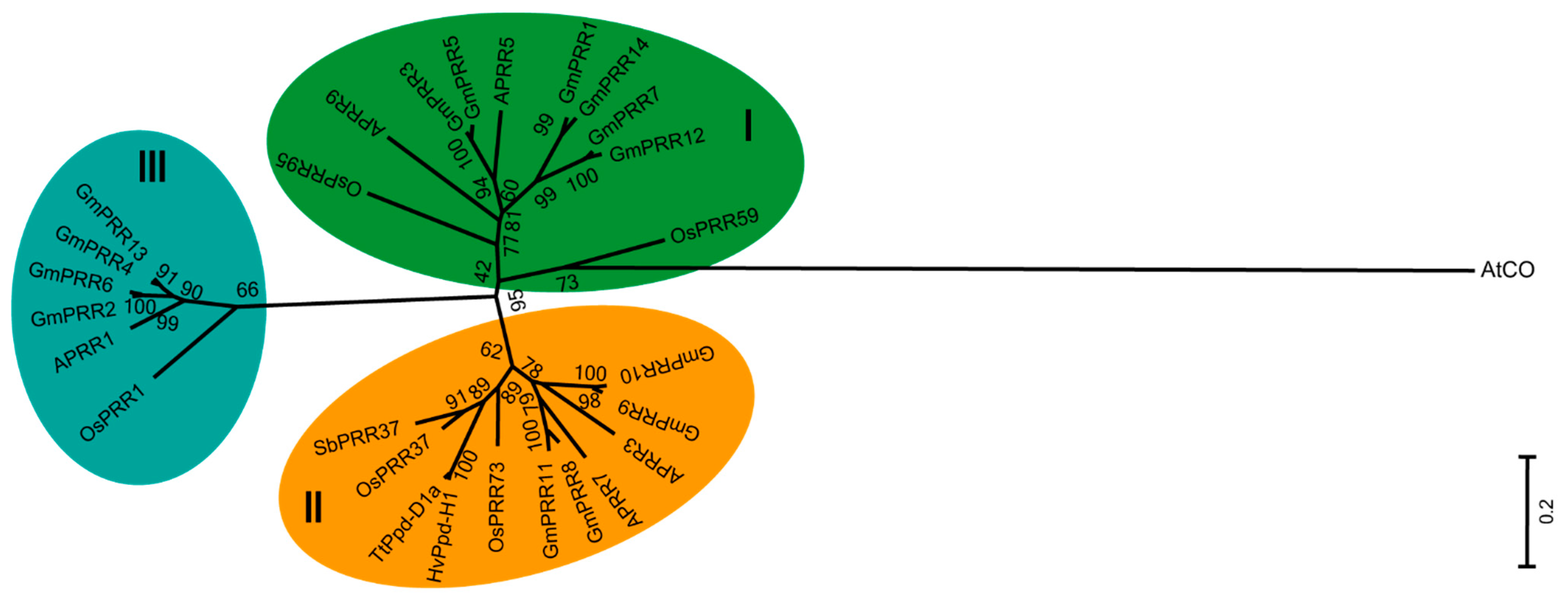
2.2. Evolutionary Analysis of the PRR Gene Family in Soybean
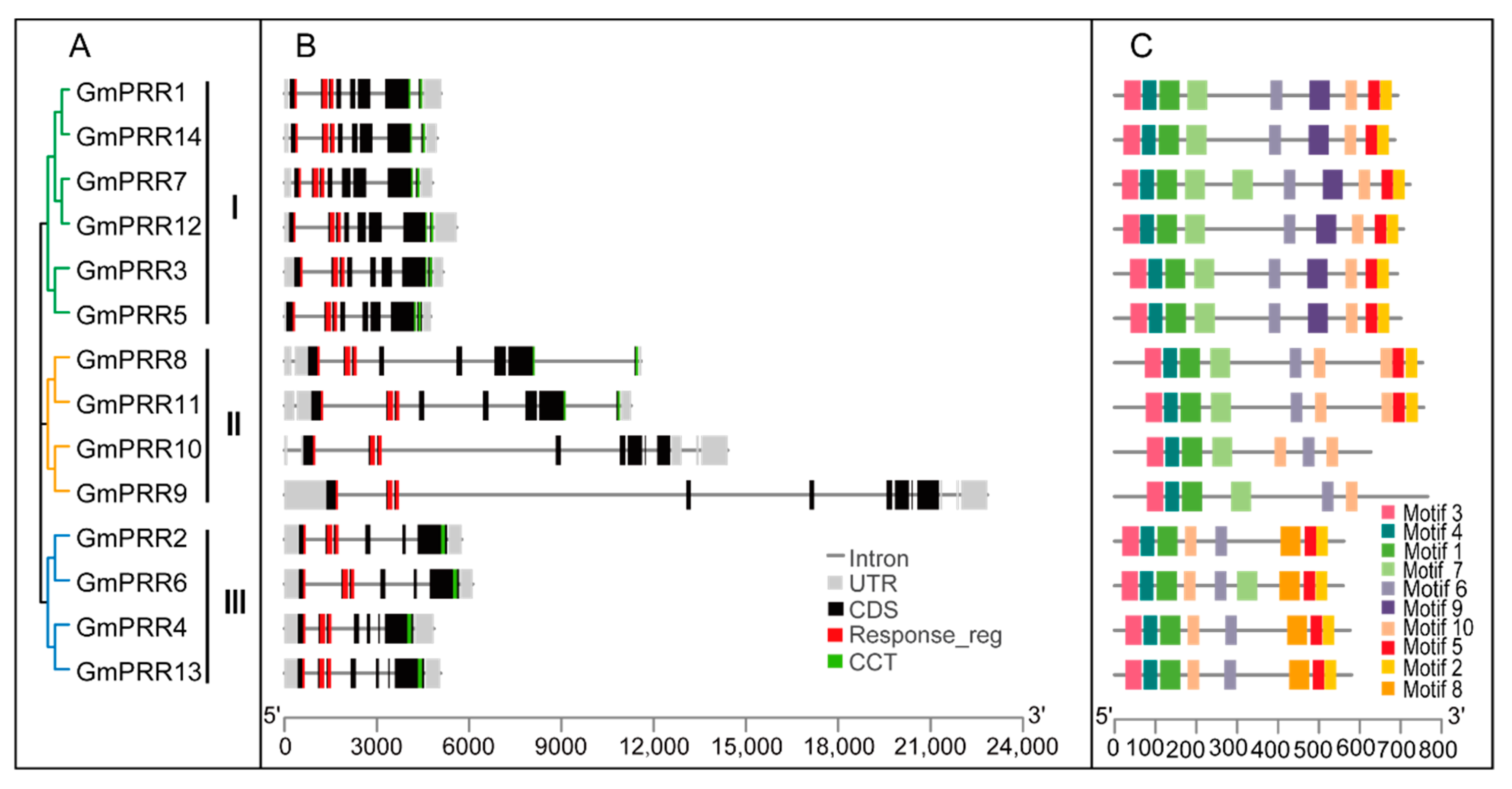
2.3. Gene Structure, Motif Composition and Promoter Characterization of Soybean PRR Gene Family
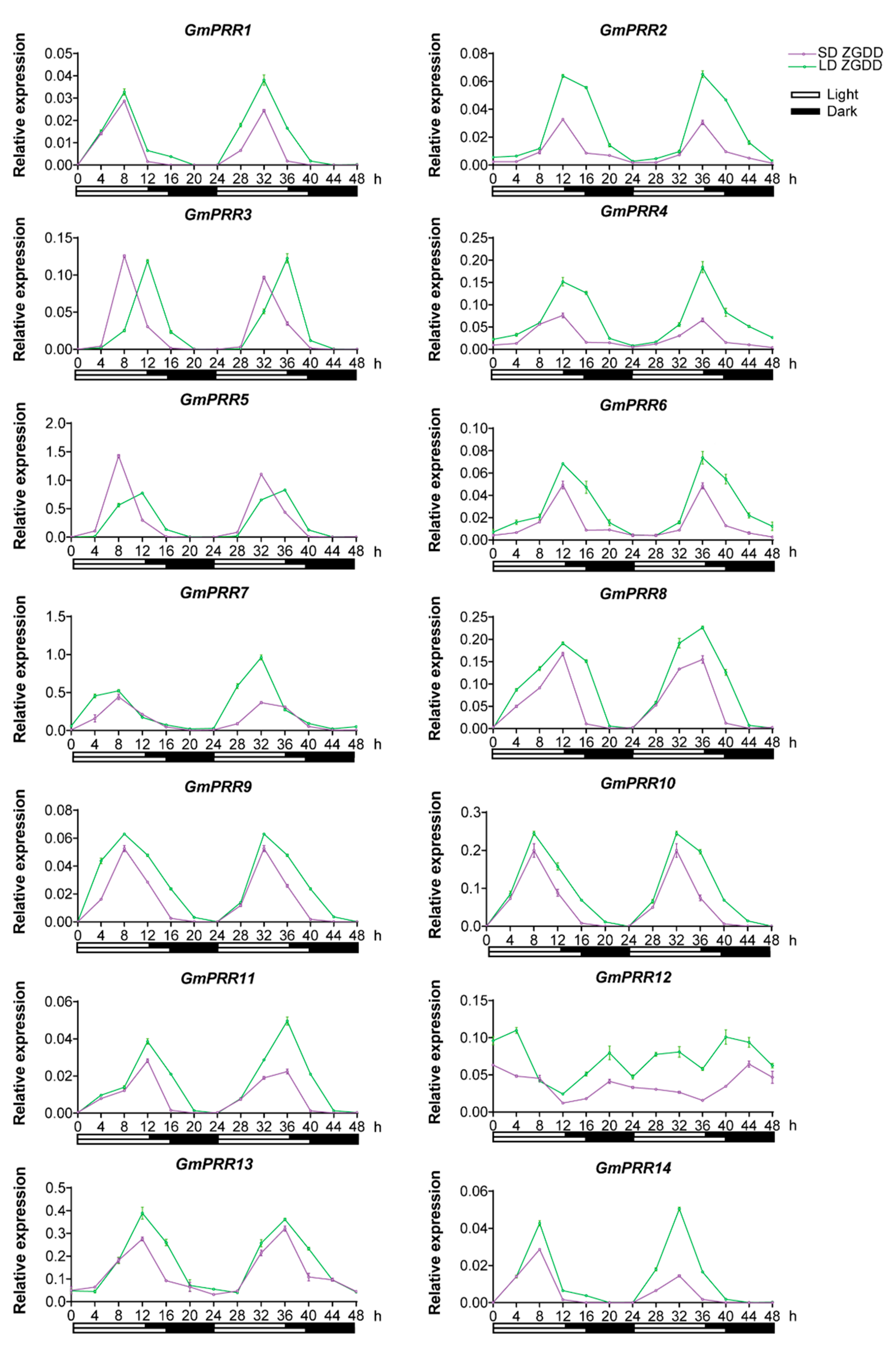
2.4. The Expression Pattern of GmPRR Genes
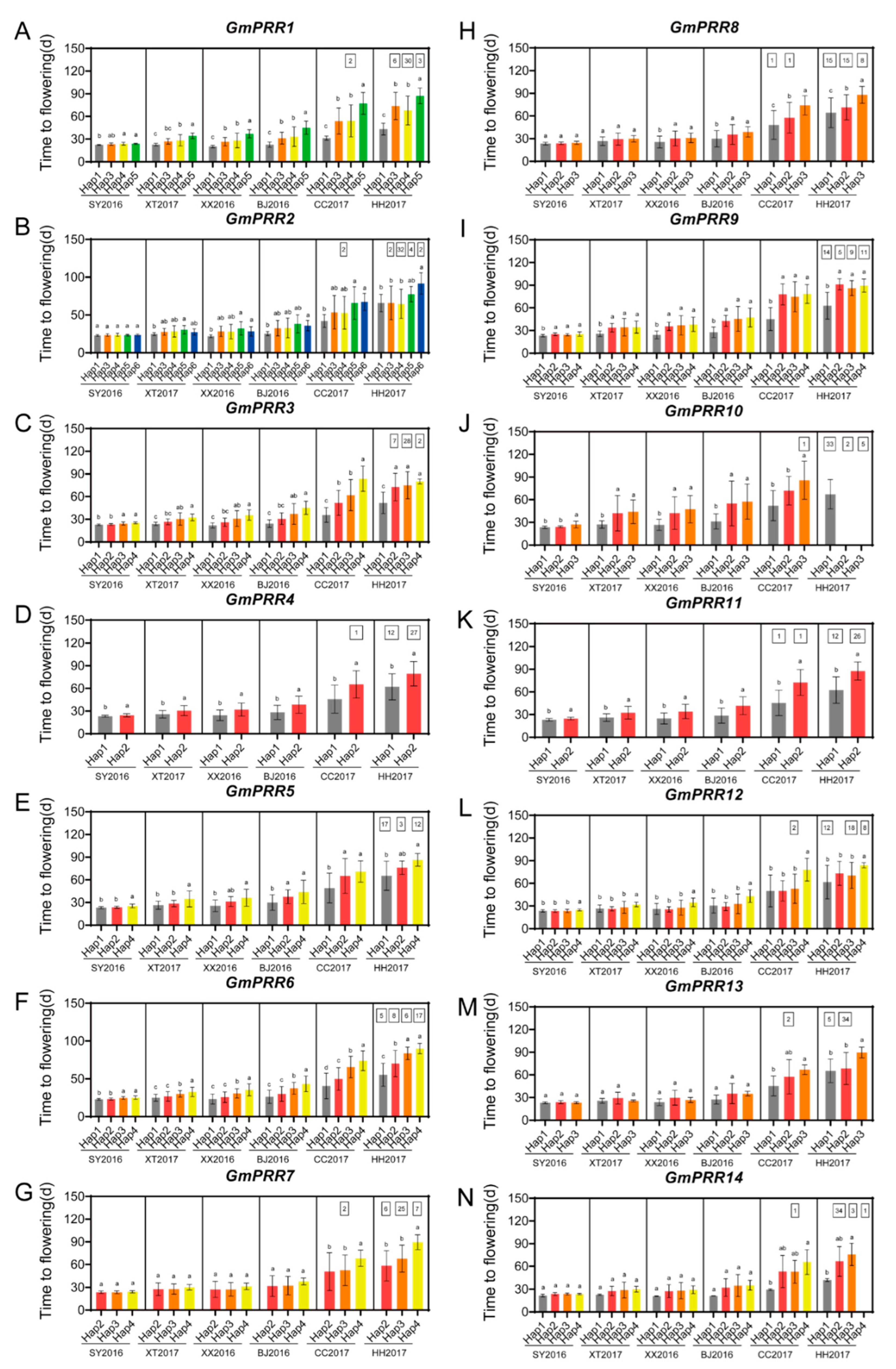
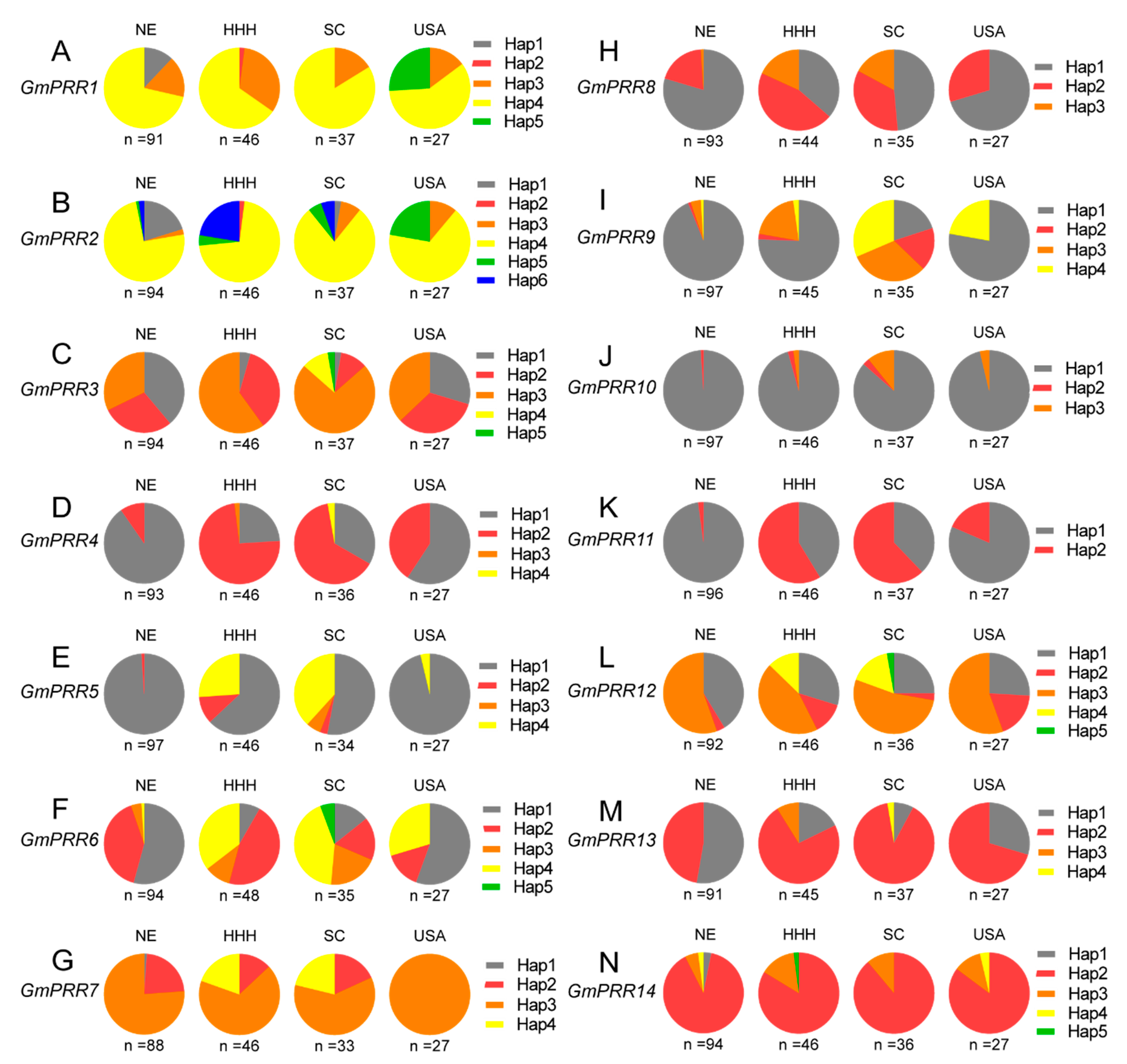
2.5. Haplotype Analysis of the 14 Soybean PRR Family Genes in Soybean Germplasm with Diverse Geographical Origins
2.6. Haplotype Combinations of GmPRRs in 207 Soybean Varieties
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Photoperiod Treatments and Multiple-Site Experiments
4.2. Identification, Phylogenetic and Bioinformatic Analysis of GmPRRs
4.3. Gene Structure, Conserved Motif and Promoter Sequence Analyses
4.4. RNA Extraction and Quantitative Real-Time PCR
4.5. Haplotype Analysis of Soybean PRR Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garner, W.W.; Allard, H.A. Effect of the Relative Length of Day and Night and Other Factors of the Environment on Growth and Reproduction in Plants. J. Agric. Res. 1920, 18, 553–606. [Google Scholar] [CrossRef]
- Garner, W.W.; Allard, H.A. Further Studies in Photoperiodism: The Response of the Plant to Relative Length of Day and Night. J. Agric. Res. 1923, 23, 871–920. [Google Scholar]
- Wang, J.; Wu, Y.; Wu, H.; Sun, S. Analysis on Photoperiod Ecotypes of Cultivated Soybean Originating from Different Locations of China. Acta. Agric. Sin. 1956, 7, 169–180. (In Chinese) [Google Scholar]
- Han, T.; Wang, J.; Fan, B.; Yao, W.; Yang, Q. Effects of Post-Flowering Daylength on Agronomic Characters of Soybean. Chin. J. Appl. Ecol. 1996, 7, 167–173, (In Chinese with English Abstract). [Google Scholar]
- Watanabe, S.; Harada, K.; Abe, J. Genetic and Molecular Bases of Photoperiod Responses of Flowering in Soybean. Breed Sci. 2012, 61, 531–543. [Google Scholar] [CrossRef]
- Cao, D.; Takeshima, R.; Zhao, C.; Liu, B.; Jun, A.; Kong, F. Molecular Mechanisms of Flowering under Long Days and Stem Growth Habit in Soybean. J. Exp. Bot. 2017, 68, 1873–1884. [Google Scholar] [CrossRef]
- Fu, M.; Wang, Y.; Ren, H.; Du, W.; Wang, D.; Bao, R.; Yang, X.; Tian, Z.; Fu, L.; Cheng, Y.; et al. Genetic Dynamics of Earlier Maturity Group Emergence in South-to-North Extension of Northeast China Soybeans. Theor. Appl. Genet. 2020, 133, 1839–1857. [Google Scholar] [CrossRef]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional Cloning and Characterization Reveal the Molecular Basis for Soybean Maturity Locus E1 that Regulates Photoperiodic Flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A Map-based Cloning Strategy Employing a Residual Heterozygous Line Reveals that the GIGANTEA Gene Is Involved in Soybean Maturity and Flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Ishimoto, M.; Anai, T.; et al. Map-based Cloning of the Gene Associated with the Soybean Maturity Locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic Redundancy in Soybean Photoresponses Associated with Duplication of the Phytochrome a Gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional Diversification of Flowering Locus T Homologs in Soybean: GmFT1a and GmFT2a/5a Have Opposite Roles in Controlling Flowering and Maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Bonato, E.R.; Vello, N.A. E6, a Dominant Gene Conditioning Early Flowering and Maturity in Soybeans. Genet. Mol. Biol. 1999, 22, 229–232. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Jia, Z.; Cao, D.; Jiang, B.; Wu, C.; Hou, W.; Liu, Y.; Fei, Z.; Zhao, D.; Han, T. GmFT2a, a Soybean Homolog of FLOWERING LOCUS T, Is Involved in Flowering Transition and Maintenance. PLoS ONE 2011, 6, e29238. [Google Scholar] [CrossRef]
- Kong, F.; Nan, H.; Cao, F.; Wang, J.; Yuan, S. A New Dominant Gene E9 Conditions Early Flowering and Maturity in Soybean. Crop Sci. 2014, 54, 2529–2535. [Google Scholar] [CrossRef]
- Kong, F.; Liu, B.; Xia, Z.; Sato, S.; Kim, B.M.; Watanabe, S.; Yamada, T.; Tabata, S.; Kanazawa, A.; Harada, K.; et al. Two Coordinately Regulated Homologs of FLOWERING LOCUS T Are Involved in the Control of Photoperiodic Flowering in Soybean. Plant Physiol. 2010, 154, 1220–1231. [Google Scholar] [CrossRef]
- Zhao, C.; Takeshima, R.; Zhu, J.; Xu, M.; Sato, M.; Watanabe, S.; Kanazawa, A.; Liu, B.; Kong, F.; Yamada, T.; et al. A Recessive Allele for Delayed Flowering at the Soybean Maturity Locus E9 Is a Leaky Allele of FT2a, a FLOWERING LOCUS T Ortholog. BMC Plant Biol. 2016, 16, 20. [Google Scholar] [CrossRef]
- Samanfar, B.; Molnar, S.J.; Charette, M.; Schoenrock, A.; Dehne, F.; Golshani, A.; Belzile, F.; Cober, E.R. Mapping and Identification of a Potential Candidate Gene for a Novel Maturity Locus, E10, in Soybean. Theor. Appl. Genet. 2017, 130, 377–390. [Google Scholar] [CrossRef]
- Takeshima, R.; Hayashi, T.; Zhu, J.; Zhao, C.; Xu, M.; Yamaguchi, N.; Sayama, T.; Ishimoto, M.; Kong, L.; Shi, X.; et al. A Soybean Quantitative Trait Locus that Promotes Flowering under Long Days Is Identified as FT5a, a FLOWERING LOCUS T Ortholog. J. Exp. Bot. 2016, 67, 5247–5258. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural Variation at the Soybean J Locus Improves Adaptation to the Tropics and Enhances Yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, N.; Jiang, B.; Li, M.; Wang, H.; Jiang, Z.; Pan, H.; Xia, Q.; Ma, Q.; Han, T.; et al. A Single Nucleotide Deletion in J Encoding GmELF3 Confers Long Juvenility and Is Associated with Adaption of Tropic Soybean. Mol. Plant. 2017, 10, 656–658. [Google Scholar] [CrossRef]
- Bu, T.; Lu, S.; Wang, K.; Dong, L.; Li, S.; Xie, Q.; Xu, X.; Cheng, Q.; Chen, L.; Fang, C.; et al. A Critical Role of the Soybean Evening Complex in the Control of Photoperiod Sensitivity and Adaptation. Proc. Natl. Acad. Sci. USA 2021, 23, e2010241118. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Y.; Wu, H.; Hu, B.; Zhai, H.; Yang, J.; Xia, Z. Positional Cloning of the Flowering Time QTL qFT12-1 Reveals the Link between the Clock Related PRR Homolog with Photoperiodic Response in Soybeans. Front. Plant Sci. 2019, 10, 1303. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L.; et al. Natural Variation and CRISPR/Cas9-Mediated Mutation in GmPRR37 Affect Photoperiodic Flowering and Contribute to Regional Adaptation of Soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef]
- Li, C.; Li, Y.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant. 2020, 13, 745–759. [Google Scholar] [CrossRef]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise Selection on Homeologous PRR Genes Controlling Flowering and Maturity during Soybean Domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef]
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic Basis and Adaptation Trajectory of Soybean from Its Temperate Origin to Tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar] [CrossRef]
- Cheng, Q.; Dong, L.; Su, T.; Li, T.; Gan, Z.; Nan, H.; Lu, S.; Fang, C.; Kong, L.; Li, H.; et al. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmLHY Genes Alters Plant Height and Internode Length in Soybean. BMC Plant Biol. 2019, 19, 562. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light and Temperature Entrainable Circadian Clock in Soybean Development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef]
- Creux, N.; Harmer, S. Circadian Rhythms in Plants. Cold Spring Harb. Perspect. Biol. 2019, 11, a034611. [Google Scholar] [CrossRef]
- Farré, E.M.; Liu, T. The PRR Family of Transcriptional Regulators Reflects the Complexity and Evolution of Plant Circadian Clocks. Curr. Opin. Plant Biol. 2013, 16, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.E.; Park, C.M. Thermal Adaptation and Plasticity of the Plant Circadian Clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef] [PubMed]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian Waves of Expression of the APRR1/TOC1 Family of Pseudo-Response Regulators in Arabidopsis Thaliana: Insight into the Plant Circadian Clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Matsushika, A.; Makino, S.; Kojima, M.; Yamashino, T.; Mizuno, T. The APRR1/TOC1 Quintet Implicated in Circadian Rhythms of Arabidopsis Thaliana: II. Characterization with CCA1-Overexpressing Plants. Plant Cell Physiol. 2002, 43, 118–122. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis Clock-Associated Pseudo-response Regulators PRR9, PRR7 and PRR5 Coordinately and Positively Regulate Flowering Time through the Canonical CONSTANS-Dependent Photoperiodic Pathway. Plant Cell Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef]
- Koo, B.H.; Yoo, S.C.; Park, J.W.; Kwon, C.T.; Lee, B.D.; An, G.; Zhang, Z.; Li, J.; Li, Z.; Paek, N.C. Natural Variation in OsPRR37 Regulates Heading Date and Contributes to Rice Cultivation at a Wide Range of Latitudes. Mol. Plant 2013, 6, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Liu, H.; Zhou, X.; Li, Q.; Zhang, J.; Lu, L.; Liu, T.; Liu, H.; Zhang, C.; Zhang, Z.; et al. Natural Variation in Ghd7.1 Plays an Important Role in Grain Yield and Adaptation in Rice. Cell Res. 2013, 23, 969–971. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jin, M.; Zheng, X.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to Heading 7, a Major Quantitative Locus Determining Photoperiod Sensitivity and Regional Adaptation in Rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef]
- Beales, J.; Turner, A.; Griffiths, S.; Snape, J.W.; Laurie, D.A. A Pseudo-Response Regulator is Mis-expressed in the Photoperiod Insensitive Ppd-D1a Mutant of Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 115, 721–733. [Google Scholar] [CrossRef]
- Turner, A.; Beales, J.; Faure, S.; Dunford, R.P.; Laurie, D.A. The Pseudo-Response Regulator Ppd-H1 Provides Adaptation to Photoperiod in Barley. Science 2005, 310, 1031–1034. [Google Scholar] [CrossRef]
- Murphy, R.L.; Klein, R.R.; Morishige, D.T.; Brady, J.A.; Rooney, W.L.; Miller, F.R.; Dugas, D.V.; Klein, P.E.; Mullet, J.E. Coincident Light and Clock Regulation of Pseudo-Response Regulator Protein 37 (PRR37) Controls Photoperiodic Flowering in Sorghum. Proc. Natl. Acad. Sci. USA 2011, 108, 16469–16474. [Google Scholar] [CrossRef]
- Yang, J.; Huang, X. A New High-Quality Genome Sequence in Soybean. Sci. China Life Sci. 2018, 61, 1604–1605. [Google Scholar] [CrossRef]
- Shen, Y.; Du, H.; Liu, Y.; Ni, L.; Wang, Z.; Liang, C.; Tian, Z. Update Soybean Zhonghuang 13 Genome to a Golden Reference. Sci. China Life Sci. 2019, 62, 1257–1260. [Google Scholar] [CrossRef]
- Wang, L.; Sun, J.; Wang, L.; Li, B.; Zhao, R. Breeding and Application of Widely Adapted, High-yield and High-protein Soybean Variety Zhonghuang 13. Soybean Sci. 2019, 38, 1–6, (In Chinese with English Abstract). [Google Scholar]
- Bendix, C.; Marshall, C.; Harmon, F. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis Clock Gene TOC1, an Autoregulatory Response Regulator Homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef]
- Salomé, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 Are Partially Redundant Genes Essential for the Temperature Responsiveness of the Arabidopsis Circadian Clock. Plant Cell 2005, 17, 791–803. [Google Scholar] [CrossRef]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The Evolutionarily Conserved OsPRR Quintet: Rice Pseudo-Response Regulators Implicated in Circadian Rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef]
- Shu, Y.; Yu, D.; Wang, D.; Guo, D.; Guo, C. Genome-wide Survey and Expression Analysis of the MADS-Box Gene Family in Soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis Circadian Clock Protein, TOC1, Is a DNA-binding Transcription Factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional Repressor PRR5 Directly Regulates Clock-Output Pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef]
- Michael, T.P.; Salomé, P.A.; Yu, H.J.; Spencer, T.R.; Sharp, E.L.; McPeek, M.A.; Alonso, J.M.; Ecker, J.R.; McClung, C.R. Enhanced Fitness Conferred by Naturally Occurring Variation in the Circadian Clock. Science 2003, 302, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.A.; Wijnen, C.L.; Srinivasan, A.; Ryngajllo, M.; Ofner, I.; Lin, T.; Ranjan, A.; West, D.; Maloof, J.N.; Sinha, N.R.; et al. Domestication Selected for Deceleration of the Circadian Clock in Cultivated Tomato. Nat. Genet. 2016, 48, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Greenham, K.; Lou, P.; Puzey, J.R.; Kumar, G.; Arnevik, C.; Farid, H.; Willis, J.H.; McClung, C.R. Geographic Variation of Plant Circadian Clock Function in Natural and Agricultural Settings. J. Biol. Rhythm. 2017, 32, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Muranaka, T.; Ito, S.; Kudoh, H.; Oyama, T. Circadian-Period Variation Underlies the Local Adaptation of Photoperiodism in the Short-Day Plant Lemna aequinoctialis. iScience 2022, 25, 104634. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Pozzoli, U.; Cagliani, R.; Tresoldi, C.; Menozzi, G.; Riva, S.; Guerini, F.R.; Comi, G.P.; Bolognesi, E.; Bresolin, N.; et al. Genetic Adaptation of the Human Circadian Clock to Day-Length Latitudinal Variations and Relevance for Affective Disorders. Genome Biol. 2014, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Lemay, M.A.; Russello, M.A. Latitudinal Cline in Allele Length Provides Evidence for Selection in a Circadian Rhythm Gene. Biol. J. Linn. Soc. 2014, 111, 869–877. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, T.; Wang, L.; Jiang, B.; Zhen, C.; Yuan, S.; Hou, W.; Wu, C.; Han, T.; Sun, S. A Combined Linkage and GWAS Analysis Identifies QTLs Linked to Soybean Seed Protein and Oil Content. Int. J. Mol. Sci. 2019, 20, 5915. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Qu, M.; Wang, L.; Sun, H.; Jiang, B.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; et al. Soybean Adaption to High-latitude Regions Is Associated with Natural Variations of GmFT2b, An Ortholog of FLOWERING LOCUS T. Plant Cell Environ. 2020, 43, 934–944. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamure, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Fehr, W.; Caviness, C.; Burmood, D.; Pennington, J. Stage of Development Descriptions for Soybeans, Glycine Max (L.) Merrill. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Wang, L.; Zhang, L.; Wu, T.; Sun, B.; Zhang, J.; Sapey, E.; Yuan, S.; Jiang, B.; Chen, F.; et al. Genomic Dissection and Diurnal Expression Analysis Reveal the Essential Roles of the PRR Gene Family in Geographical Adaptation of Soybean. Int. J. Mol. Sci. 2022, 23, 9970. https://doi.org/10.3390/ijms23179970
Wang P, Wang L, Zhang L, Wu T, Sun B, Zhang J, Sapey E, Yuan S, Jiang B, Chen F, et al. Genomic Dissection and Diurnal Expression Analysis Reveal the Essential Roles of the PRR Gene Family in Geographical Adaptation of Soybean. International Journal of Molecular Sciences. 2022; 23(17):9970. https://doi.org/10.3390/ijms23179970
Chicago/Turabian StyleWang, Peiguo, Liwei Wang, Lixin Zhang, Tingting Wu, Baiquan Sun, Junquan Zhang, Enoch Sapey, Shan Yuan, Bingjun Jiang, Fulu Chen, and et al. 2022. "Genomic Dissection and Diurnal Expression Analysis Reveal the Essential Roles of the PRR Gene Family in Geographical Adaptation of Soybean" International Journal of Molecular Sciences 23, no. 17: 9970. https://doi.org/10.3390/ijms23179970




