Potential Therapeutic Agents against Paclitaxel—And Sorafenib-Resistant Papillary Thyroid Carcinoma
Abstract
1. Introduction
2. Results
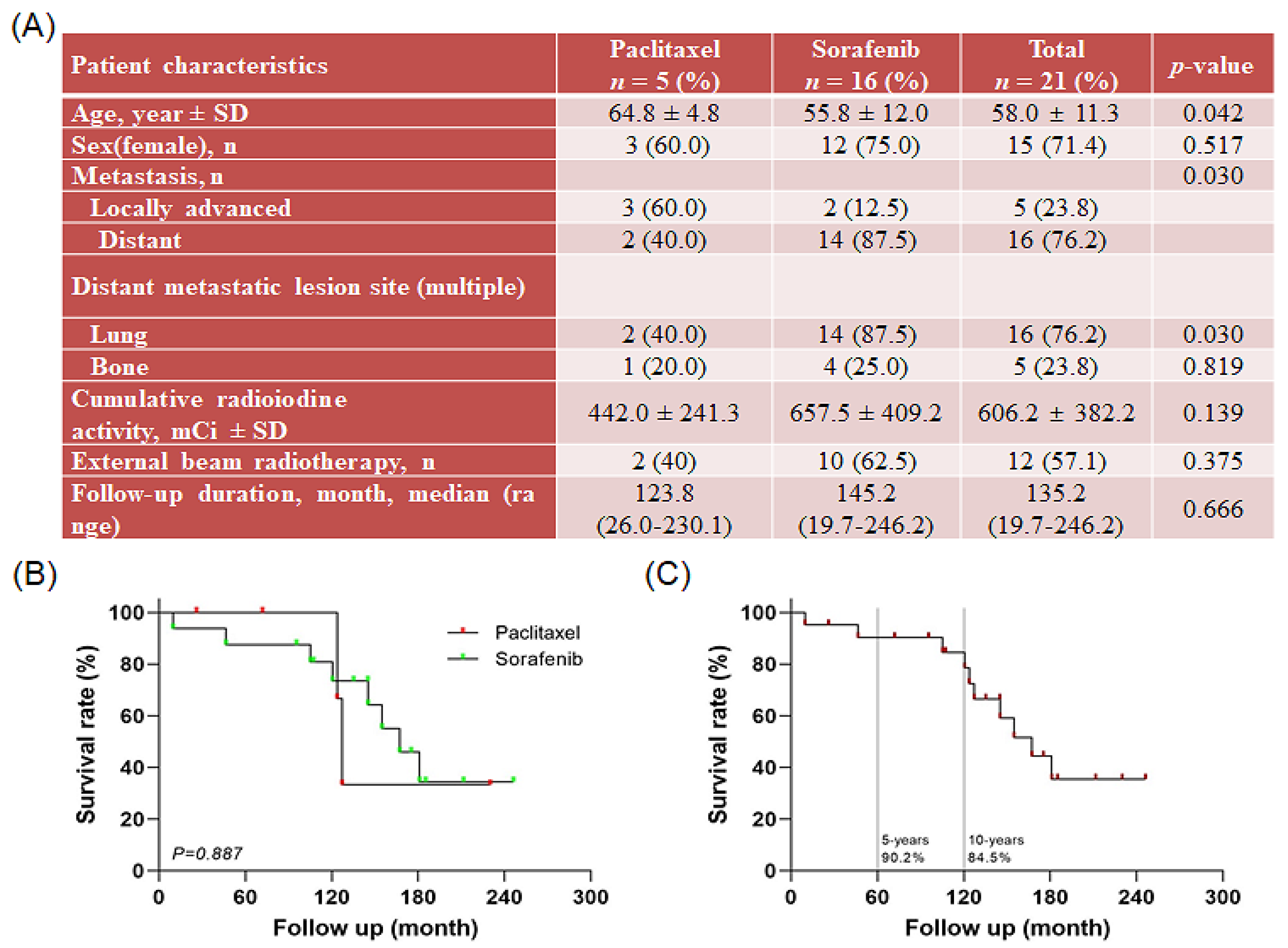
2.1. Patient Disease Characteristics
2.2. Characteristics of Patient-Derived Drug-Resistant PTC Cell Lines
2.3. Discovery of Candidates 7 and 13 as Specific Inhibitors of SERCA Using In Silico Screening
2.4. Inhibitory Effect of Novel Candidates 7 and 13 on SERCA and Their Effect on The Survival of Paclitaxel- and Sorafenib-Resistant PTC Cells
2.5. The Effect of the Co-Treatment of Paclitaxel or Sorafenib and SERCA Inhibitors on ER Stress-Mediated Apoptosis in YUMC-R-P1, -P2, and -P3 Cells
2.6. In Vivo Assessment of the Anticancer Efficacy of Candidates 7 and 13 in a Patient-Derived Drug-Resistant PTC Cell Mouse Xenograft Model
3. Discussion
4. Materials and Methods
4.1. Study Design and Ethical Considerations
4.2. Patients
4.2.1. Patient 1
4.2.2. Patient 2
4.2.3. Patient 3
4.2.4. Patient 4
4.3. Patient Tissue Specimens
4.4. Tumor Cell Isolation and Primary Culture
4.5. mRNA-Seq Data
4.6. Statistical Aanalysis of Gene Expression Level
4.7. Hierarchical Clustering
4.8. Cell Culture
4.9. Cell Viability Assay
4.10. Cell CycleAnalysis Using Flow Cytometry
4.11. Immunoblot Analysis
4.12. Human PTC Cell Xenograft
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nikiforov, Y.E. Thyroid carcinoma: Molecular pathways and therapeutic targets. Mod. Pathol. 2008, 21 (Suppl. S2), S37–S43. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.; Lee, E.J.; Huang, M.G.; Park, Y.I.; Khullar, A.; Plodkowski, R.A. Diagnosis and treatment of patients with thyroid cancer. Am. Health Drug Benefits 2015, 8, 30–40. [Google Scholar] [PubMed]
- Fahiminiya, S.; de Kock, L.; Foulkes, W.D. Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med. 2016, 375, 2306–2307. [Google Scholar] [PubMed]
- Raue, F.; Frank-Raue, K. Thyroid cancer: Risk-stratified management and individualized therapy. Clin. Cancer Res. 2016, 22, 5012–5021. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimpasic, T.; Ghossein, R.; Shah, J.P.; Ganly, I. Poorly differentiated carcinoma of the thyroid gland: Current status and future prospects. Thyroid 2019, 29, 311–321. [Google Scholar] [CrossRef]
- Xu, B.; Ghossein, R. Poorly differentiated thyroid carcinoma. Semin. Diagn. Pathol. 2020, 37, 243–247. [Google Scholar] [CrossRef]
- Asa, S.L. The current histologic classification of thyroid cancer. Endocrinol. Metab. Clin. N. Am. 2019, 48, 1–22. [Google Scholar] [CrossRef]
- Eloy, C.; Ferreira, L.; Salgado, C.; Soares, P.; Sobrinho-Simoes, M. Poorly differentiated and undifferentiated thyroid carcinomas. Turk. Patoloji. Derg. 2015, 31 (Suppl. S1), 48–59. [Google Scholar] [CrossRef][Green Version]
- Aashiq, M.; Silverman, D.A.; Na’ara, S.; Takahashi, H.; Amit, M. Radioiodine-refractory thyroid cancer: Molecular basis of redifferentiation therapies, management, and novel therapies. Cancers 2019, 11, 1382. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Lin, Y.; Liang, J. Radioactive iodine-refractory differentiated thyroid cancer and redifferentiation therapy. Endocrinol. Metab. 2019, 34, 215–225. [Google Scholar] [CrossRef]
- D’Andrea, A.D. Mechanisms of parp inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Lei, Y.H.; Yao, N.; Wang, C.R.; Hu, N.; Ye, W.C.; Zhang, D.M.; Chen, Z.S. Autophagy and multidrug resistance in cancer. Chin. J. Cancer 2017, 36, 52. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Yu, M.; Yin, D.; Sun, D.; Zhu, Y.; Bu, Y.; Sang, M. Impact of runx2 on drug-resistant human pancreatic cancer cells with p53 mutations. BMC Cancer 2018, 18, 309. [Google Scholar] [CrossRef] [PubMed]
- He, Y. Systematic response of staurosporine scaffold-based inhibitors to drug-resistant cancer kinase mutations. Arch. Pharm. 2020, 353, e1900320. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.D.; Kaufman, M.D.; Lu, W.P.; Gupta, A.; Leary, C.B.; Wise, S.C.; Rutkoski, T.J.; Ahn, Y.M.; Al-Ani, G.; Bulfer, S.L.; et al. Ripretinib (dcc-2618) is a switch control kinase inhibitor of a broad spectrum of oncogenic and drug-resistant kit and pdgfra variants. Cancer Cell 2019, 35, 738–751.e739. [Google Scholar] [CrossRef]
- Otte, J.; Dizdar, L.; Behrens, B.; Goering, W.; Knoefel, W.T.; Wruck, W.; Stoecklein, N.H.; Adjaye, J. Fgf signalling in the self-renewal of colon cancer organoids. Sci. Rep. 2019, 9, 17365. [Google Scholar] [CrossRef]
- Brown, W.S.; Akhand, S.S.; Wendt, M.K. Fgfr signaling maintains a drug persistent cell population following epithelial-mesenchymal transition. Oncotarget 2016, 7, 83424–83436. [Google Scholar] [CrossRef]
- Grygielewicz, P.; Dymek, B.; Bujak, A.; Gunerka, P.; Stanczak, A.; Lamparska-Przybysz, M.; Wieczorek, M.; Dzwonek, K.; Zdzalik, D. Epithelial-mesenchymal transition confers resistance to selective fgfr inhibitors in snu-16 gastric cancer cells. Gastric Cancer 2016, 19, 53–62. [Google Scholar] [CrossRef]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Brose, M.S.; Cabanillas, M.E.; Cohen, E.E.; Wirth, L.J.; Riehl, T.; Yue, H.; Sherman, S.I.; Sherman, E.J. Vemurafenib in patients with braf(v600e)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: A non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1272–1282. [Google Scholar] [CrossRef]
- Miftari, R.; Topciu, V.; Nura, A.; Haxhibeqiri, V. Management of the patient with aggressive and resistant papillary thyroid carcinoma. Med. Arch. 2016, 70, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Brini, M.; Carafoli, E. Calcium pumps in health and disease. Physiol. Rev. 2009, 89, 1341–1378. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Merritt, R.; Fu, L.; Pan, Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B 2017, 7, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Denmeade, S.R.; Isaacs, J.T. The serca pump as a therapeutic target: Making a “smart bomb” for prostate cancer. Cancer Biol. Ther. 2005, 4, 14–22. [Google Scholar] [CrossRef]
- Xu, S.; Nam, S.M.; Kim, J.H.; Das, R.; Choi, S.K.; Nguyen, T.T.; Quan, X.; Choi, S.J.; Chung, C.H.; Lee, E.Y.; et al. Palmitate induces er calcium depletion and apoptosis in mouse podocytes subsequent to mitochondrial oxidative stress. Cell Death. Dis. 2015, 6, e1976. [Google Scholar] [CrossRef]
- Chemaly, E.R.; Troncone, L.; Lebeche, D. Serca control of cell death and survival. Cell Calcium. 2018, 69, 46–61. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef]
- Pimentel, A.A.; Benaim, G. Ca2+ and sphingolipids as modulators for apoptosis and cancer. Investig. Clin. 2012, 53, 84–110. [Google Scholar]
- Seo, J.A.; Kim, B.; Dhanasekaran, D.N.; Tsang, B.K.; Song, Y.S. Curcumin induces apoptosis by inhibiting sarco/endoplasmic reticulum Ca2+ atpase activity in ovarian cancer cells. Cancer Lett. 2016, 371, 30–37. [Google Scholar] [CrossRef]
- Lim, J.H.; Park, K.; Choi, K.H.; Kim, C.W.; Lee, J.H.; Weicker, R.; Pan, C.H.; Kim, S.M.; Park, K.C. Drug discovery using evolutionary similarities in chemical binding to inhibit patient-derived hepatocellular carcinoma. Int. J. Mol. Sci. 2022, 23, 7971. [Google Scholar] [CrossRef]
- Bergdorf, K.; Ferguson, D.C.; Mehrad, M.; Ely, K.; Stricker, T.; Weiss, V.L. Papillary thyroid carcinoma behavior: Clues in the tumor microenvironment. Endocr. Relat. Cancer 2019, 26, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Lewinski, A.; Adamczewski, Z. Papillary thyroid carcinoma: A cancer with an extremely diverse genetic background and prognosis. Pol. Arch. Intern. Med. 2017, 127, 388–389. [Google Scholar] [CrossRef] [PubMed]
- Zelinskaya, A. Immunocytochemical characteristics of thyrocytes in radioiodine refractory metastases of papillary thyroid cancer. Exp. Oncol. 2019, 41, 342–345. [Google Scholar] [CrossRef]
- Stassi, G.; Todaro, M.; Zerilli, M.; Ricci-Vitiani, L.; Di Liberto, D.; Patti, M.; Florena, A.; Di Gaudio, F.; Di Gesu, G.; De Maria, R. Thyroid cancer resistance to chemotherapeutic drugs via autocrine production of interleukin-4 and interleukin-10. Cancer Res. 2003, 63, 6784–6790. [Google Scholar]
- Wilson, M.M.; Weinberg, R.A.; Lees, J.A.; Guen, V.J. Emerging mechanisms by which emt programs control stemness. Trends Cancer 2020, 6, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Kang, Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 2019, 49, 361–374. [Google Scholar] [CrossRef]
- Tsubakihara, Y.; Moustakas, A. Epithelial-mesenchymal transition and metastasis under the control of transforming growth factor beta. Int. J. Mol. Sci. 2018, 19, 3672. [Google Scholar] [CrossRef]
- Sarkar, F.H.; Li, Y.; Wang, Z.; Kong, D. Pancreatic cancer stem cells and emt in drug resistance and metastasis. Minerva Chir. 2009, 64, 489–500. [Google Scholar]
- Singh, A.; Settleman, J. Emt, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef]
- Chang, J.C. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine 2016, 95, S20–S25. [Google Scholar] [CrossRef]
- de Groot, J.W.; Links, T.P.; Plukker, J.T.; Lips, C.J.; Hofstra, R.M. Ret as a diagnostic and therapeutic target in sporadic and hereditary endocrine tumors. Endocr. Rev. 2006, 27, 535–560. [Google Scholar]
- Ivan, M.; Bond, J.A.; Prat, M.; Comoglio, P.M.; Wynford-Thomas, D. Activated ras and ret oncogenes induce over-expression of c-met (hepatocyte growth factor receptor) in human thyroid epithelial cells. Oncogene 1997, 14, 2417–2423. [Google Scholar] [CrossRef] [PubMed]
- Kerr, A.J.; Dodwell, D.; McGale, P.; Holt, F.; Duane, F.; Mannu, G.; Darby, S.C.; Taylor, C.W. Adjuvant and neoadjuvant breast cancer treatments: A systematic review of their effects on mortality. Cancer Treat. Rev. 2022, 105, 102375. [Google Scholar] [CrossRef] [PubMed]
- Ettrich, T.J.; Sturm, N.; Guthle, M.; Huttner, F.J.; Perkhofer, L. Pancreatic cancer: Current multimodality treatment options and the future impact of molecular biological profiling. Visc. Med. 2022, 38, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, V.; Stintzing, S. Neoadjuvant and adjuvant therapy of resectable colon cancer—Current standards and developments. Dtsch. Med. Wochenschr. 2021, 146, 1457–1467. [Google Scholar]
- Friedlaender, A.; Naidoo, J.; Banna, G.L.; Metro, G.; Forde, P.; Addeo, A. Role and impact of immune checkpoint inhibitors in neoadjuvant treatment for nsclc. Cancer Treat. Rev. 2022, 104, 102350. [Google Scholar] [CrossRef] [PubMed]
- Akateh, C.; Black, S.M.; Conteh, L.; Miller, E.D.; Noonan, A.; Elliott, E.; Pawlik, T.M.; Tsung, A.; Cloyd, J.M. Neoadjuvant and adjuvant treatment strategies for hepatocellular carcinoma. World J. Gastroenterol. 2019, 25, 3704–3721. [Google Scholar] [CrossRef]
- Foerster, F.; Galle, P.R. The current landscape of clinical trials for systemic treatment of hcc. Cancers 2021, 13, 1962. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Xavier, C.P.; Pesic, M.; Vasconcelos, M.H. Understanding cancer drug resistance by developing and studying resistant cell line models. Curr. Cancer Drug Targets 2016, 16, 226–237. [Google Scholar] [CrossRef]
- Nedeljkovic, M.; Damjanovic, A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells 2019, 8, 975. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, M.; Pavlopoulou, A.; Georgakilas, A.G.; Kyrodimos, E. The challenge of drug resistance in cancer treatment: A current overview. Clin. Exp. Metastasis 2018, 35, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- van Zyl, B.; Tang, D.; Bowden, N.A. Biomarkers of platinum resistance in ovarian cancer: What can we use to improve treatment. Endocr. Relat. Cancer 2018, 25, R303–R318. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Wang, J.; Ji, X.; Cao, H.; Zhu, J.; Chen, Y.; Yang, J.; Zhao, Z.; Ren, T.; Xing, J. Mcur1 facilitates epithelial-mesenchymal transition and metastasis via the mitochondrial calcium dependent ros/nrf2/notch pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 136. [Google Scholar] [CrossRef]
- Sun, J.; Ailiman, M. Regulation of calcium pump through notch/jagged/hes signaling pathway in canine model of chronic atrial fibrillation. Int. J. Clin. Exp. Pathol. 2019, 12, 4034–4040. [Google Scholar] [PubMed]
- Pagliaro, L.; Marchesini, M.; Roti, G. Targeting oncogenic notch signaling with serca inhibitors. J. Hematol. Oncol. 2021, 14, 8. [Google Scholar] [CrossRef]
- Marchesini, M.; Gherli, A.; Montanaro, A.; Patrizi, L.; Sorrentino, C.; Pagliaro, L.; Rompietti, C.; Kitara, S.; Heit, S.; Olesen, C.E.; et al. Blockade of oncogenic notch1 with the serca inhibitor cad204520 in t cell acute lymphoblastic leukemia. Cell Chem. Biol. 2020, 27, 678–697.e13. [Google Scholar] [CrossRef]
- Park, K.C.; Kim, S.W.; Jeon, J.Y.; Jo, A.R.; Choi, H.J.; Kim, J.; Lee, H.G.; Kim, Y.; Mills, G.B.; Noh, S.H.; et al. Survival of cancer stem-like cells under metabolic stress via camk2alpha-mediated upregulation of sarco/endoplasmic reticulum calcium atpase expression. Clin. Cancer Res. 2018, 24, 1677–1690. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of rna-seq experiments with hisat, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-M.; Park, K.; Lim, J.H.; Yun, H.J.; Kim, S.Y.; Choi, K.H.; Kim, C.W.; Lee, J.H.; Weicker, R.; Pan, C.-H.; et al. Potential Therapeutic Agents against Paclitaxel—And Sorafenib-Resistant Papillary Thyroid Carcinoma. Int. J. Mol. Sci. 2022, 23, 10378. https://doi.org/10.3390/ijms231810378
Kim S-M, Park K, Lim JH, Yun HJ, Kim SY, Choi KH, Kim CW, Lee JH, Weicker R, Pan C-H, et al. Potential Therapeutic Agents against Paclitaxel—And Sorafenib-Resistant Papillary Thyroid Carcinoma. International Journal of Molecular Sciences. 2022; 23(18):10378. https://doi.org/10.3390/ijms231810378
Chicago/Turabian StyleKim, Seok-Mo, Keunwan Park, Jin Hong Lim, Hyeok Jun Yun, Sang Yong Kim, Kyung Hwa Choi, Chan Wung Kim, Jae Ha Lee, Raymond Weicker, Cheol-Ho Pan, and et al. 2022. "Potential Therapeutic Agents against Paclitaxel—And Sorafenib-Resistant Papillary Thyroid Carcinoma" International Journal of Molecular Sciences 23, no. 18: 10378. https://doi.org/10.3390/ijms231810378
APA StyleKim, S.-M., Park, K., Lim, J. H., Yun, H. J., Kim, S. Y., Choi, K. H., Kim, C. W., Lee, J. H., Weicker, R., Pan, C.-H., & Park, K. C. (2022). Potential Therapeutic Agents against Paclitaxel—And Sorafenib-Resistant Papillary Thyroid Carcinoma. International Journal of Molecular Sciences, 23(18), 10378. https://doi.org/10.3390/ijms231810378





