Transcriptome Analysis Reveals Critical Genes and Pathways in Carbon Metabolism and Ribosome Biogenesis in Poplar Fertilized with Glutamine
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Data Analysis and Quality Evaluation of the cDNA Libraries
2.2. DEGs Obtained under Gln Treatment
2.3. DEGs Classification and GO Functional Annotation
2.4. DEGs Involved in Ribosome
2.5. Sugar and Carbon Metabolism in Response to Gln Treatment
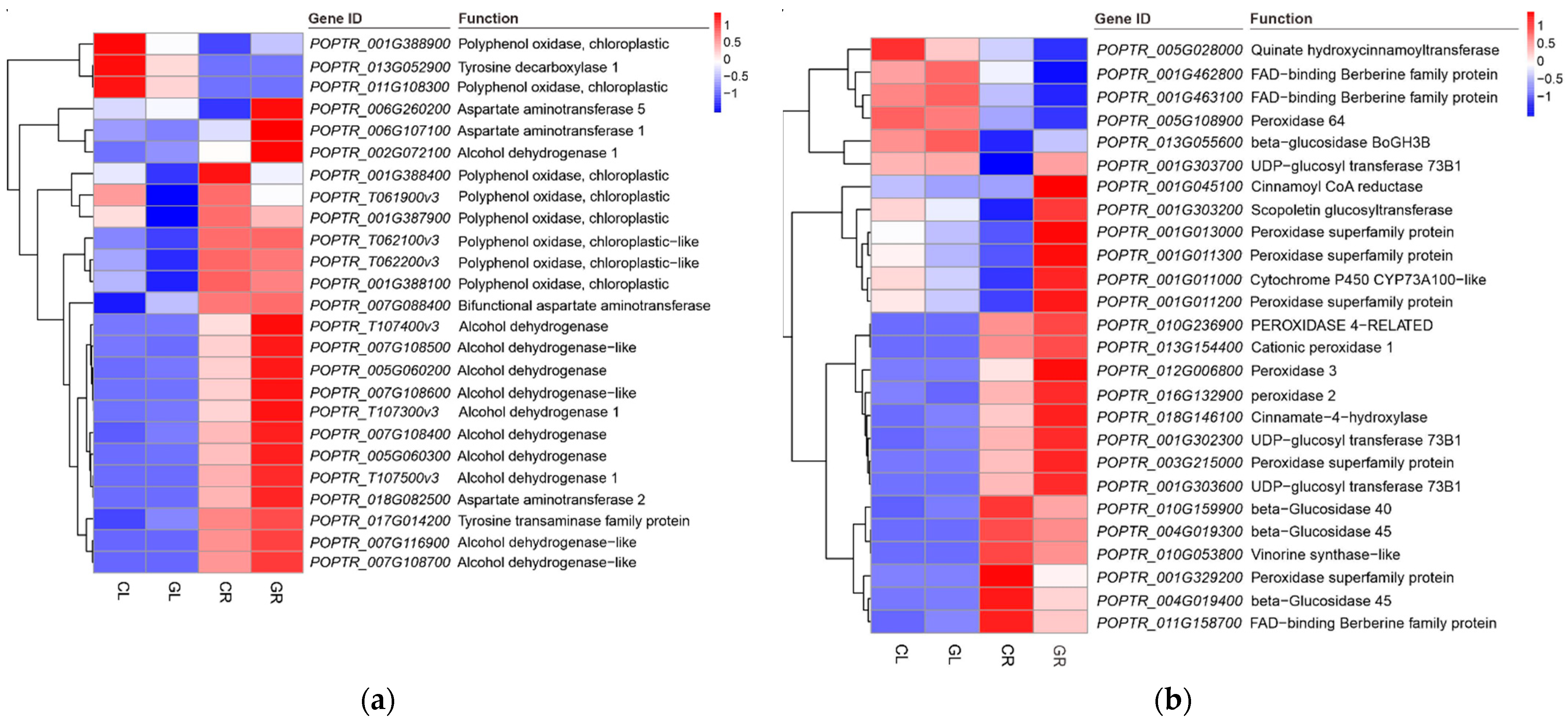
2.6. Secondary Metabolism in Response to Gln Treatment
2.7. Plant Hormone Signal Transduction
2.8. Validation of RNA-Seq Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Gln Treatments
4.2. Total RNA Extraction and cDNA Library Construction
4.3. Reads Mapping to the Reference Genome
4.4. Differential Expression Gene Analysis
4.5. GO and KEGG Enrichment
4.6. Quantitative Real-Time Validation of DEGs
Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Camargo, E.L.O.; Nascimento, L.C.; Soler, M.; Salazar, M.M.; Lepikson-Neto, J.; Marques, W.L.; Alves, A.; Teixeira, P.J.P.L.; Mieczkowski, P.; Carazzolle, M.F.; et al. Contrasting Nitrogen Fertilization Treatments Impact Xylem Gene Expression and Secondary Cell Wall Lignification in Eucalyptus. BMC Plant Biol. 2014, 14, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, Allocation and Signaling of Nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Dalmau, J.; Berbel, J.; Ordóñez-Fernández, R. Nitrogen Fertilization. A Review of the Risks Associated with the Inefficiency of Its Use and Policy Responses. Sustainability 2021, 13, 5625. [Google Scholar] [CrossRef]
- Cañas, R.; de la Torre, F.; Pascual, M.; Avila, C.; Cánovas, F. Nitrogen Economy and Nitrogen Environmental Interactions in Conifers. Agronomy 2016, 6, 26. [Google Scholar] [CrossRef]
- Taranet, P.; Harper, S.; Kirchhof, G.; Fujinuma, R.; Menzies, N. Growth and Yield Response of Glasshouse- and Field-Grown Sweetpotato to Nitrogen Supply. Nutr. Cycl. Agroecosyst. 2017, 108, 309–321. [Google Scholar] [CrossRef]
- Jørgensen, H.; Hecke, J.; Zhang, H.; Malik, P.L.; Felby, C.; Schjoerring, J.K. Wheat as a Dual Crop for Biorefining: Straw Quality Parameters and Their Interactions with Nitrogen Supply in Modern Elite Cultivars. GCB Bioenergy 2019, 11, 400–415. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, B.; Yang, X.; Zhang, Z.; Li, B. The Sucrose Synthase Gene Family in Populus: Structure, Expression, and Evolution. Tree Genet. Genomes 2011, 7, 443–456. [Google Scholar] [CrossRef]
- Yan, J.; Xu, C.; Wei, H. Response of Carbon and Nitrogen Allocation in Two Hybrid Poplar Clones to Soil Nitrogen in Northern China. J. Food Agric. Environ. 2013, 11, 1050–1054. [Google Scholar]
- Plavcová, L.; Hacke, U.G.; Almeida-Rodriguez, A.M.; Li, E.; Douglas, C.J. Gene Expression Patterns Underlying Changes in Xylem Structure and Function in Response to Increased Nitrogen Availability in Hybrid Poplar: Effects of Nitrogen Availability on Xylogenesis. Plant Cell Environ. 2013, 36, 186–199. [Google Scholar] [CrossRef]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray Analysis of the Nitrate Response in Arabidopsis Roots and Shoots Reveals over 1,000 Rapidly Responding Genes and New Linkages to Glucose, Trehalose-6-Phosphate, Iron, and Sulfate Metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, T.; Huang, Y.; Wang, J.; Chen, Y.; Kistler, H.C.; Ma, Z.; Yin, Y. A Fungal ABC Transporter FgAtm1 Regulates Iron Homeostasis via the Transcription Factor Cascade FgAreA-HapX. PLoS Pathog 2019, 15, e1007791. [Google Scholar] [CrossRef] [PubMed]
- Franklin, O.; Cambui, C.A.; Gruffman, L.; Palmroth, S.; Oren, R.; Näsholm, T. The Carbon Bonus of Organic Nitrogen Enhances Nitrogen Use Efficiency of Plants. Plant Cell Environ. 2017, 40, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, J.; Svennerstam, H.; Moritz, T.; Egertsdotter, U.; Ganeteg, U. Nitrogen Uptake and Assimilation in Proliferating Embryogenic Cultures of Norway Spruce-Investigating the Specific Role of Glutamine. PLoS ONE 2017, 12, e0181785. [Google Scholar] [CrossRef] [PubMed]
- Nasholm, T. Qualitative and Quantitative Changes in Plant Nitrogen Acquisition Induced by Anthropogenic Nitrogen Deposition. New Phytol. 1998, 139, 87–90. [Google Scholar] [CrossRef]
- Svennerstam, H.; Ganeteg, U.; Näsholm, T. Root Uptake of Cationic Amino Acids by Arabidopsis Depends on Functional Expression of Amino Acid Permease 5. New Phytol. 2008, 180, 620–630. [Google Scholar] [CrossRef]
- Couturier, J.; de Faÿ, E.; Fitz, M.; Wipf, D.; Blaudez, D.; Chalot, M. PtAAP11, a High Affinity Amino Acid Transporter Specifically Expressed in Differentiating Xylem Cells of Poplar. J. Exp. Bot. 2010, 61, 1671–1682. [Google Scholar] [CrossRef]
- Yao, X.; Nie, J.; Bai, R.; Sui, X. Amino Acid Transporters in Plants: Identification and Function. Plants 2020, 9, 972. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Foster, J.; Chen, J.; Voll, L.M.; Weber, A.P.M.; Tegeder, M. AAP1 Transports Uncharged Amino Acids into Roots of Arabidopsis. Plant J. 2007, 50, 305–319. [Google Scholar] [CrossRef]
- Näsholm, T.; Kielland, K.; Ganeteg, U. Uptake of Organic Nitrogen by Plants. New Phytol. 2009, 182, 31–48. [Google Scholar] [CrossRef]
- Dolev, N.; Katz, Z.; Ludmer, Z.; Ullmann, A.; Brauner, N.; Goikhman, R. Natural Amino Acids as Potential Chelators for Soil Remediation. Environ. Res. 2020, 183, 109140. [Google Scholar] [CrossRef]
- Popko, M.; Michalak, I.; Wilk, R.; Gramza, M.; Chojnacka, K.; Górecki, H. Effect of the New Plant Growth Biostimulants Based on Amino Acids on Yield and Grain Quality of Winter Wheat. Molecules 2018, 23, 470. [Google Scholar] [CrossRef] [Green Version]
- Hassan, M.U.; Islam, M.M.; Wang, R.; Guo, J.; Luo, H.; Chen, F.; Li, X. Glutamine Application Promotes Nitrogen and Biomass Accumulation in the Shoot of Seedlings of the Maize Hybrid ZD958. Planta 2020, 251, 66. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Chen, Y.; Ma, C.; Qin, J.; Nguyen, T.H.N.; Liu, D.; Gan, H.; Ding, S.; Luo, Z.-B. Phenylalanine as a Nitrogen Source Induces Root Growth and Nitrogen-Use Efficiency in Populus × Canescens. Tree Physiol. 2018, 38, 66–82. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to Clinic: Glutamine Metabolism to Cancer Therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Xu, M.; Wang, S.; Wu, L.; Sun, S.; Su, T. Effects of Exogenous L-Glutamine as a Sole Nitrogen Source on Physiological Characteristics and Nitrogen Use Efficiency of Poplar. Plant Physiol. Biochem. 2022, 172, 1–13. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in Plants: Metabolism, Regulation, and Signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Kan, C.C.; Chung, T.Y.; Juo, Y.A.; Hsieh, M.H. Glutamine Rapidly Induces the Expression of Key Transcription Factor Genes Involved in Nitrogen and Stress Responses in Rice Roots. BMC Genom. 2015, 16, 731. [Google Scholar] [CrossRef]
- Amin, A.A.; Gharib, F.A.E.; El-Awadi, M.; Rashad, E.-S.M. Physiological Response of Onion Plants to Foliar Application of Putrescine and Glutamine. Sci. Hortic. 2011, 129, 353–360. [Google Scholar] [CrossRef]
- Souri, M.K.; Hatamian, M. Aminochelates in Plant Nutrition: A Review. J. Plant Nutr. 2019, 42, 67–78. [Google Scholar] [CrossRef]
- Aghaye Noroozlo, Y.; Souri, M.K.; Delshad, M. Effects of Soil Application of Amino Acids, Ammonium, and Nitrate on Nutrient Accumulation and Growth Characteristics of Sweet Basil. Commun. Soil Sci. Plant Anal. 2019, 50, 2864–2872. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine Reliance in Cell Metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, A.; Selvaraj, N.; Ganapathi, A.; Kasthurirengan, S.; Anbazhagan, V.; Manickavasagam, M. Glutamine: A Suitable Nitrogen Source for Enhanced Shoot Multiplication in Cucumis Sativus L. Biol. Plant. 2004, 48, 125–128. [Google Scholar] [CrossRef]
- Hamasaki, R.M.; Purgatto, E.; Mercier, H. Glutamine enhances competence for organogenesis in pineapple leaves cultivated in vitro. Braz. J. Plant Physiol. 2005, 17, 383–389. [Google Scholar] [CrossRef]
- Husin, N.; Jalil, M.; Othman, R.Y.; Khalid, N. Enhancement of Regeneration Efficiency in Banana (Musa Acuminata cv. Berangan) by Using Proline and Glutamine. Sci. Hortic. 2014, 168, 33–37. [Google Scholar] [CrossRef]
- Forsum, O.; Svennerstam, H.; Ganeteg, U.; Näsholm, T. Capacities and Constraints of Amino Acid Utilization in Arabidopsis. New Phytol. 2008, 179, 1058–1069. [Google Scholar] [CrossRef]
- Zhang, Y.; Mulpuri, S.; Liu, A. Exogenous Glutamine Increases Lipid Accumulation in Developing Seeds of Castor Bean (Ricinus Communis L.) Cultured in Vitro. Arch. Biol. Sci. 2015, 67, 1137–1149. [Google Scholar] [CrossRef]
- Sarropoulou, V.; Chatzissavvidis, C.; Dimassi-Theriou, K.; Therios, I. Effect of Asparagine, Cysteine, Citrulline, and Glutamine on in Vitro Rooting and Biochemical Constituents in Cherry Rootstocks. Biol. Plant. 2016, 60, 1–12. [Google Scholar] [CrossRef]
- Salih, K.J.; Duncan, O.; Li, L.; O’Leary, B.; Fenske, R.; Trösch, J.; Millar, A.H. Impact of Oxidative Stress on the Function, Abundance, and Turnover of the Arabidopsis 80S Cytosolic Ribosome. Plant J. 2020, 103, 128–139. [Google Scholar] [CrossRef]
- Rudra, D.; Warner, J.R. What Better Measure than Ribosome Synthesis? Genes Dev. 2004, 18, 2431–2436. [Google Scholar] [CrossRef]
- Mager, W.H. Control of Ribosomal Protein Gene Expression. BBA-Gene Struct. Expr. 1988, 949, 1–15. [Google Scholar] [CrossRef]
- Schmollinger, S.; Mühlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, S.; Anderson, P. Reprogramming mRNA Translation during Stress. Curr. Opin. Cell Biol. 2008, 20, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Hummel, M.; Cordewener, J.H.G.; de Groot, J.C.M.; Smeekens, S.; America, A.H.P.; Hanson, J. Dynamic Protein Composition of Arabidopsis Thaliana Cytosolic Ribosomes in Response to Sucrose Feeding as Revealed by Label Free MSE Proteomics. Proteomics 2012, 12, 1024–1038. [Google Scholar] [CrossRef]
- Li, Y.; Xu, S.; Wang, Z.; He, L.; Xu, K.; Wang, G. Glucose Triggers Stomatal Closure Mediated by GIN1, GIN2, PYR/RCAR, ROS, NO, CDPK6 and SLAC1 in Arabidposis. J. Exp. Bot. 2018, 69, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yang, Y.; Liu, C.; Sun, Y.; Zhang, T.; Hou, M.; Huang, S.; Yuan, H. The Evolutionary History of the Sucrose Synthase Gene Family in Higher Plants. BMC Plant Biol. 2019, 19, 566. [Google Scholar] [CrossRef]
- Dong, X.; Duan, S.; Wang, H.; Jin, H. Plastid Ribosomal Protein LPE2 Is Involved in Photosynthesis and the Response to C/N Balance in Arabidopsis Thaliana. J. Integr. Plant Biol. 2020, 62, 1418–1432. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, N.; Keller, F. Allocation of Raffinose Family Oligosaccharides to Transport and Storage Pools in Ajuga Reptans: The Roles of Two Distinct Galactinol Synthases. Plant J. 2000, 21, 249–258. [Google Scholar] [CrossRef]
- Peterbauer, T.; Richter, A. Biochemistry and Physiology of Raffinose Family Oligosaccharides and Galactosyl Cyclitols in Seeds. Seed Sci. Res. 2001, 11, 185–197. [Google Scholar]
- Taji, T.; Ohsumi, C.; Iuchi, S.; Seki, M.; Kasuga, M.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Important Roles of Drought- and Cold-Inducible Genes for Galactinol Synthase in Stress Tolerance in Arabidopsis Thaliana. Plant J. 2002, 29, 417–426. [Google Scholar] [CrossRef]
- Vinson, C.C.; Mota, A.P.Z.; Porto, B.N.; Oliveira, T.N.; Sampaio, I.; Lacerda, A.L.; Danchin, E.G.J.; Guimaraes, P.M.; Williams, T.C.R.; Brasileiro, A.C.M. Characterization of Raffinose Metabolism Genes Uncovers a Wild Arachis Galactinol Synthase Conferring Tolerance to Abiotic Stresses. Sci. Rep. 2020, 10, 15258. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Ishizaki, T.; Valencia, M.; Ogawa, S.; Dedicova, B.; Ogata, T.; Yoshiwara, K.; Maruyama, K.; Kusano, M.; Saito, K.; et al. Overexpression of an Arabidopsis Thaliana Galactinol Synthase Gene Improves Drought Tolerance in Transgenic Rice and Increased Grain Yield in the Field. Plant Biotechnol. J. 2017, 15, 1465–1477. [Google Scholar] [CrossRef]
- Sun, Z.; Qi, X.; Wang, Z.; Li, P.; Wu, C.; Zhang, H.; Zhao, Y. Overexpression of TsGOLS2, a Galactinol Synthase, in Arabidopsis Thaliana Enhances Tolerance to High Salinity and Osmotic Stresses. Plant Physiol. Biochem. 2013, 69, 82–89. [Google Scholar] [CrossRef]
- Xu, J.-J.; Fang, X.; Li, C.-Y.; Yang, L.; Chen, X.Y. General and Specialized Tyrosine Metabolism Pathways in Plants. Abiotech 2020, 1, 97–105. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The Polyphenol Oxidase Gene Family in Land Plants: Lineage-Specific Duplication and Expansion. BMC Genom. 2012, 13. [Google Scholar] [CrossRef]
- Chen, X.; Yang, B.; Huang, W.; Wang, T.; Li, Y.; Zhong, Z.; Yang, L.; Li, S.; Tian, J. Comparative Proteomic Analysis Reveals Elevated Capacity for Photosynthesis in Polyphenol Oxidase Expression-Silenced Clematis Terniflora DC. Leaves. Int. J. Mol. Sci. 2018, 19, 3897. [Google Scholar] [CrossRef]
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M.; et al. Novel Roles for the Polyphenol Oxidase Enzyme in Secondary Metabolism and the Regulation of Cell Death in Walnut. Plant Physiol. 2014, 164, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Steffens, J.C. Overexpression of Polyphenol Oxidase in Transgenic Tomato Plants Results in Enhanced Bacterial Disease Resistance. Planta 2002, 215, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Thipyapong, P.; Hunt, M.D.; Steffens, J.C. Antisense Downregulation of Polyphenol Oxidase Results in Enhanced Disease Susceptibility. Planta 2004, 220, 105–117. [Google Scholar] [CrossRef]
- Wang, J.; Constabel, C.P. Polyphenol Oxidase Overexpression in Transgenic Populus Enhances Resistance to Herbivory by Forest Tent Caterpillar (Malacosoma Disstria). Planta 2004, 220, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Boeckx, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol Oxidase in Leaves: Is There Any Significance to the Chloroplastic Localization? J. Exp. Bot. 2015, 66, 3571–3579. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Assmann, S.M. The Glycolytic Enzyme, Phosphoglycerate Mutase, Has Critical Roles in Stomatal Movement, Vegetative Growth, and Pollen Production in Arabidopsis Thaliana. J. Exp. Bot. 2011, 62, 5179–5189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosa-Téllez, S.; Anoman, A.D.; Flores-Tornero, M.; Toujani, W.; Alseek, S.; Fernie, A.R.; Nebauer, S.G.; Muñoz-Bertomeu, J.; Segura, J.; Ros, R. Phosphoglycerate Kinases Are Co-Regulated to Adjust Metabolism and to Optimize Growth. Plant Physiol. 2018, 176, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Baier, M.; Dietz, K.-J. Chloroplasts as Source and Target of Cellular Redox Regulation: A Discussion on Chloroplast Redox Signals in the Context of Plant Physiology. J. Exp. Bot. 2005, 56, 1449–1462. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Min, J.; Cao, D.; Zhai, G.; Zhou, H.; Li, N.; Li, M. Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations. Forests 2019, 10, 449. [Google Scholar] [CrossRef] [Green Version]
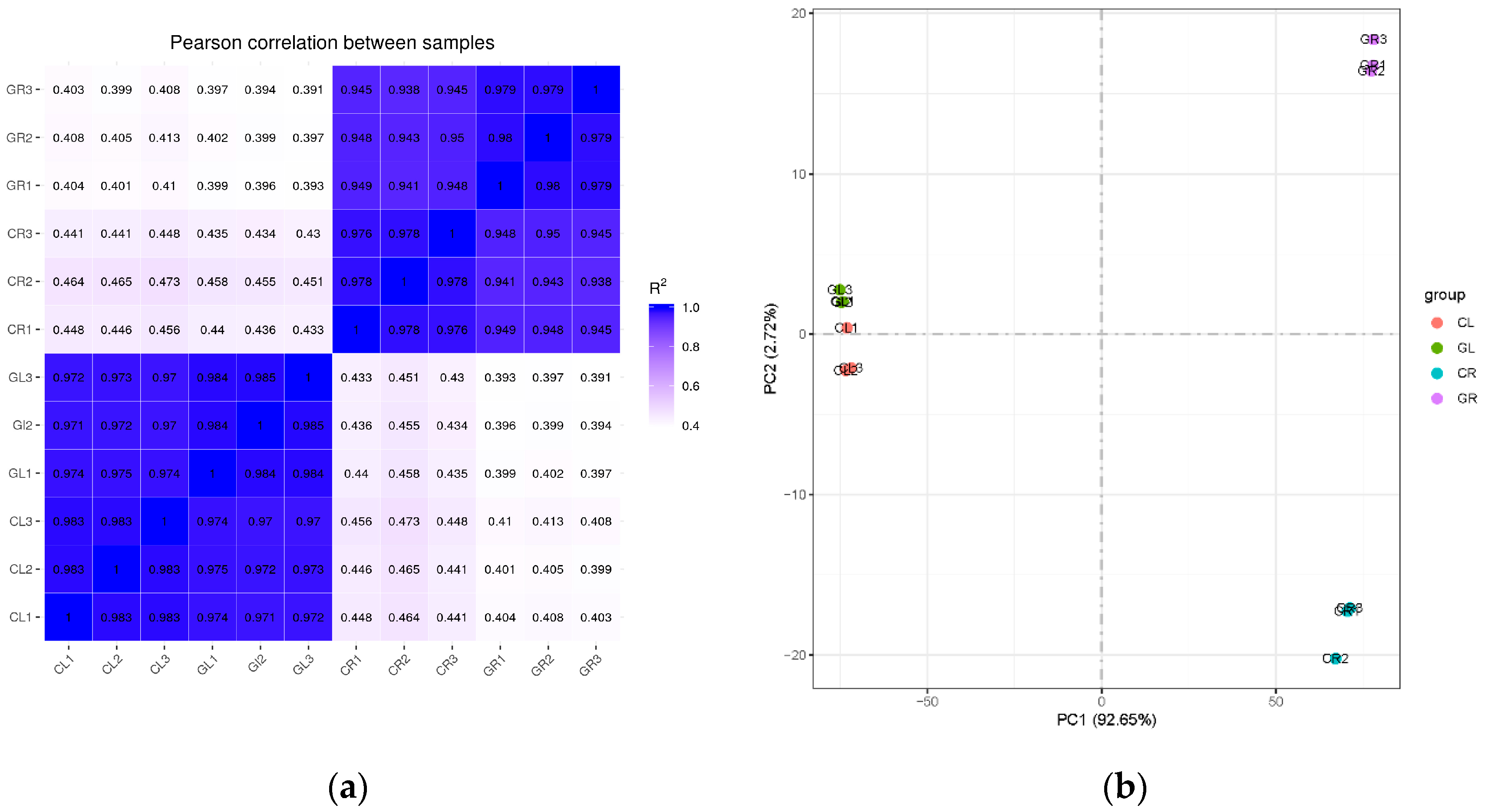
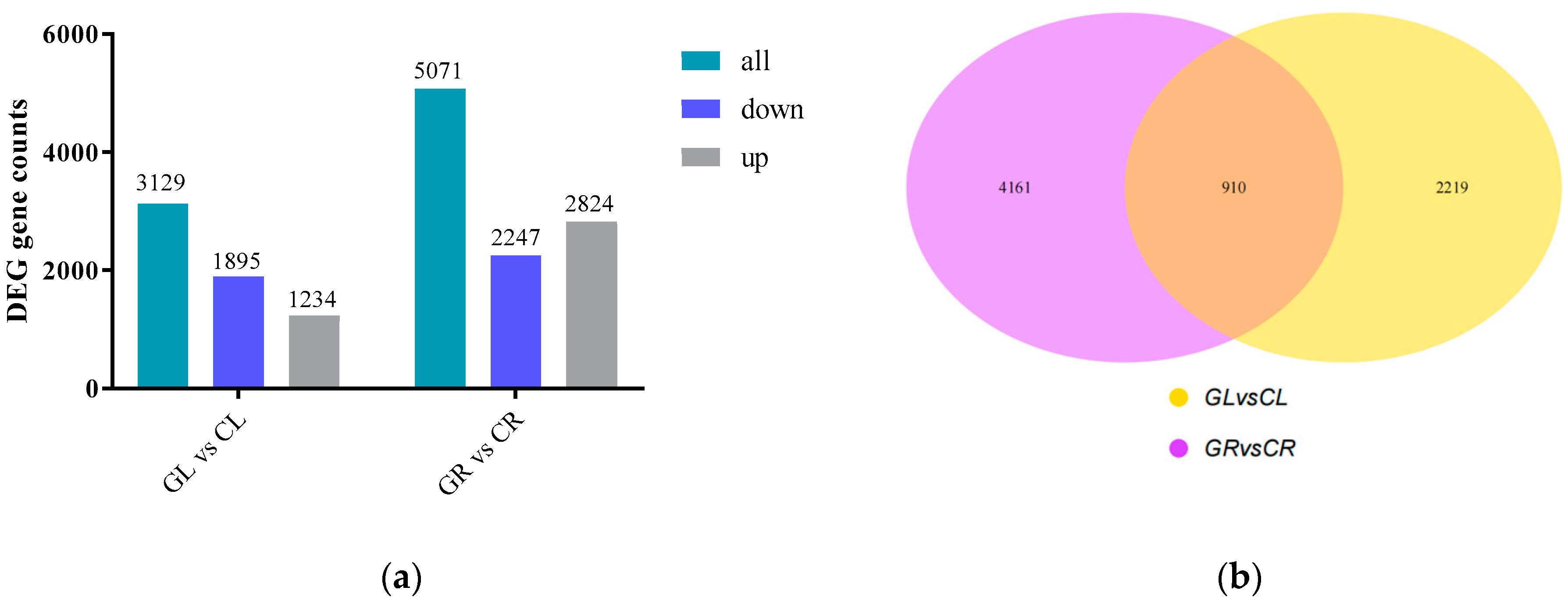

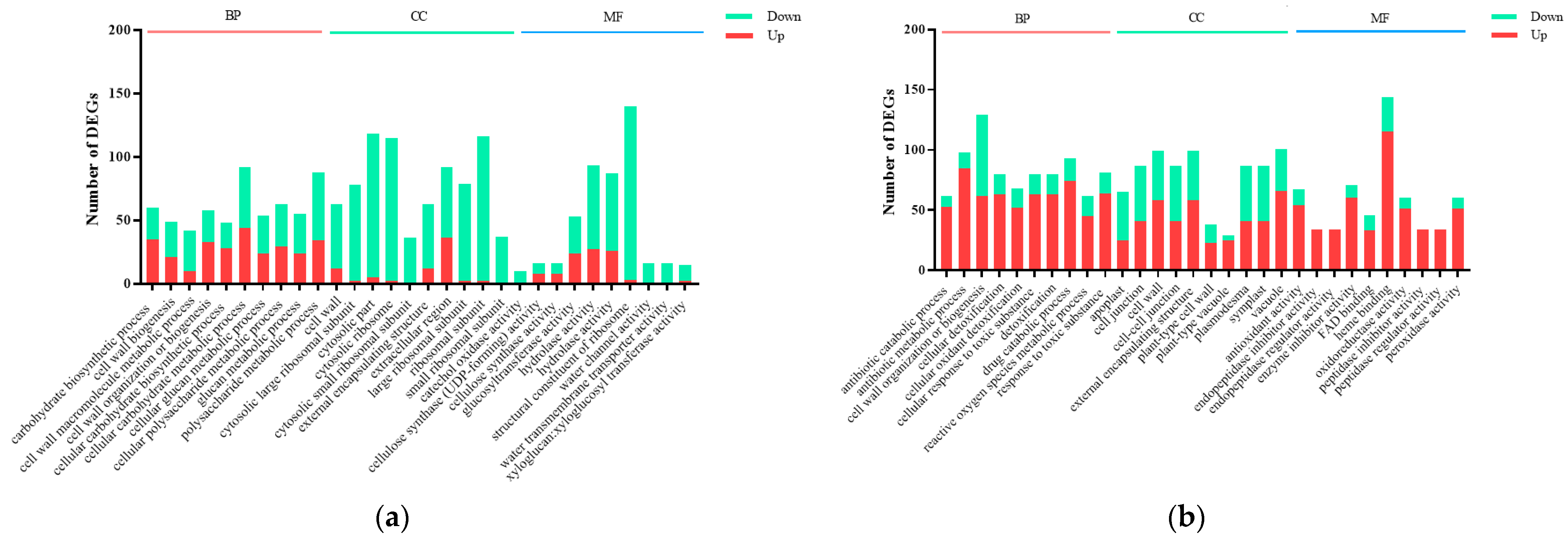


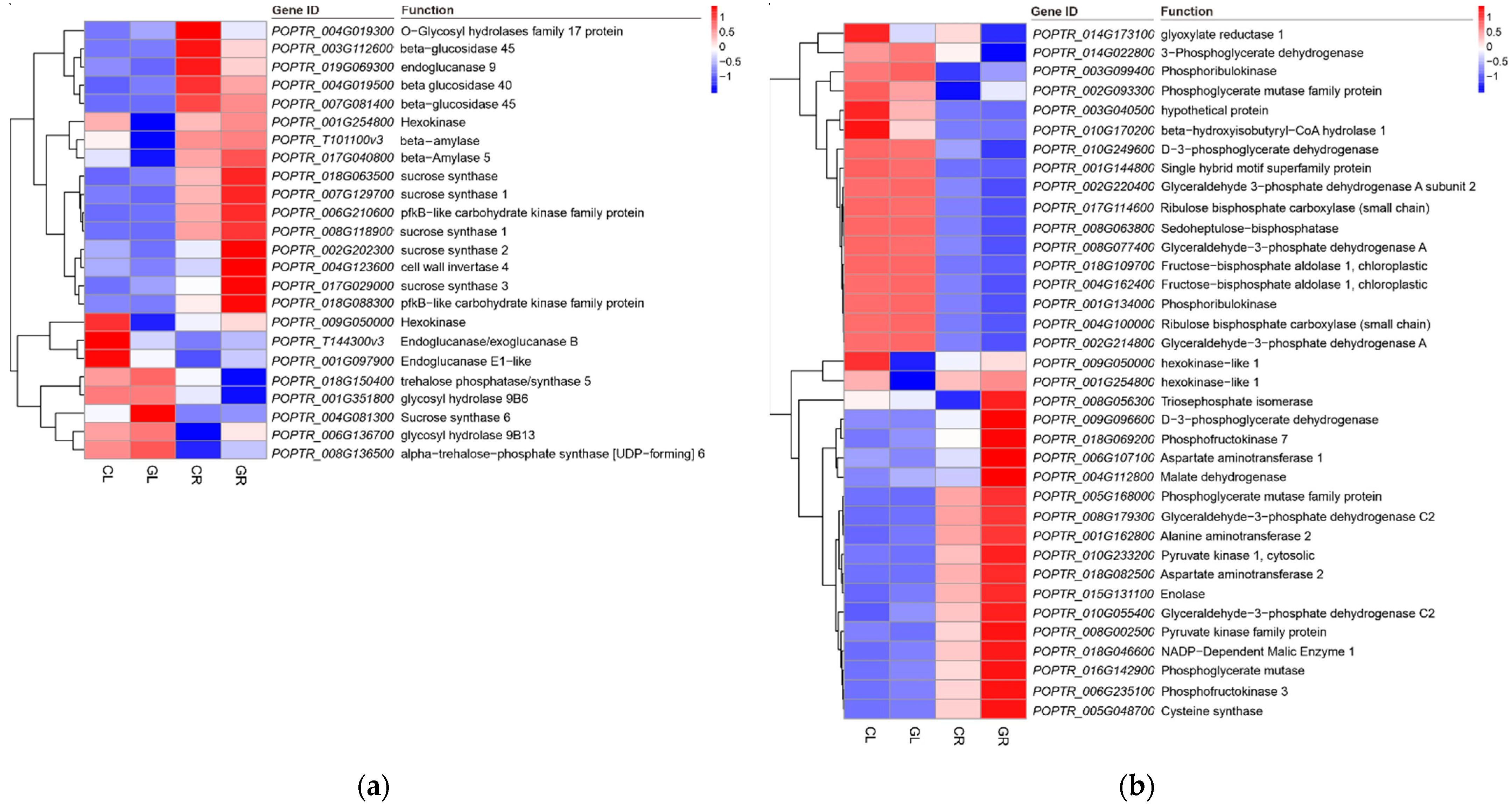




| Sample | Raw Reads | Clean Reads | Clean Bases | Error Rate (%) | Q30 (%) | GC (%) | Total Mapping (%) | Unique Mapping (%) |
|---|---|---|---|---|---|---|---|---|
| CL | 44,993,545 | 43,578,975 | 6.53G | 0.03 | 93.75 | 44.29 | 87.48 | 85.31 |
| GL | 44,483,713 | 43,239,443 | 6.48G | 0.03 | 93.83 | 44.18 | 88.02 | 85.77 |
| CR | 43,680,084 | 41,972,709 | 6.29G | 0.03 | 93.71 | 43.82 | 85.51 | 83.11 |
| GR | 43,732,617 | 41,306,527 | 6.16G | 0.03 | 93.43 | 44.20 | 74.47 | 72.35 |
| Gene | Number of DEGs in CL vs. GL | Number of DEGs in CR vs. GR | ||
|---|---|---|---|---|
| Upregulated | Sum | Upregulated | Sum | |
| sucrose synthase | 2 | 2 | 3 | 3 |
| probable sucrose phosphate synthase | 2 | 2 | 1 | 1 |
| starch synthase | 2 | 3 | 0 | 1 |
| granule-bound starch synthase | 2 | 3 | - | - |
| probable fructokinase | - | - | 3 | 3 |
| alpha-1,4 glucan phosphorylase | 1 | 1 | 2 | 2 |
| 1,4-alpha-glucan-branching enzyme | 2 | 2 | - | - |
| 4-alpha-glucanotransferase | - | - | 1 | 2 |
| endoglucanase | 1 | 5 | 2 | 5 |
| beta-amylase | 1 | 2 | 1 | 1 |
| ectonucleotide pyrophosphatase | - | - | 1 | 1 |
| hexokinase | 1 | 3 | 1 | 2 |
| beta-glucosidase | 0 | 3 | 3 | 7 |
| probable trehalose phosphate phosphatase | 2 | 4 | 5 | 5 |
| trehalose phosphate synthase | 1 | 3 | 2 | 3 |
| alpha-amylase | 1 | 1 | 2 | 2 |
| phosphoglucomutase | 1 | 1 | 1 | 1 |
| acid beta-fructofuranosidase | 0 | 2 | 0 | 1 |
| glucose-6-phosphate isomerase | 1 | 1 | 1 | 1 |
| glucan endo-1,3-beta-glucosidase | 0 | 1 | 0 | 3 |
| beta-fructofuranosidase | - | - | 1 | 1 |
| Gene | Number of Upregulated | Number of Downregulated | Sum |
|---|---|---|---|
| galactinol synthase | 6 | 1 | 7 |
| raffinose synthase | 3 | 2 | 5 |
| beta-galactosidase | 1 | 1 | 2 |
| hexokinase | 1 | 2 | 3 |
| alpha-galactosidase | 0 | 2 | 2 |
| stachyose synthase | 1 | 0 | 1 |
| phosphoglucomutase | 1 | 0 | 1 |
| aldose 1-epimerase | 0 | 1 | 1 |
| acid beta-fructofuranosidase | 0 | 2 | 2 |
| Gene | Number of Upregulated | Number of Dowregulated | Sum |
|---|---|---|---|
| polyphenol oxidase | 1 | 9 | 10 |
| primary amine oxidase isoform | 2 | 0 | 2 |
| tyrosine aminotransferase | 2 | 0 | 2 |
| bifunctional aspartate aminotransferase | 2 | 0 | 2 |
| tyrosine decarboxylase | 0 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, M.; Xu, M.; Su, T.; Wang, S.; Wu, L.; Feng, J.; Ding, C. Transcriptome Analysis Reveals Critical Genes and Pathways in Carbon Metabolism and Ribosome Biogenesis in Poplar Fertilized with Glutamine. Int. J. Mol. Sci. 2022, 23, 9998. https://doi.org/10.3390/ijms23179998
Han M, Xu M, Su T, Wang S, Wu L, Feng J, Ding C. Transcriptome Analysis Reveals Critical Genes and Pathways in Carbon Metabolism and Ribosome Biogenesis in Poplar Fertilized with Glutamine. International Journal of Molecular Sciences. 2022; 23(17):9998. https://doi.org/10.3390/ijms23179998
Chicago/Turabian StyleHan, Mei, Mingyue Xu, Tao Su, Shizhen Wang, Liangdan Wu, Junhu Feng, and Changjun Ding. 2022. "Transcriptome Analysis Reveals Critical Genes and Pathways in Carbon Metabolism and Ribosome Biogenesis in Poplar Fertilized with Glutamine" International Journal of Molecular Sciences 23, no. 17: 9998. https://doi.org/10.3390/ijms23179998
APA StyleHan, M., Xu, M., Su, T., Wang, S., Wu, L., Feng, J., & Ding, C. (2022). Transcriptome Analysis Reveals Critical Genes and Pathways in Carbon Metabolism and Ribosome Biogenesis in Poplar Fertilized with Glutamine. International Journal of Molecular Sciences, 23(17), 9998. https://doi.org/10.3390/ijms23179998








