Histidine-Rich Glycoprotein Suppresses the S100A8/A9-Mediated Organotropic Metastasis of Melanoma Cells
Abstract
:1. Introduction
2. Results
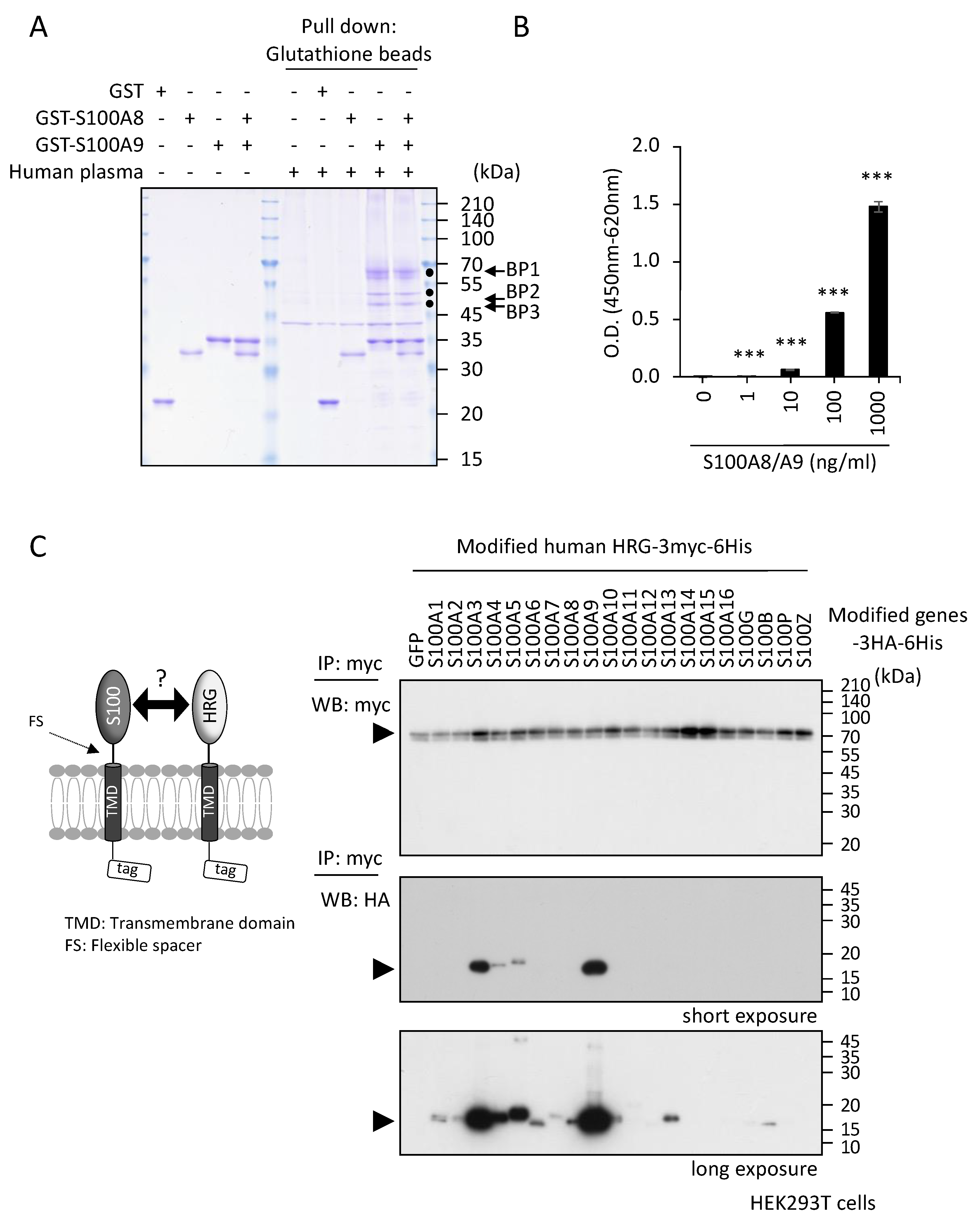
2.1. Identification of S100A8/A9 Binding Protein(s) in Human Plasma
2.2. Inhibitory Effects of HRG on S100A8/A9-Mediated Melanoma Cell Behaviors In Vitro
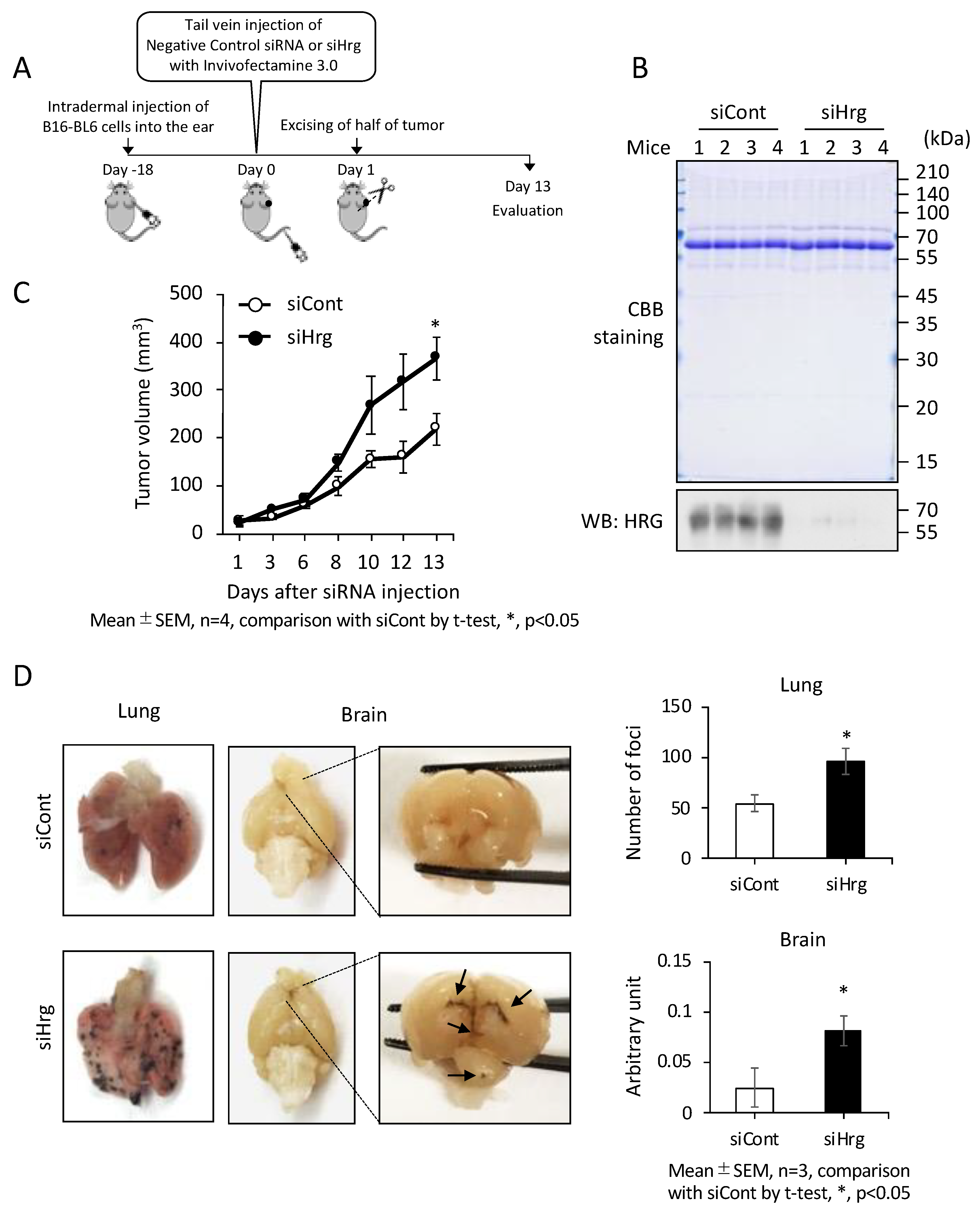
2.3. Preventive Roles of HRG on Cancer Growth and Metastasis In Vivo
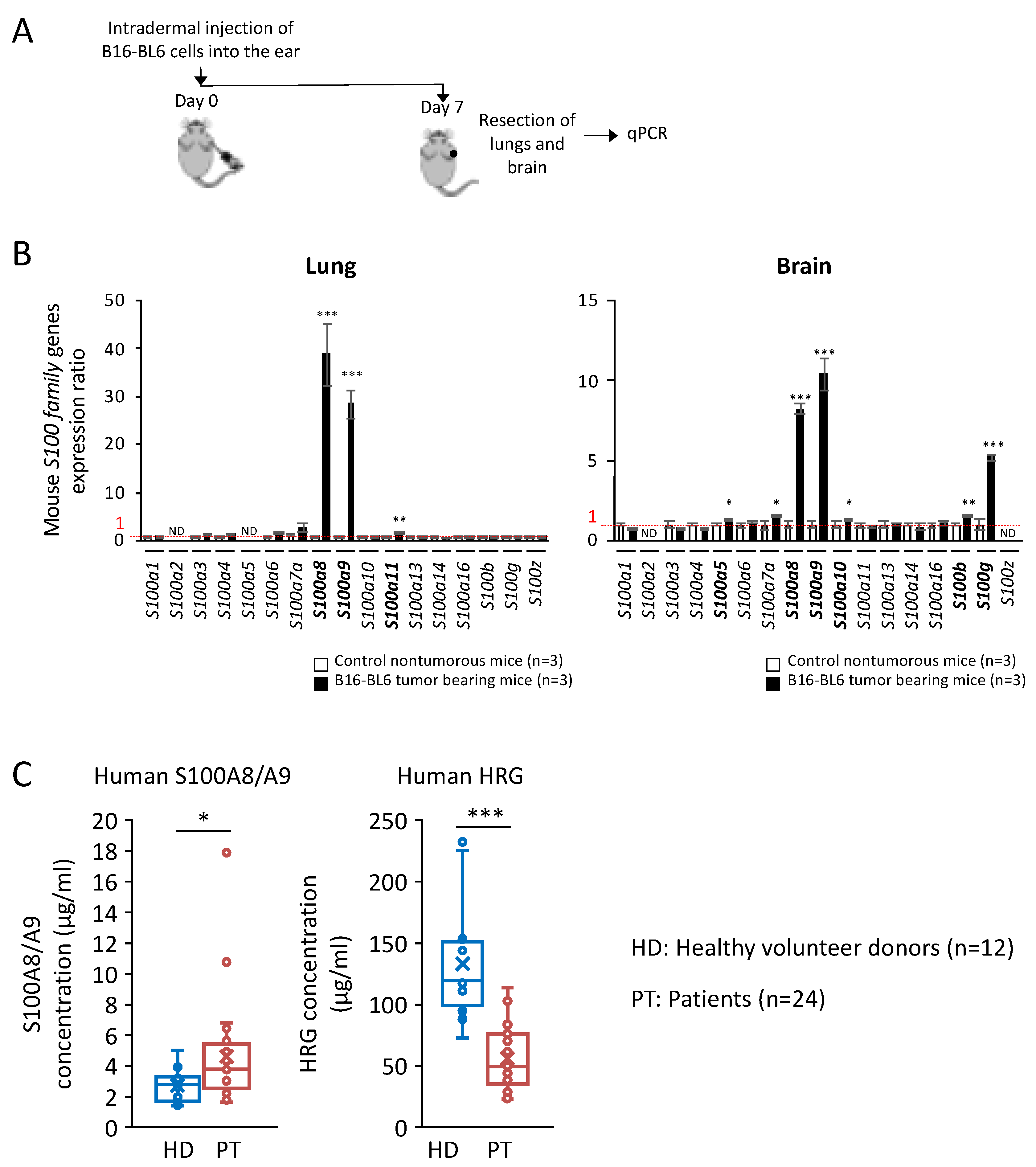
2.4. Significant Increase in S100A8/A9 at Premetastatic Stage in the Lungs and Brain of Melanoma-Bearing Mice
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Liquid Chromatography-Mass Spectrometry (LC-Ms/Ms)
4.3. Enzyme-Linked Immunosorbent Assay (ELISA)
4.4. Recombinant Proteins
4.5. Plasmids
4.6. Immunoprecipitation (IP)
4.7. Western Blotting (WB)
4.8. Migration and Invasion Assays
4.9. Quantitative Real-Time PCR
4.10. Animal Experiment
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kinoshita, R.; Sato, H.; Yamauchi, A.; Takahashi, Y.; Inoue, Y.; Sumardika, I.W.; Chen, Y.; Tomonobu, N.; Araki, K.; Shien, K.; et al. Newly developed anti-S100A8/A9 monoclonal antibody efficiently prevents lung tropic cancer metastasis. Int. J. Cancer 2019, 145, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, S.; Watanabe, A.; Aburatani, H.; Maru, Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 2006, 8, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, S.; Watanabe, A.; Sakurai, Y.; Takamura, S.A.; Ishibashi, S.; Miyake, K.; Shibuya, M.; Akira, S.; Aburatani, H.; Maru, Y. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat. Cell Biol. 2008, 10, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Lee, Y.C.; Zhang, Z.; Chandra, G.; Su, S.B.; Mukherjee, A.B. Lack of an endogenous anti-inflammatory protein in mice enhances colonization of B16F10 melanoma cells in the lungs. J. Biol. Chem. 2010, 285, 10822–10831. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Riehl, A.; Durchdewald, M.; Németh, J.; Fürstenberger, G.; Decker, K.M.; Enk, A.; Arnold, B.; Bierhaus, A.; Nawroth, P.P.; et al. RAGE signaling sustains inflammation and promotes tumor development. J. Exp. Med. 2008, 205, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.I.; Murata, H.; Putranto, E.W.; Kataoka, K.; Motoyama, A.; Hibino, T.; Inoue, Y.; Sakaguchi, M.; Huh, N.H. DOCK7 is a critical regulator of the RAGE-Cdc42 signaling axis that induces formation of dendritic pseudopodia in human cancer cells. Oncol. Rep. 2013, 29, 1073–1079. [Google Scholar] [CrossRef]
- Hibino, T.; Sakaguchi, M.; Miyamoto, S.; Yamamoto, M.; Motoyama, A.; Hosoi, J.; Shimokata, T.; Ito, T.; Tsuboi, R.; Huh, N.H. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013, 73, 172–183. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Yamamoto, M.; Miyai, M.; Maeda, T.; Hiruma, J.; Murata, H.; Kinoshita, R.; Ruma, I.M.W.; Putranto, E.W.; Inoue, Y.; et al. Identification of an S100A8 receptor neuroplastin-β and its heterodimer formation with EMMPRIN. J. Investig. Dermatol. 2016, 136, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Ruma, I.M.W.; Putranto, E.W.; Kondo, E.; Murata, H.; Watanabe, M.; Huang, P.; Kinoshita, R.; Futami, J.; Inoue, Y.; Yamauchi, A.; et al. MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-κB and ROS formation upon ligand binding. Clin. Exp. Metastasis 2016, 33, 609–627. [Google Scholar] [CrossRef]
- Tomonobu, N.; Kinoshita, R.; Sakaguchi, M. S100 soil sensor receptors and molecular targeting therapy against them in cancer metastasis. Transl. Oncol. 2020, 13, 100753. [Google Scholar] [CrossRef]
- Fu, X.; Liu, B.; Wang, Y.; Li, J.; Zhu, P.; Li, T.; Tse, K.; Chou, J.; Yin, C.; Bai, J.; et al. Activation of STAT3 is a key event in TLR4 signaling-mediated melanoma progression. Cell Death Dis. 2020, 11, 246. [Google Scholar] [CrossRef]
- Caudron, A.; Battistella, M.; Feugeas, J.P.; Pages, C.; Basset-Seguin, N.; Dorval, S.M.; Brentano, E.F.; Sadoux, A.; Podgorniak, M.P.; Menashi, S.; et al. EMMPRIN/CD147 is an independent prognostic biomarker in cutaneous melanoma. Exp. Dermatol. 2016, 25, 618–622. [Google Scholar] [CrossRef]
- Chen, Y.; Sumardika, I.W.; Tomonobu, N.; Ruma, I.M.W.; Kinoshita, R.; Kondo, E.; Inoue, Y.; Sato, H.; Yamauchi, A.; Murata, H.; et al. Melanoma cell adhesion molecule is the driving force behind the dissemination of melanoma upon S100A8/A9 binding in the original skin lesion. Cancer Lett. 2019, 452, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sumardika, I.W.; Tomonobu, N.; Kinoshita, R.; Inoue, Y.; Iioka, H.; Mitsui, Y.; Saito, K.; Ruma, I.M.W.; Sato, H.; et al. Critical role of the MCAM-ETV4 axis triggered by extracellular S100A8/A9 in breast cancer aggressiveness. Neopalsia 2019, 21, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Sumardika, I.W.; Chen, Y.; Tomonobu, N.; Kinoshita, R.; Ruma, I.M.W.; Sato, H.; Kondo, E.; Inoue, Y.; Yamauchi, A.; Murata, H.; et al. Neuroplastin-β mediates S100A8/A9-induced lung cancer disseminative progression. Mol. Carcinog. 2019, 58, 980–995. [Google Scholar] [CrossRef]
- Edgeworth, J.; Gorman, M.; Bennett, R.; Freemont, P.; Hogg, N. Identification of p8, 14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J. Biol. Chem. 1991, 266, 7706–7713. [Google Scholar] [CrossRef]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Kinoshita, R.; Tomonobu, N.; Gohara, Y.; Tomida, S.; Takahashi, Y.; Senoo, S.; Taniguchi, A.; Itano, J.; Yamamoto, K.I.; et al. The heterodimer S100A8/A9 is potent therapeutic target for idiopathic pulmonary fibrosis. J. Mol. Med. 2021, 99, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Patel, K.K.; Davis, D.S.; Parish, C.R.; Hulett, M.D. Histidine-rich glycoprotein: The Swiss Army knife of mammalian plasma. Blood 2011, 117, 2093–2101. [Google Scholar] [CrossRef]
- Nishibori, M. Novel aspects of sepsis pathophysiology: NETs, plasma glycoproteins, endotheliopathy and COVID-19. J. Pharmacol. Sci. 2022, 150, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wake, H.; Sakaguchi, M.; Wang, D.; Takahashi, Y.; Teshigawara, K.; Zhong, H.; Mori, S.; Liu, K.; Takahashi, H.; et al. Histidine-Rich Glycoprotein inhibits High-Mobility Group Box-1-mediated pathways in vascular endothelial cells through CLEC-1A. iScience 2020, 23, 101180. [Google Scholar] [CrossRef]
- Wake, H.; Mori, S.; Liu, K.; Morioka, Y.; Teshigawara, K.; Sakaguchi, M.; Kuroda, K.; Gao, Y.; Takahashi, H.; Ohtsuka, A.; et al. Histidine-Rich Glycoprotein prevents septic lethality through regulation of immunothrombosis and inflammation. EBioMedicine 2016, 9, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, J.J., Jr.; Jeter, M.A.; Bruck, D.; Feinberg, W.M. Histidine-rich glycoprotein levels in children: The effect of age. Thromb. Res. 1990, 59, 681–686. [Google Scholar] [CrossRef]
- Koide, T.; Foster, D.; Yoshitake, S.; Davie, E.W. Amino acid sequence of human histidine-rich glycoprotein derived from the nucleotide sequence of its cDNA. Biochemistry 1986, 25, 2220–2225. [Google Scholar] [CrossRef]
- Simantov, R.; Febbraio, M.; Crombie, R.; Asch, A.S.; Nachman, R.L.; Silverstein, R.L. Histidine-rich glycoprotein inhibits the antiangiogenic effect of thrombospondin-1. J. Clin. Investig. 2001, 107, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.L.; Hulett, M.D.; Parish, C.R. Histidine-rich glycoprotein: A novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol. Cell Biol. 2005, 83, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, S.; Koide, T. Histidine-rich glycoprotein: A possible modulator of coagulation and fibrinolysis. Semin. Thromb. Hemost. 2011, 37, 389–394. [Google Scholar] [CrossRef]
- Rydengärd, V.; Olsson, A.K.; Mörgelin, M.; Schmidtchen, A. Histidine-rich glycoprotein exerts antibacterial activity. FEBS J. 2007, 274, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Rydengärd, V.; Shannon, O.; Lundqvist, K.; Kacprzyk, L.; Chalupka, A.; Olsson, A.K.; Mörgelin, M.; Jahnen-Dechent, W.; Malmsten, M.; Schmidtchen, A. A Histidine-rich glycoprotein protects from systemic Candida infection. PLoS Pathog. 2008, 4, e1000116. [Google Scholar] [CrossRef] [PubMed]
- Shannon, O.; Rydengärd, V.; Schmidtchen, A.; Mörgelin, M.; Alm, P.; Sørensen, O.E.; Björck, L. Histidine-rich glycoprotein promotes bacterial entrapment in clots and decreases mortality in a mouse model of sepsis. Blood 2010, 116, 365–2372. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Denstaedt, S.; Spencer-Segal, J.L.; Newstead, M.W.; Laborc, K.; Zhao, A.P.; Hjelmaas, A.; Zeng, X.; Akil, H.; Standiford, T.J.; Singer, B.H. S100A8/A9 drives neuroinflammatory priming and protects against anxiety-like behavior after sepsis. J. Immunol. 2018, 200, 3188–3200. [Google Scholar] [CrossRef]
- Schluesener, H.J.; Kremsner, P.G.; Meyermann, R. Widespread expression of MRP8 and MRP14 in human cerebral malaria by microglial cells. Acta Neropathol. 1998, 96, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Sevko, A.; Jiang, H.; Lichtenberger, R.; Reith, M.; Tarnanidis, K.; Holland-Letz, T.; Umansky, L.; Beckhove, P.; Sucker, A.; et al. Myeloid cells and related chronic inflammatory factors as novel predictive markers in melanoma treatment with Ipilimumab. Clin. Cancer Res. 2015, 21, 5453–5459. [Google Scholar] [CrossRef] [PubMed]
- Boye, K.; Mælandsmo, G.M. S100A4 and Metastasis. Am. J. Pathol. 2010, 176, 528–535. [Google Scholar] [CrossRef]
- Takahashi, Y.; Wake, H.; Sakaguchi, M.; Yoshii, Y.; Teshigawara, K.; Wang, D.; Nishibori, M. Histidine-Rich Glycoprotein stimulates human neutrophil phagocytosis and prolongs survival through CLEC1A. J. Immunol. 2021, 206, 737–750. [Google Scholar] [CrossRef]
- Langiu, M.; Palacios-Acedo, A.L.; Crescence, L.; Mege, D.; Dubois, C.; Panicot-Dubois, L. Neutrophils, Cancer and Thrombosis: The new bermuda triangle in cancer research. Int. J. Mol. Sci. 2022, 23, 1257. [Google Scholar] [CrossRef] [PubMed]
- Denning, N.; Aziz, M.; Gurien, S.D.; Wang, P. DAMPs and NETs in Sepsis. Front. Immunol. 2019, 10, 2536. [Google Scholar] [CrossRef]
- Sprenkeler, E.G.G.; Zandstra, J.; van Kleef, N.D.; Goetschalckx, I.; Verstegen, B.; Aarts, C.E.M.; Janssen, H.; Tool, A.T.J.; van Mierlo, G.; van Bruggen, R.; et al. S100A8/A9 Is a marker for the release of neutrophil extracellular traps and induces neutrophil activation. Cells 2022, 11, 236. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Lü, M.; Fan, Y.; Cao, Y.; Zhang, Z.; Yang, S. Macrophages in tumor microenvironments and the progression of tumors. J. Immunol. Res. 2012, 2012, 948098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolny, C.; Mazzone, M.; Tugues, S.; Laoui, D.; Johansson, I.; Coulon, C.; Squadrito, M.L.; Segura, I.; Li, X.; Knevels, E.; et al. HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Cancer Cell 2011, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Wake, H.; Teshigawara, K.; Wang, D.; Sakaguchi, M.; Otsuka, F.; Nishibori, M. Histidine-rich glycoprotein augments natural killer cell function by modulating PD-1 expression via CLEC-1B. Pharmacol. Res. Perspect. 2019, 7, e00481. [Google Scholar] [CrossRef]
- Futami, J.; Atago, Y.; Azuma, A.; Putranto, E.W.; Kinoshita, R.; Murata, H.; Sakaguchi, M. An efficient method for the preparation of preferentially heterodimerized recombinant S100A8/A9 coexpressed in Escherichia coli. Biochem. Biophys. Rep. 2016, 6, 94–100. [Google Scholar] [CrossRef]
- Nukui, T.; Ehama, R.; Sakaguchi, M.; Sonegawa, H.; Katagiri, C.; Hibino, T.; Huh, N.H. S100A8/A9, a key mediator for positive feedback growth stimulation of normal human keratinocytes. J. Cell Biochem. 2007, 104, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M.; Watanabe, M.; Kinoshita, R.; Kaku, H.; Ueki, H.; Futami, J.; Murata, H.; Inoue, Y.; Li, S.; Huang, P.; et al. Dramatic increase in expression of a transgene by insertion of promoters downstream of the cargo gene. Mol. Biotechnol. 2014, 56, 621–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Plasma | Healthy Volunteer Donors | Patients | |
|---|---|---|---|
| n | 12 | 24 | |
| Sex | Male/Female | 9/3 | 13/11 |
| Age, years | Median (range) | 35 (24–72) | 67 (46–78) |
| Histology of lung cancer | Ad/Sq/NEC | 0/0/0 | 19/2/3 |
| Stage of lung cancer | 1A/1B/2A/2B/3A/3B/4A/4B | 1/0/0/0/2/4/10/7 |
| Target | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| Tbp | gggagaatcatggaccagaa | gatgggaattccaggagtca |
| S100a1 | ctttctggcttcctggatgt | accagcacaacatactccttga |
| S100a2 | gggagataaaggagcttttgc | tcttcaccttctcatcatctacgtt |
| S100a3 | ccagtcggagctcaagga | ttgtagtcacactcccggaac |
| S100a4 | tcagcacttcctctctcttgg | tttgtggaaggtggacacaa |
| S100a5 | ggagttgatcaagacagagctga | tccaggctcttcatcaagttatc |
| S100a6 | aggaaggtgacaagcacacc | agccttgcaatttcagcatc |
| S100a7a | tgcaccaagagcaacagact | gtctggcatgactgatggac |
| S100a8 | tccttgcgatggtgataaaa | ggccagaagctctgctactc |
| S100a9 | gacaccctgacaccctgag | tgagggcttcatttctcttctc |
| S100a10 | gttccctgggtttttggaa | aagcccactttgccatctc |
| S100a11 | gggaaggatggaaacaacact | caccaggatccttctggttc |
| S100a13 | ccttgcctggtgcttataaactt | tgatgtccacacagattgacc |
| S100a14 | atgggacagtgtcggtcag | gtgtctcaatggccctctct |
| S100a16 | gatcagcaagtccagcttcc | ccaggttctggatgagcttg |
| S100b | aacaacgagctctctcacttcc | cgtctccatcactttgtcca |
| S100g | gctctccaaggaggagctaaa | cagctccttaaagagattgtcca |
| S100z | actcacagagttcctcacatgc | tccacttcgttgtctttattgg |
| Hrg | caccaactgtgatgcttctga | agtagtagactgtggccgttcc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomonobu, N.; Kinoshita, R.; Wake, H.; Inoue, Y.; Ruma, I.M.W.; Suzawa, K.; Gohara, Y.; Komalasari, N.L.G.Y.; Jiang, F.; Murata, H.; et al. Histidine-Rich Glycoprotein Suppresses the S100A8/A9-Mediated Organotropic Metastasis of Melanoma Cells. Int. J. Mol. Sci. 2022, 23, 10300. https://doi.org/10.3390/ijms231810300
Tomonobu N, Kinoshita R, Wake H, Inoue Y, Ruma IMW, Suzawa K, Gohara Y, Komalasari NLGY, Jiang F, Murata H, et al. Histidine-Rich Glycoprotein Suppresses the S100A8/A9-Mediated Organotropic Metastasis of Melanoma Cells. International Journal of Molecular Sciences. 2022; 23(18):10300. https://doi.org/10.3390/ijms231810300
Chicago/Turabian StyleTomonobu, Nahoko, Rie Kinoshita, Hidenori Wake, Yusuke Inoue, I Made Winarsa Ruma, Ken Suzawa, Yuma Gohara, Ni Luh Gede Yoni Komalasari, Fan Jiang, Hitoshi Murata, and et al. 2022. "Histidine-Rich Glycoprotein Suppresses the S100A8/A9-Mediated Organotropic Metastasis of Melanoma Cells" International Journal of Molecular Sciences 23, no. 18: 10300. https://doi.org/10.3390/ijms231810300
APA StyleTomonobu, N., Kinoshita, R., Wake, H., Inoue, Y., Ruma, I. M. W., Suzawa, K., Gohara, Y., Komalasari, N. L. G. Y., Jiang, F., Murata, H., Yamamoto, K.-i., Sumardika, I. W., Chen, Y., Futami, J., Yamauchi, A., Kuribayashi, F., Kondo, E., Toyooka, S., Nishibori, M., & Sakaguchi, M. (2022). Histidine-Rich Glycoprotein Suppresses the S100A8/A9-Mediated Organotropic Metastasis of Melanoma Cells. International Journal of Molecular Sciences, 23(18), 10300. https://doi.org/10.3390/ijms231810300








