Characterization of Recombinant Antimicrobial Peptide BMGlv2 Heterologously Expressed in Trichoderma reesei
Abstract
:1. Introduction
2. Results
2.1. Construction of Recombinant Expression Vector PCBHG-BMGlv2
2.2. Expression and Purification of BMGlv2 in T. reesei
2.3. Antimicrobial Activity of BMGlv2
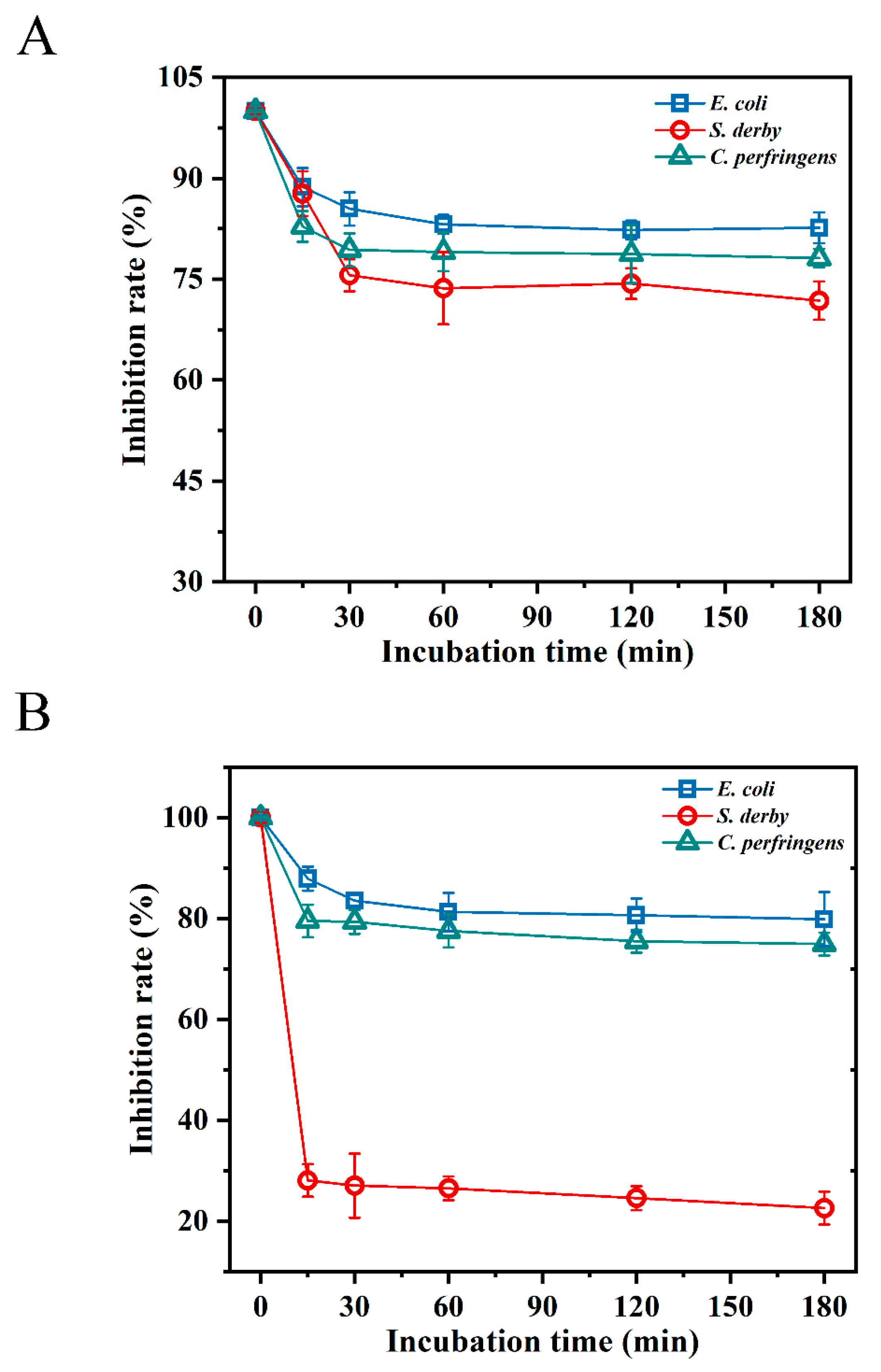
2.4. Temperature and Salt Sensitivity
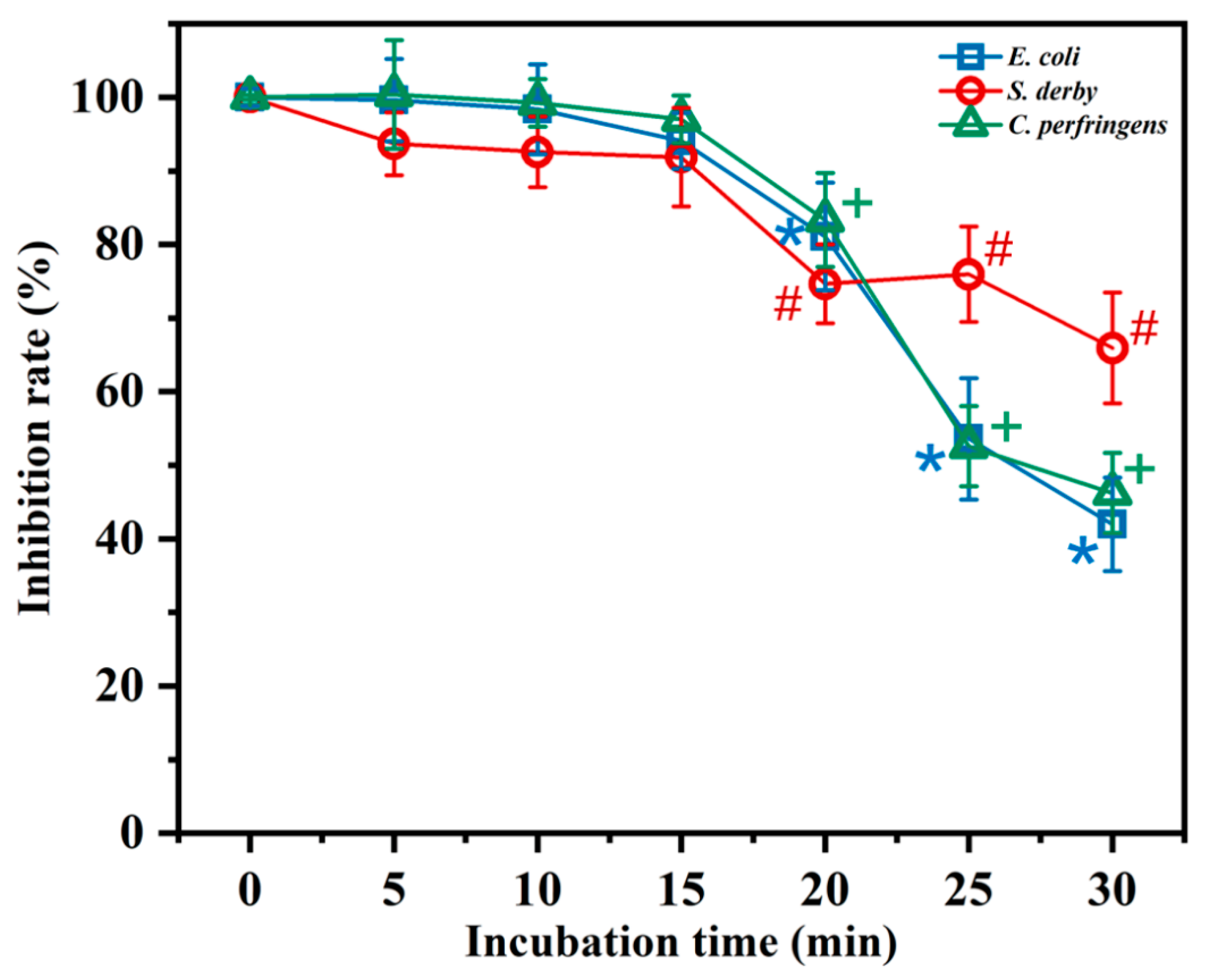
2.5. Resistance to Digestive Enzymes
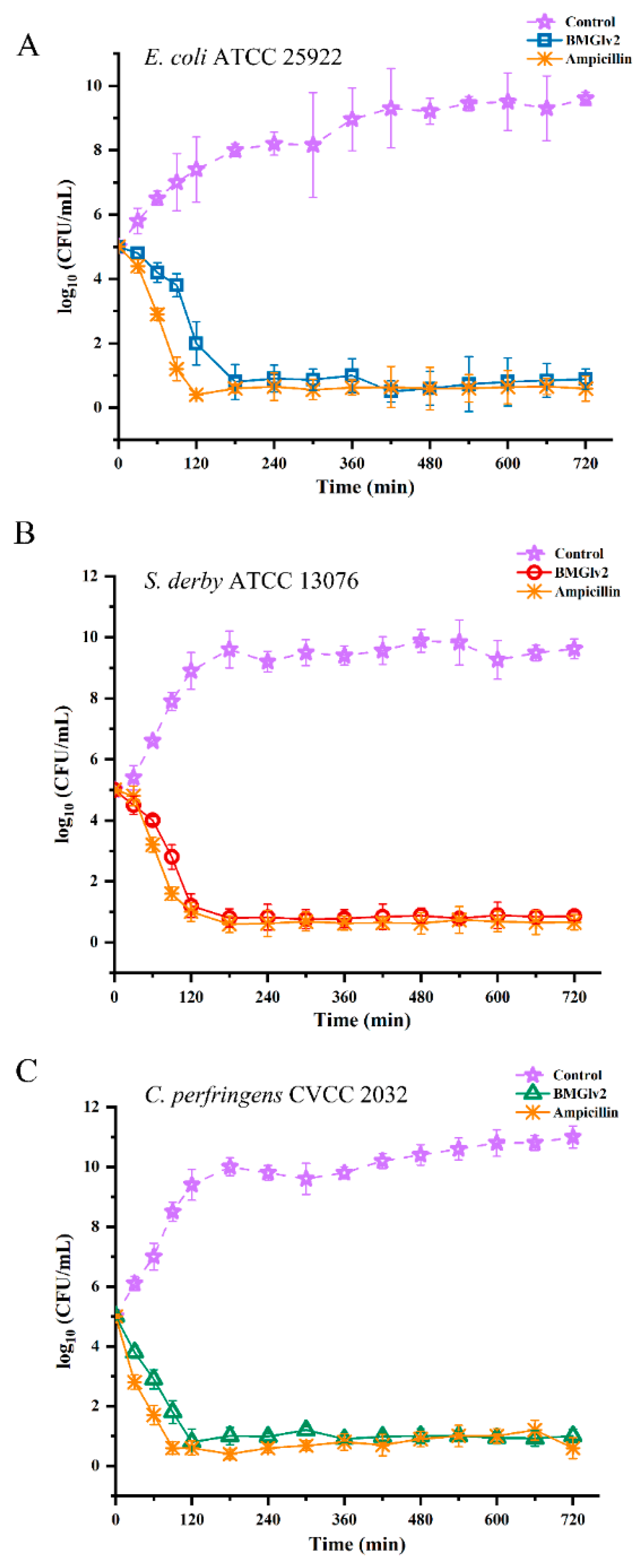
2.6. Bactericidal Kinetic Assays
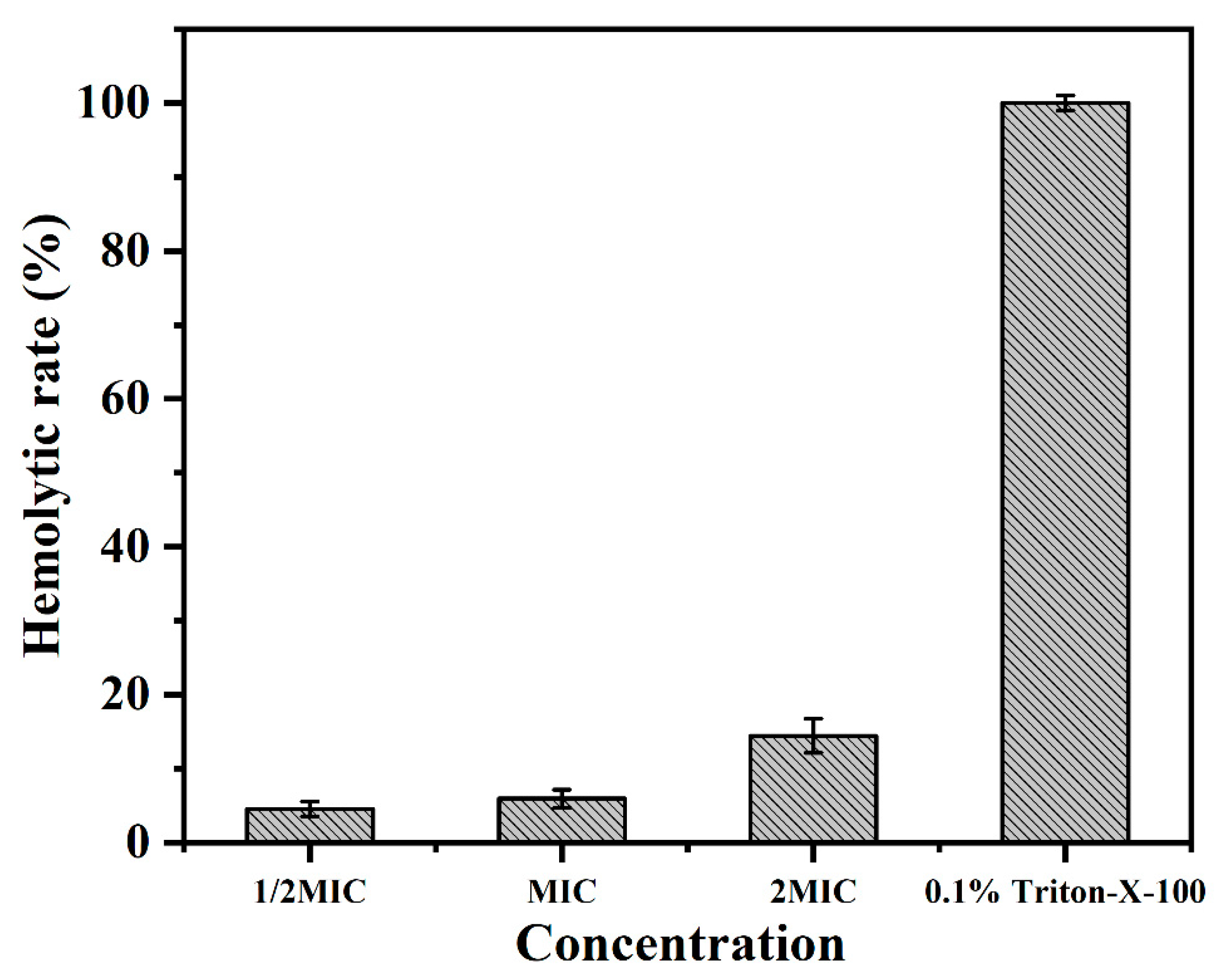
2.7. Hemolytic Activity Assays
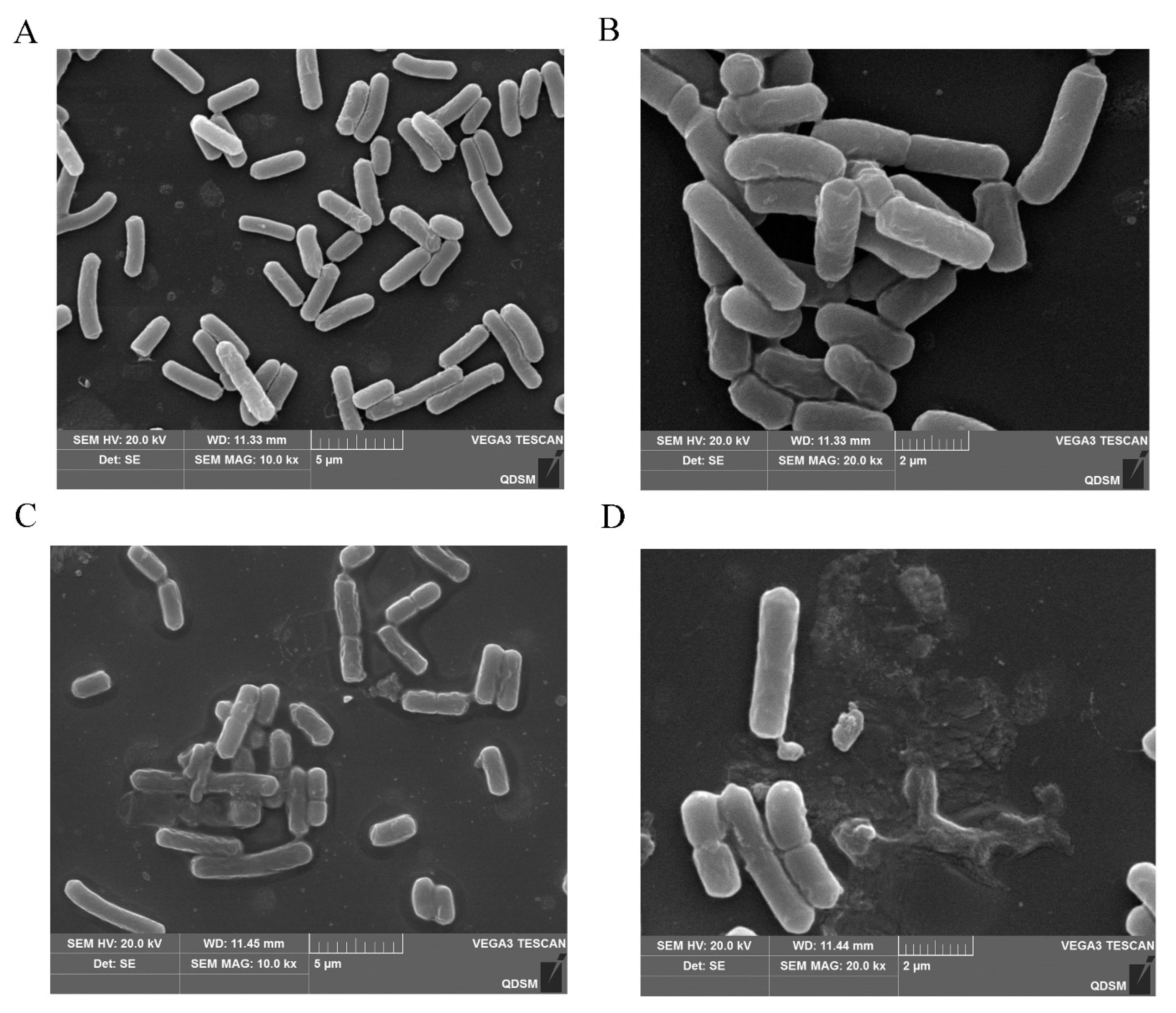
2.8. Action Mechanism of BMGlv2 against C. perfringens
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Construction of the Expression Vector PCBHG-BMGlv2
4.3. Transformation and Expression of PCBHG-BMGlv2 in T. reesei and Purification of BMGlv2
4.4. Measurement of Antimicrobial Activity of BMGlv2
4.5. Sensitivity Analysis to Temperature and Salinity
4.6. Evaluation of Resistance to Digestive Enzymes
4.7. Bactericidal Kinetic Assays
4.8. Measurement of Hemolytic Activity
4.9. Antibacterial Mechanism against C. perfringens
4.10. SEM Observations
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, C.; Song, P.; Fan, P.; Hou, C.; Thacker, P.; Ma, X. Dietary sodium butyrate decreases postweaning diarrhea by modulating intestinal permeability and changing the bacterial communities in weaned piglets. J. Nutr. 2015, 145, 2774–2780. [Google Scholar] [CrossRef]
- Volynets, V.; Rings, A.; Bárdos, G.; Ostaff, M.J.; Wehkamp, J.; Bischoff, S.C. Intestinal barrier analysis by assessment of mucins, tight junctions, and α-defensins in healthy C57BL/6J and BALB/cJ mice. Tissue Barriers 2016, 4, e1208468. [Google Scholar] [CrossRef]
- Yi, H.; Hu, W.; Chen, S.; Lu, Z.; Wang, Y. Cathelicidin-WA improves intestinal epithelial barrier function and enhances host defense against enterohemorrhagic Escherichia coli O157:H7 infection. J. Immunol. 2017, 198, 1696–1705. [Google Scholar] [CrossRef]
- Huang, G.; Li, X.; Lu, D.; Liu, S.; Suo, X.; Li, Q.; Li, N. Lysozyme improves gut performance and protects against enterotoxigenic Escherichia coli infection in neonatal piglets. Vet. Res. 2018, 49, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kylla, H.; Dutta, T.K.; Roychoudhury, P.; Subudhi, P.K. Coinfection of diarrheagenic bacterial and viral pathogens in piglets of Northeast region of India. Vet. World 2019, 12, 224–230. [Google Scholar] [CrossRef]
- Songer, J.G. Clostridia as agents of zoonotic disease. Vet. Microbiol. 2010, 140, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.; Zhang, W. Oral immunization of a live attenuated Escherichia coli strain expressing a holotoxin-structured adhesin-toxoid fusion (1FaeG-FedF-LTA2:5LTB) protected young pigs against enterotoxigenic E. coli (ETEC) infection. Vaccine 2013, 31, 1458–1463. [Google Scholar] [CrossRef]
- Stanley, T.H.; Port, J.D.; van der Maaten, J.; Kimball, J. Treatment of stress hyperthermia in elk with ketanserin, a serotonin receptor blocker. Vet. Surg. 1986, 15, 214–217. [Google Scholar] [CrossRef]
- Qi, M.; Cao, Z.; Shang, P.; Zhang, H.; Hussain, R.; Mehmood, K.; Chang, Z.; Wu, Q.; Dong, H. Comparative analysis of fecal microbiota composition diversity in Tibetan piglets suffering from diarrheagenic Escherichia coli (DEC). Microb. Pathog. 2021, 158, 105106. [Google Scholar] [CrossRef] [PubMed]
- Fischer Walker, C.L.; Rudan, I.; Liu, L.; Nair, H.; Theodoratou, E.; Bhutta, Z.A.; O’Brien, K.L.; Campbell, H.; Black, R.E. Global burden of childhood pneumonia and diarrhoea. Lancet 2013, 381, 1405–1416. [Google Scholar] [CrossRef]
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2161. [Google Scholar] [CrossRef]
- Hu, W.; Yang, Y.; Li, Z.; Lu, Z.; Wang, F.; Wang, Y. Antibacterial, cytotoxicity and mechanism of the antimicrobial peptide KR-32 in weaning piglets. Int. J. Pept. Res. Ther. 2020, 26, 943–953. [Google Scholar] [CrossRef]
- Gholamiandehkordi, A.; Eeckhaut, V.; Lanckriet, A.; Timbermont, L.; Bjerrum, L.; Ducatelle, R.; Haesebrouck, F.; Van Immerseel, F. Antimicrobial resistance in Clostridium perfringens isolates from broilers in Belgium. Vet. Res. Commun. 2009, 33, 1031–1037. [Google Scholar] [CrossRef]
- Park, M.; Rooney, A.P.; Hecht, D.W.; Li, J.; McClane, B.A.; Nayak, R.; Paine, D.D.; Rafii, F. Phenotypic and genotypic characterization of tetracycline and minocycline resistance in Clostridium perfringens. Arch. Microbiol. 2010, 192, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut microbiota dysbiosis in postweaning piglets: Understanding the keys to health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [CrossRef]
- Kesselheim, A.S.; Outterson, K. Fighting antibiotic resistance: Marrying new financial incentives to meeting public health goals. Health Aff. 2010, 29, 1689–1696. [Google Scholar] [CrossRef]
- Xu, B.C.; Fu, J.; Zhu, L.Y.; Li, Z.; Wang, Y.Z.; Jin, M.L. Overall assessment of antimicrobial peptides in piglets: A set of meta-analyses. Animal 2020, 14, 2463–2471. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, Z.; Zou, T.; Chen, J.; Li, G.; Zheng, L.; Li, S.; You, J. Bacteriophage as an alternative to antibiotics promotes growth performance by regulating intestinal inflammation, intestinal barrier function and gut microbiota in weaned piglets. Front. Vet. Sci. 2021, 8, 1–14. [Google Scholar] [CrossRef]
- Wibowo, D.; Zhao, C.X. Recent achievements and perspectives for large-scale recombinant production of antimicrobial peptides. Appl. Microbiol. Biotechnol. 2019, 103, 659–671. [Google Scholar] [CrossRef]
- Hammami, R.; Hamida, J.B.; Vergoten, G.; Lacroix, J.M.; Slomianny, M.C.; Mohamed, N.; Fliss, I. A new antimicrobial peptide isolated from Oudneya africana seeds. Microbiol. Immunol. 2009, 53, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Kim, D.H.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides: Their physicochemical properties and therapeutic application. Arch. Pharm. Res. 2012, 35, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Philip, A.T.; Malcolm, W.; Qiao, S.Y. Functions of antimicrobial peptides in gut homeostasis. Curr. Protein Pept. Sc. 2015, 16, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004, 75, 39–48. [Google Scholar] [CrossRef]
- McPhee, J.B.; Lewenza, S.; Hancock, R.E.W. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 205–217. [Google Scholar] [CrossRef]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.M.; Bechinger, B.; Naas, T. Antimicrobial peptides: A potent alternative to antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.; Maximiano, M.R.; Franco, O.L. Antimicrobial peptides used as growth promoters in livestock production. Appl. Microbiol. Biot. 2021, 105, 7115–7121. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150290. [Google Scholar] [CrossRef]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Geng, T.; Hou, C.; Huang, Y.; Qin, G.; Guo, X. Bombyx mori cecropin A has a high antifungal activity to entomopathogenic fungus Beauveria bassiana. Gene 2016, 583, 29–35. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, P.; Wang, Z.; Liu, H.; Zhang, Y.; Jiang, S.; Han, W.; Xia, Q.; Zhao, P. Antibacterial mechanism of gloverin2 from silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, H.Y.; Deng, X.J.; Yang, W.Y.; Zhou, C.Z.; Cao, Y.; Yu, X.Q. Gloverins of the silkworm Bombyx mori: Structural and binding properties and activities. Insect. Biochem. Mol. Biol. 2013, 43, 612–625. [Google Scholar] [CrossRef]
- Wohlschlager, L.; Csarman, F.; Chang, H.; Fitz, E.; Seiboth, B.; Ludwig, R. Heterologous expression of Phanerochaete chrysosporium cellobiose dehydrogenase in Trichoderma reesei. Microb. Cell Fact. 2021, 20, 1–12. [Google Scholar] [CrossRef]
- Qin, L.; Jiang, X.; Dong, Z.; Huang, J.; Chen, X. Identification of two integration sites in favor of transgene expression in Trichoderma reesei. Biotechnol. Biofuels. 2018, 11, 1–15. [Google Scholar] [CrossRef]
- Li, Y. Recombinant production of antimicrobial peptides in Escherichia coli: A review. Protein Expr. Purif. 2011, 80, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Wohlschlager, L.; Csarman, F.; Zrilić, M.; Seiboth, B.; Ludwig, R. Comparative characterization of glyoxal oxidase from Phanerochaete chrysosporium expressed at high levels in Pichia pastoris and Trichoderma reesei. Enzyme Microb. Technol. 2021, 145, 109748. [Google Scholar] [CrossRef] [PubMed]
- De Beeck, M.O.; Persson, P.; Tunlid, A. Fungal extracellular polymeric substance matrices—Highly specialized microenvironments that allow fungi to control soil organic matter decomposition reactions. Soil. Biol. Biochem. 2021, 159, 108304. [Google Scholar] [CrossRef]
- Subramanian, V.; Schuster, L.A.; Moore, K.T.; Taylor, L.E.; Baker, J.O.; Vander Wall, T.A.; Linger, J.G.; Himmel, M.E.; Decker, S.R. A versatile 2A peptide-based bicistronic protein expressing platform for the industrial cellulase producing fungus, Trichoderma reesei. Biotechnol. Biofuels. 2017, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Qian, Y.; Wang, W.; Zhong, Y.; Dai, M. Heterologous expression of Talaromyces emersonii cellobiohydrolase Cel7A in Trichoderma reesei increases the efficiency of corncob residues saccharification. Biotechnol. Lett. 2018, 40, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Axen, A.; Carlsson, A.; Engstrom, A.; Bennich, H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur. J. Biochem. 1997, 247, 614–619. [Google Scholar] [CrossRef]
- Kaneko, Y.; Furukawa, S.; Tanaka, H.; Yamakawa, M. Expression of antimicrobial peptide genes encoding Enbocin and Gloverin isoforms in the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2007, 71, 2233–2241. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Guo, P.; Wang, Q.; Zhang, Y.; Xu, H.; Zhao, P. Overexpression of Gloverin2 in the Bombyx mori silk gland enhances cocoon/silk antimicrobial activity. Dev. Comp. Immunol. 2019, 98, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Ding, J.; Li, H.; Li, L.; Zhao, R.; Shen, Z.; Fan, X.; Xi, T. Effects of cations and pH on antimicrobial activity of thanatin and s-thanatin against Escherichia coli ATCC25922 and B. subtilis ATCC 21332. Curr. Microbiol. 2008, 57, 552–557. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, M.J.; Riva, S.L.; Burges, S.C.M.; Fanning, S.; Duffy, G. Inhibition of verocytotoxigenic Escherichia coli by antimicrobial peptides caseicin A and B and the factors affecting their antimicrobial activities. Int. J. Food Microbiol. 2012, 153, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, L.; Wang, J.; Ma, Z.; Xu, W.; Li, J.; Shan, A. Characterization of antimicrobial activity and mechanisms of low amphipathic peptides with different α-helical propensity. Acta Biomater. 2015, 18, 155–167. [Google Scholar] [CrossRef]
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 2014, 10, 244–257. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, Y.; Mao, R.; Teng, D.; Wang, X.; Wang, J. Design and recombination expression of a novel plectasin-derived peptide MP1106 and its properties against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 2649–2662. [Google Scholar] [CrossRef]
- Cruz, J.; Ortiz, C.; Guzman, F.; Cardenas, C.; Fernandez-Lafuente, R.; Torres, R. Design and activity of novel lactoferrampin analogues against O157:H7 enterohemorrhagic Escherichia coli. Biopolymers 2014, 101, 319–328. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Teng, D.; Tian, Z.; Wang, S.; Wang, J. Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr. Purif. 2011, 78, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tian, Z.; Teng, D.; Zhang, J.; Wang, J.; Wang, J. High-level production of a candidacidal peptide lactoferrampin in Escherichia coli by fusion expression. J. Biotechnol. 2009, 139, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Teng, D.; Wang, X.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Mechanism of action of a novel recombinant peptide, MP1102, against Clostridium perfringens type C. Appl. Microbiol. Biotechnol. 2016, 100, 5045–5057. [Google Scholar] [CrossRef]
- Sørensen, O.; Bratt, T.; Johnsen, A.H.; Madsen, M.T.; Borregaard, N. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J. Biol. Chem. 1999, 274, 22445–22451. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta-Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]
- Zhang, L.; Dhillon, P.; Yan, H.; Farmer, S.; Hancock, R.E.W. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3317–3321. [Google Scholar] [CrossRef] [PubMed]
- Schuster, A.; Bruno, K.S.; Collett, J.R.; Baker, S.E.; Seiboth, B.; Kubicek, C.P.; Schmoll, M. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol. Biofuels. 2012, 5, 1–10. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z.; Zhou, S.; Mou, H.; Zhang, R. Cloning and expression of a β-mannanase gene from Bacillus sp. MK-2 and its directed evolution by random mutagenesis. Enzyme Microb. Technol. 2019, 124, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.M.; Lv, Y.J.; Zhao, J.F.; Liu, Q.Y.; Shi, L.Y.; Wang, J.P.; Yang, Y.H.; Fan, Z.C. Efficient production of a recombinant Venerupis philippinarum defensin (VpDef) in Pichia pastoris and characterization of its antibacterial activity and stability. Protein Expr. Purif. 2018, 147, 78–84. [Google Scholar] [CrossRef]
- Meng, D.M.; Zhao, J.F.; Ling, X.; Dai, H.X.; Guo, Y.J.; Gao, X.F.; Dong, B.; Zhang, Z.Q.; Meng, X.; Fan, Z.C. Recombinant expression, purification and antimicrobial activity of a novel antimicrobial peptide PaDef in Pichia pastoris. Protein Expr. Purif. 2017, 130, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Lee, D.G. The characteristic region of arenicin-1 involved with a bacterial membrane targeting mechanism. Biochem. Biophys. Res. Commun. 2011, 405, 422–427. [Google Scholar] [CrossRef]
- Gök, Y.; Akkoç, S.; Erdoğan, H.; Albayrak, S. In vitro antimicrobial studies of new benzimidazolium salts and silver N-heterocyclic carbene complexes. J. Enzyme Inhib. Med. Chem. 2016, 31, 1322–1327. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, K.B.W.R.; Lim, S.I.; Ahn, D.H. Antibacterial mechanism of Myagropsis myagroides extract on Listeria monocytogenes. Food Control. 2014, 42, 23–28. [Google Scholar] [CrossRef]
- Li, M.; Mou, H.; Kong, Q.; Zhang, T.; Fu, X. Bacteriostatic effect of lipopeptides from Bacillus subtilis N-2 on Pseudomonas putida using soybean meal by solid-state fermentation. Mar. Life Sci. Technol. 2020, 2, 172–180. [Google Scholar] [CrossRef]
- Lin, L.; Gu, Y.; Li, C.; Vittayapadung, S.; Cui, H. Antibacterial mechanism of ε-Poly-lysine against Listeria monocytogenes and its application on cheese. Food Control. 2018, 91, 76–84. [Google Scholar] [CrossRef]
- Sharma, A.; Bajpai, V.K.; Baek, K.H. Determination of antibacterial mode of action of Allium sativum essential oil against foodborne pathogens using membrane permeability and surface characteristic parameters. J. Food Saf. 2013, 33, 197–208. [Google Scholar] [CrossRef]
- Yang, X.; Huang, E.; Yousef, A.E. Brevibacillin, a cationic lipopeptide that binds to lipoteichoic acid and subsequently disrupts cytoplasmic membrane of Staphylococcus aureus. Microbiol. Res. 2017, 195, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhao, C.; Lin, L. The specific antibacterial activity of liposome-encapsulated Clove oil and its application in tofu. Food Control. 2015, 56, 128–134. [Google Scholar] [CrossRef]
- Asad, M.; Arshad, M.N.; Oves, M.; Khalid, M.; Khan, S.A.; Asiri, A.M.; Rehan, M.; Dzudzevic-Cancar, H. N-Trifluoroacetylated pyrazolines: Synthesis, characterization and antimicrobial studies. Bioorg. Chem. 2020, 99, 103842. [Google Scholar] [CrossRef] [PubMed]

| Sample | Volume (mL) | Activity (AU/mL) | Total Protein (mg) | Total Activity (AU) | Specific Activity (AU/mg) | Yield (%) | Purification (Fold) |
|---|---|---|---|---|---|---|---|
| Culture supernatant | 50 | 150 | 9.3 | 7500 | 806.5 | 100 | 1 |
| Purified BMGlv2 | 14 | 292 | 1.6 | 4088 | 2555 | 54.5 | 3.2 |
| Bacteria | MIC (μg/mL) | MBC (μg/mL) | ||
|---|---|---|---|---|
| BMGlv2 | Ampicillin | BMGlv2 | Ampicillin | |
| Gram-negative | ||||
| E. coli ATCC 25922 | 43.75 | 3.75 | 43.75 | 3.75 |
| S. derby ATCC 13076 | 43.75 | 7.50 | 43.75 | 7.50 |
| Gram-positive | ||||
| C. perfringens CVCC 2032 | 21.86 | 3.75 | 43.75 | 3.75 |
| Strains | Control | NaCl (150 mM) | KCl (4.5 mM) | NH4Cl (6 μM) | MgCl2 (1 mM) | ZnCl2 (8 μM) | FeCl3 (4 μM) |
|---|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | 43.75 | 21.86 | 43.75 | 21.86 | 43.75 | 21.86 | 21.86 |
| S. derby ATCC 13076 | 43.75 | 43.75 | 43.75 | 43.75 | 43.75 | 43.75 | 43.75 |
| C. perfringens CVCC 2032 | 21.86 | 10.93 | 21.86 | 21.86 | 10.93 | 10.93 | 10.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Q.; Cao, L.; Zhu, C.; Kong, Q.; Sun, H.; Zhang, F.; Mou, H.; Liu, Z. Characterization of Recombinant Antimicrobial Peptide BMGlv2 Heterologously Expressed in Trichoderma reesei. Int. J. Mol. Sci. 2022, 23, 10291. https://doi.org/10.3390/ijms231810291
Liang Q, Cao L, Zhu C, Kong Q, Sun H, Zhang F, Mou H, Liu Z. Characterization of Recombinant Antimicrobial Peptide BMGlv2 Heterologously Expressed in Trichoderma reesei. International Journal of Molecular Sciences. 2022; 23(18):10291. https://doi.org/10.3390/ijms231810291
Chicago/Turabian StyleLiang, Qingping, Linyuan Cao, Changliang Zhu, Qing Kong, Han Sun, Fang Zhang, Haijin Mou, and Zhemin Liu. 2022. "Characterization of Recombinant Antimicrobial Peptide BMGlv2 Heterologously Expressed in Trichoderma reesei" International Journal of Molecular Sciences 23, no. 18: 10291. https://doi.org/10.3390/ijms231810291
APA StyleLiang, Q., Cao, L., Zhu, C., Kong, Q., Sun, H., Zhang, F., Mou, H., & Liu, Z. (2022). Characterization of Recombinant Antimicrobial Peptide BMGlv2 Heterologously Expressed in Trichoderma reesei. International Journal of Molecular Sciences, 23(18), 10291. https://doi.org/10.3390/ijms231810291





