Common and Rare PCSK9 Variants Associated with Low-Density Lipoprotein Cholesterol Levels and the Risk of Diabetes Mellitus: A Mendelian Randomization Study
Abstract
1. Introduction
2. Results
2.1. Regional Plot Association Analysis for the PCSK9 Region
2.2. Association between Rare PCSK9 Nonsynonymous Exonic Mutations and LDL-C Levels
2.3. Stepwise Linear Regression Analysis
2.4. Association between LDL-C Levels and DM Status
2.5. GWAS Analysis for LDL-C Levels
2.6. Association between LDL-C-Level-Associated PCSK9 Genotypes and Clinical and Laboratory Parameters
2.7. MR Analysis along with 2SLS IV Regression for Determining the Association of Genetic Determinants of LDL-C Levels with DM Status
2.8. Scatter Plots and Test for Heterogeneity
2.9. Sensitivity Analysis for Causal Inference from Standard Mendelian Randomization with Multiple Genetic Variants Determining LDL-C Levels
3. Discussion
3.1. Ethnic Heterogeneity of PCSK9 Gain-of-Function and Loss-of-Function Mutations
3.2. Role of Noncoding PCSK9 Variants
3.3. Mendelian Randomization for LDL-C and DM
3.4. Limitations
4. Methods and Materials
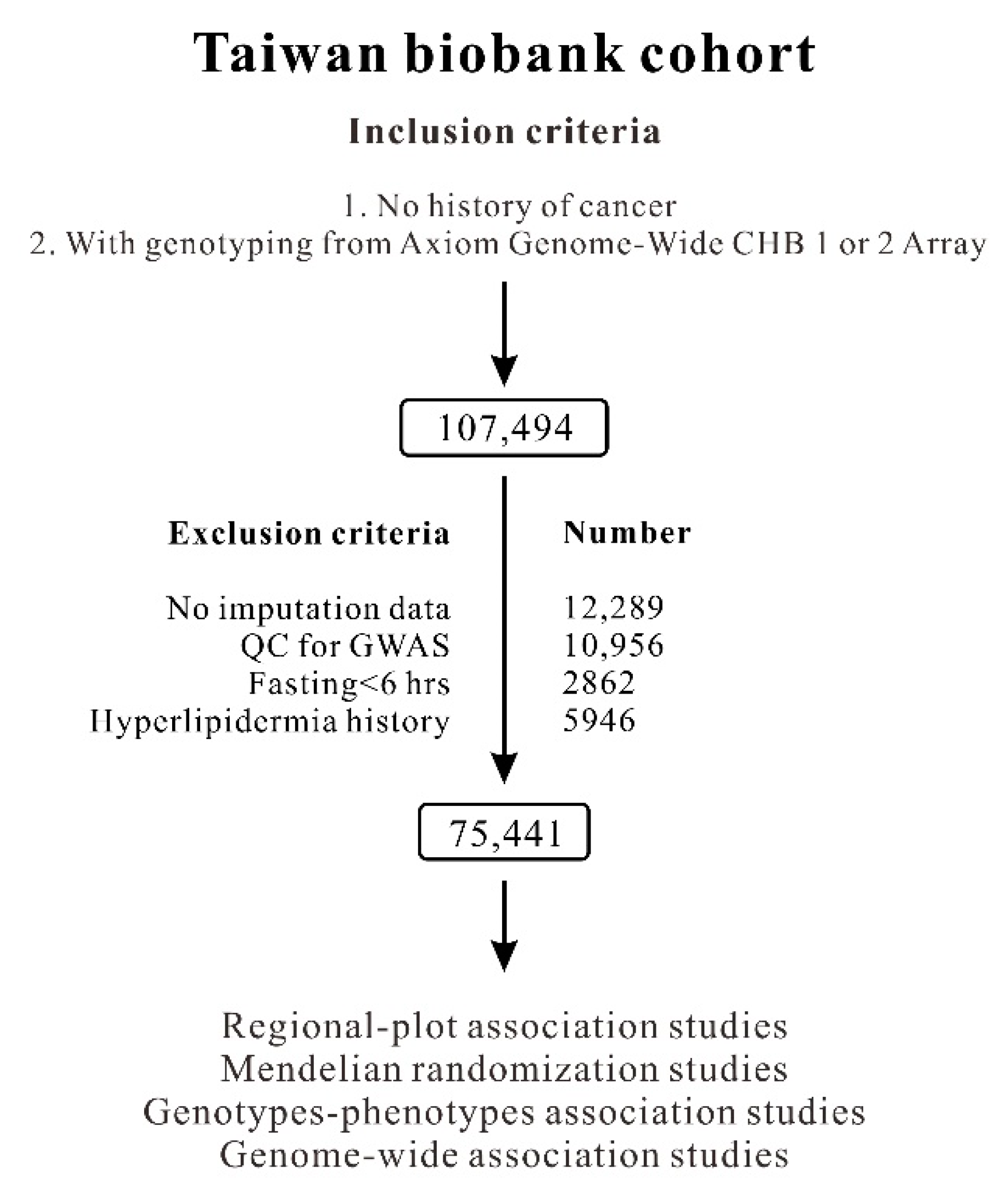
4.1. TWB Participants
4.2. Genomic DNA Extraction and Genotyping
4.3. Clinical Phenotypes and Laboratory Examinations
4.4. Regional Plot Analysis and GWAS
4.5. Statistical Analysis
4.6. MR Analysis
4.7. Scatterplots and Testing for Heterogeneity
4.8. Sensitivity Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomas, H.; Diamond, J.; Vieco, A.; Chaudhuri, S.; Shinnar, E.; Cromer, S.; Perel, P.; Mensah, G.A.; Narula, J.; Johnson, C.O.; et al. Global Atlas of Cardiovascular Disease 2000–2016: The Path to Prevention and Control. Glob. Heart 2018, 13, 143–163. [Google Scholar] [CrossRef]
- Mihaylova, B.; Emberson, J.; Blackwell, L.; Keech, A.; Simes, J.; Barnes, E.H.; Voysey, M.; Gray, A.; Collins, R.; Baigent, C. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: Meta-analysis of individual data from 27 randomised trials. Lancet 2012, 380, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Baigent, C.; Blackwell, L.; Emberson, J.; Holland, L.E.; Reith, C.; Bhala, N.; Peto, R.; Barnes, E.H.; Keech, A.; Simes, J.; et al. Efficacy and safety of more intensive lowering of LDL cholesterol: A meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet 2010, 376, 1670–1681. [Google Scholar] [CrossRef] [PubMed]
- Boekholdt, S.M.; Arsenault, B.J.; Mora, S.; Pedersen, T.R.; LaRosa, J.C.; Nestel, P.J.; Simes, R.J.; Durrington, P.; Hitman, G.A.; Welch, K.M.; et al. Association of LDL cholesterol, non-HDL cholesterol, and apolipoprotein B levels with risk of cardiovascular events among patients treated with statins: A meta-analysis. JAMA 2012, 307, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, J.; O’Connell, R.; Voysey, M.; Emberson, J.; Blackwell, L.; Mihaylova, B.; Simes, J.; Collins, R.; Kirby, A.; Colhoun, H.; et al. Efficacy and safety of LDL-lowering therapy among men and women: Meta-analysis of individual data from 174,000 participants in 27 randomised trials. Lancet 2015, 385, 1397–1405. [Google Scholar] [CrossRef]
- Luirink, I.K.; Wiegman, A.; Kusters, D.M.; Hof, M.H.; Groothoff, J.W.; de Groot, E.; Kastelein, J.J.P.; Hutten, B.A. 20-Year Follow-up of Statins in Children with Familial Hypercholesterolemia. N. Engl. J. Med. 2019, 381, 1547–1556. [Google Scholar] [CrossRef]
- Silverman, M.G.; Ference, B.A.; Im, K.; Wiviott, S.D.; Giugliano, R.P.; Grundy, S.M.; Braunwald, E.; Sabatine, M.S. Association Between Lowering LDL-C and Cardiovascular Risk Reduction Among Different Therapeutic Interventions: A Systematic Review and Meta-analysis. JAMA 2016, 316, 1289–1297. [Google Scholar] [CrossRef]
- Yusuf, S.; Lonn, E.; Pais, P.; Bosch, J.; López-Jaramillo, P.; Zhu, J.; Xavier, D.; Avezum, A.; Leiter, L.A.; Piegas, L.S.; et al. Blood-Pressure and Cholesterol Lowering in Persons without Cardiovascular Disease. N. Engl. J. Med. 2016, 374, 2032–2043. [Google Scholar] [CrossRef]
- Ference, B.A.; Robinson, J.G.; Brook, R.D.; Catapano, A.L.; Chapman, M.J.; Neff, D.R.; Voros, S.; Giugliano, R.P.; Davey Smith, G.; Fazio, S.; et al. Variation in PCSK9 and HMGCR and Risk of Cardiovascular Disease and Diabetes. N. Engl. J. Med. 2016, 375, 2144–2153. [Google Scholar] [CrossRef]
- Schmidt, A.F.; Holmes, M.V.; Preiss, D.; Swerdlow, D.I.; Denaxas, S.; Fatemifar, G.; Faraway, R.; Finan, C.; Valentine, D.; Fairhurst-Hunter, Z.; et al. Phenome-wide association analysis of LDL-cholesterol lowering genetic variants in PCSK9. BMC Cardiovasc. Disord. 2019, 19, 240. [Google Scholar] [CrossRef]
- Tragante, V.; Asselbergs, F.W.; Swerdlow, D.I.; Palmer, T.M.; Moore, J.H.; de Bakker, P.I.W.; Keating, B.J.; Holmes, M.V. Harnessing publicly available genetic data to prioritize lipid modifying therapeutic targets for prevention of coronary heart disease based on dysglycemic risk. Hum. Genet. 2016, 135, 453–467. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Swerdlow, D.I.; Preiss, D.; Fairhurst-Hunter, Z.; Keating, B.J.; Asselbergs, F.W.; Sattar, N.; Humphries, S.E.; Hingorani, A.D.; Holmes, M.V. Association of Lipid Fractions With Risks for Coronary Artery Disease and Diabetes. JAMA Cardiol. 2016, 1, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Kostapanos, M.S.; Agouridis, A.P.; Elisaf, M.S. Variable effects of statins on glucose homeostasis parameters and their diabetogenic role. Diabetologia 2015, 58, 1960–1961. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Navarese, E.P.; Buffon, A.; Andreotti, F.; Kozinski, M.; Welton, N.; Fabiszak, T.; Caputo, S.; Grzesk, G.; Kubica, A.; Swiatkiewicz, I.; et al. Meta-analysis of impact of different types and doses of statins on new-onset diabetes mellitus. Am. J. Cardiol. 2013, 111, 1123–1130. [Google Scholar] [CrossRef]
- Ridker, P.M.; Danielson, E.; Fonseca, F.A.; Genest, J.; Gotto, A.M., Jr.; Kastelein, J.J.; Koenig, W.; Libby, P.; Lorenzatti, A.J.; MacFadyen, J.G.; et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N. Engl. J. Med. 2008, 359, 2195–2207. [Google Scholar] [CrossRef]
- Sattar, N.; Preiss, D.; Murray, H.M.; Welsh, P.; Buckley, B.M.; de Craen, A.J.; Seshasai, S.R.; McMurray, J.J.; Freeman, D.J.; Jukema, J.W.; et al. Statins and risk of incident diabetes: A collaborative meta-analysis of randomised statin trials. Lancet 2010, 375, 735–742. [Google Scholar] [CrossRef]
- Filippatos, T.D.; Panagiotopoulou, T.; Tzavella, E.; Elisaf, M.S. Hypolipidemic Drugs and Diabetes Mellitus-Mechanisms and Data From Genetic Trials. J. Cardiovasc. Pharmacol. Ther. 2018, 23, 187–191. [Google Scholar] [CrossRef]
- Schmidt, A.F.; Swerdlow, D.I.; Holmes, M.V.; Patel, R.S.; Fairhurst-Hunter, Z.; Lyall, D.M.; Hartwig, F.P.; Horta, B.L.; Hyppönen, E.; Power, C.; et al. PCSK9 genetic variants and risk of type 2 diabetes: A mendelian randomisation study. Lancet. Diabetes Endocrinol. 2017, 5, 97–105. [Google Scholar] [CrossRef]
- Swerdlow, D.I.; Preiss, D.; Kuchenbaecker, K.B.; Holmes, M.V.; Engmann, J.E.; Shah, T.; Sofat, R.; Stender, S.; Johnson, P.C.; Scott, R.A.; et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: Evidence from genetic analysis and randomised trials. Lancet 2015, 385, 351–361. [Google Scholar] [CrossRef]
- Lotta, L.A.; Sharp, S.J.; Burgess, S.; Perry, J.R.B.; Stewart, I.D.; Willems, S.M.; Luan, J.; Ardanaz, E.; Arriola, L.; Balkau, B.; et al. Association Between Low-Density Lipoprotein Cholesterol-Lowering Genetic Variants and Risk of Type 2 Diabetes: A Meta-analysis. JAMA 2016, 316, 1383–1391. [Google Scholar] [CrossRef]
- Pan, W.; Sun, W.; Yang, S.; Zhuang, H.; Jiang, H.; Ju, H.; Wang, D.; Han, Y. LDL-C plays a causal role on T2DM: A Mendelian randomization analysis. Aging 2020, 12, 2584–2594. [Google Scholar] [CrossRef] [PubMed]
- Monami, M.; Sesti, G.; Mannucci, E. PCSK9 inhibitor therapy: A systematic review and meta-analysis of metabolic and cardiovascular outcomes in patients with diabetes. Diabetes Obes. Metab. 2019, 21, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Barale, C.; Melchionda, E.; Morotti, A.; Russo, I. PCSK9 Biology and Its Role in Atherothrombosis. Int. J. Mol. Sci. 2021, 22, 5880. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, S.S.; Doring, Y.; van der Vorst, E.P.C. PCSK9: A Multi-Faceted Protein That Is Involved in Cardiovascular Biology. Biomedicines 2021, 9, 793. [Google Scholar] [CrossRef]
- Seidah, N.G.; Benjannet, S.; Wickham, L.; Marcinkiewicz, J.; Jasmin, S.B.; Stifani, S.; Basak, A.; Prat, A.; Chretien, M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): Liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA 2003, 100, 928–933. [Google Scholar] [CrossRef]
- Ragusa, R.; Basta, G.; Neglia, D.; De Caterina, R.; Del Turco, S.; Caselli, C. PCSK9 and atherosclerosis: Looking beyond LDL regulation. Eur. J. Clin. Investig. 2021, 51, e13459. [Google Scholar] [CrossRef]
- Norata, G.D.; Tavori, H.; Pirillo, A.; Fazio, S.; Catapano, A.L. Biology of proprotein convertase subtilisin kexin 9: Beyond low-density lipoprotein cholesterol lowering. Cardiovasc. Res. 2016, 112, 429–442. [Google Scholar] [CrossRef]
- Hori, M.; Ohta, N.; Takahashi, A.; Masuda, H.; Isoda, R.; Yamamoto, S.; Son, C.; Ogura, M.; Hosoda, K.; Miyamoto, Y.; et al. Impact of LDLR and PCSK9 pathogenic variants in Japanese heterozygous familial hypercholesterolemia patients. Atherosclerosis 2019, 289, 101–108. [Google Scholar] [CrossRef]
- Hsu, L.A.; Teng, M.S.; Ko, Y.L.; Chang, C.J.; Wu, S.; Wang, C.L.; Hu, C.F. The PCSK9 gene E670G polymorphism affects low-density lipoprotein cholesterol levels but is not a risk factor for coronary artery disease in ethnic Chinese in Taiwan. Clin. Chem. Lab. Med. 2009, 47, 154–158. [Google Scholar] [CrossRef]
- Huang, C.C.; Niu, D.M.; Charng, M.J. Genetic Analysis in a Taiwanese Cohort of 750 Index Patients with Clinically Diagnosed Familial Hypercholesterolemia. J. Atheroscler. Thromb. 2022, 29, 639–653. [Google Scholar] [CrossRef]
- Kim, H.I.; Ye, B.; Gosalia, N.; Köroğlu, Ç.; Hanson, R.L.; Hsueh, W.C.; Knowler, W.C.; Baier, L.J.; Bogardus, C.; Shuldiner, A.R.; et al. Characterization of Exome Variants and Their Metabolic Impact in 6716 American Indians from the Southwest US. Am. J. Hum. Genet. 2020, 107, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; Lee, Y.; Park, S.; Kang, S.M.; Jang, Y.; Lee, J.H.; Lee, S.H. Rare and common variants of APOB and PCSK9 in Korean patients with extremely low low-density lipoprotein-cholesterol levels. PLoS ONE 2017, 12, e0186446. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, J.; Li, H.; Chen, Y.; Wang, L.; He, M.; Wang, Y.; Sun, L.; Hu, Y.; Huang, J.; et al. Coding-sequence variants are associated with blood lipid levels in 14,473 Chinese. Hum. Mol. Genet. 2016, 25, 4107–4116. [Google Scholar] [CrossRef]
- Moon, S.; Kim, Y.J.; Han, S.; Hwang, M.Y.; Shin, D.M.; Park, M.Y.; Lu, Y.; Yoon, K.; Jang, H.M.; Kim, Y.K.; et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Sci. Rep. 2019, 9, 1382. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Zeng, P.; Li, X.; Zhang, Z.; Pan, B.; Peng, Z.Y.F.; Li, Y.; Ma, Y.; Leng, Y.; Chen, R. What is the impact of PCSK9 rs505151 and rs11591147 polymorphisms on serum lipids level and cardiovascular risk: A meta-analysis. Lipids Health Dis. 2017, 16, 111. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.S.; Zhang, H.; Cheung, C.Y.; Xu, M.; Ho, J.C.; Zhou, W.; Cherny, S.S.; Zhang, Y.; Holmen, O.; Au, K.W.; et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun. 2015, 6, 10206. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Kullo, I.J. Molecular population genetics of PCSK9: A signature of recent positive selection. Pharm. Genom. 2008, 18, 169–179. [Google Scholar] [CrossRef]
- Guella, I.; Asselta, R.; Ardissino, D.; Merlini, P.A.; Peyvandi, F.; Kathiresan, S.; Mannucci, P.M.; Tubaro, M.; Duga, S. Effects of PCSK9 genetic variants on plasma LDL cholesterol levels and risk of premature myocardial infarction in the Italian population. J. Lipid Res. 2010, 51, 3342–3349. [Google Scholar] [CrossRef]
- Musunuru, K.; Romaine, S.P.; Lettre, G.; Wilson, J.G.; Volcik, K.A.; Tsai, M.Y.; Taylor, H.A., Jr.; Schreiner, P.J.; Rotter, J.I.; Rich, S.S.; et al. Multi-ethnic analysis of lipid-associated loci: The NHLBI CARe project. PLoS ONE 2012, 7, e36473. [Google Scholar] [CrossRef]
- Chen, C.H.; Yang, J.H.; Chiang, C.W.K.; Hsiung, C.N.; Wu, P.E.; Chang, L.C.; Chu, H.W.; Chang, J.; Song, I.W.; Yang, S.L.; et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet. 2016, 25, 5321–5331. [Google Scholar] [CrossRef]
- Fan, C.T.; Lin, J.C.; Lee, C.H. Taiwan Biobank: A project aiming to aid Taiwan’s transition into a biomedical island. Pharmacogenomics 2008, 9, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Pu, T.; Zhang, Y.; Yan, H.; Yu, H.; Gao, W. The R93C Variant of PCSK9 Reduces the Risk of Premature MI in a Chinese Han Population. Front. Genet. 2022, 13, 875269. [Google Scholar] [CrossRef] [PubMed]
- Lacaze, P.; Riaz, M.; Sebra, R.; Hooper, A.J.; Pang, J.; Tiller, J.; Polekhina, G.; Tonkin, A.; Reid, C.; Zoungas, S.; et al. Protective lipid-lowering variants in healthy older individuals without coronary heart disease. Open Heart 2021, 8, e001710. [Google Scholar] [CrossRef] [PubMed]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Lye, S.H.; Chahil, J.K.; Bagali, P.; Alex, L.; Vadivelu, J.; Ahmad, W.A.; Chan, S.P.; Thong, M.K.; Zain, S.M.; Mohamed, R. Genetic polymorphisms in LDLR, APOB, PCSK9 and other lipid related genes associated with familial hypercholesterolemia in Malaysia. PLoS ONE 2013, 8, e60729. [Google Scholar] [CrossRef]
- Fall, T.; Xie, W.; Poon, W.; Yaghootkar, H.; Mägi, R.; Knowles, J.W.; Lyssenko, V.; Weedon, M.; Frayling, T.M.; Ingelsson, E. Using Genetic Variants to Assess the Relationship Between Circulating Lipids and Type 2 Diabetes. Diabetes 2015, 64, 2676–2684. [Google Scholar] [CrossRef]
- Besseling, J.; Kastelein, J.J.; Defesche, J.C.; Hutten, B.A.; Hovingh, G.K. Association between familial hypercholesterolemia and prevalence of type 2 diabetes mellitus. JAMA 2015, 313, 1029–1036. [Google Scholar] [CrossRef]
- Xu, H.; Ryan, K.A.; Jaworek, T.J.; Southam, L.; Reid, J.G.; Overton, J.D.; Baras, A.; Puurunen, M.K.; Zeggini, E.; Taylor, S.I.; et al. Familial Hypercholesterolemia and Type 2 Diabetes in the Old Order Amish. Diabetes 2017, 66, 2054–2058. [Google Scholar] [CrossRef]
- Liu, D.J.; Peloso, G.M.; Yu, H.; Butterworth, A.S.; Wang, X.; Mahajan, A.; Saleheen, D.; Emdin, C.; Alam, D.; Alves, A.C.; et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017, 49, 1758–1766. [Google Scholar] [CrossRef]
- Hsu, L.A.; Chou, H.H.; Teng, M.S.; Wu, S.; Ko, Y.L. Circulating chemerin levels are determined through circulating platelet counts in nondiabetic Taiwanese people: A bidirectional Mendelian randomization study. Atherosclerosis 2021, 320, 61–69. [Google Scholar] [CrossRef]
- Boughton, A.P.; Welch, R.P.; Flickinger, M.; VandeHaar, P.; Taliun, D.; Abecasis, G.R.; Boehnke, M. LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics 2021, 37, 3017–3018. [Google Scholar] [CrossRef] [PubMed]
- Palmer, T.M.; Lawlor, D.A.; Harbord, R.M.; Sheehan, N.A.; Tobias, J.H.; Timpson, N.J.; Davey Smith, G.; Sterne, J.A. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 2012, 21, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.Y.; Chan, K.C.; Hsu, L. Testing concordance of instrumental variable effects in generalized linear models with application to Mendelian randomization. Stat. Med. 2014, 33, 3986–4007. [Google Scholar] [CrossRef]
- Bowden, J.; Spiller, W.; Del Greco, M.F.; Sheehan, N.; Thompson, J.; Minelli, C.; Davey Smith, G. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int. J. Epidemiol. 2018, 47, 1264–1278. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith, S.C., Jr.; et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 2005, 11, 2735–2752. [Google Scholar] [CrossRef]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian Randomization Analysis with Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Johnson, T.; Uk, S. Efficient Calculation for Multi-SNP Genetic Risk Scores. 2012. Available online: https://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.398.7674 (accessed on 24 May 2022).
- Suurmond, R.; van Rhee, H.; Hak, T. Introduction, comparison, and validation of Meta-Essentials: A free and simple tool for meta-analysis. Res. Synth. Methods 2017, 8, 537–553. [Google Scholar] [CrossRef] [PubMed]

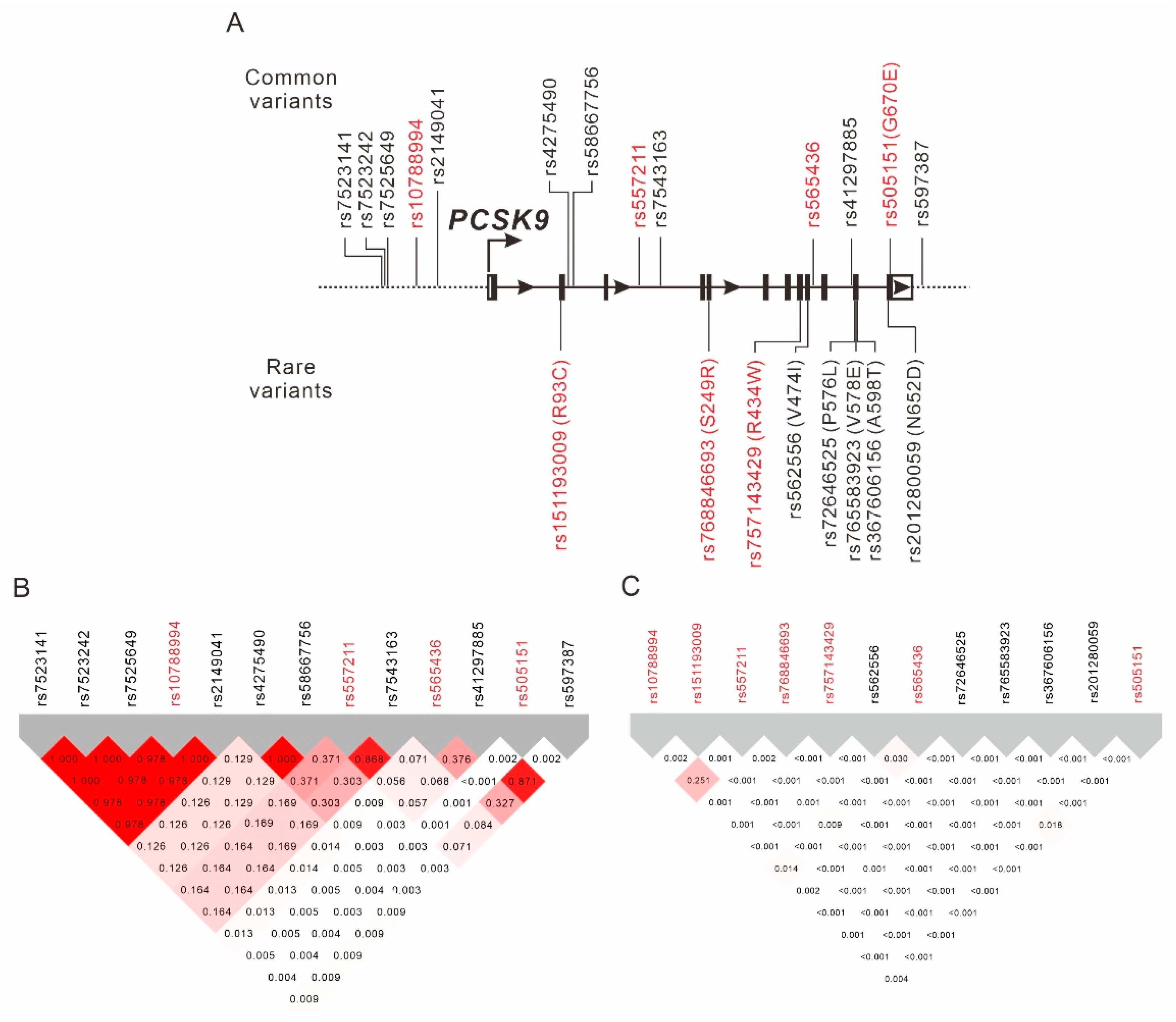
| PCSK9 Variants | Chr | Position | Ref/Alt | Func.refGene | Gene.refGene | HWE | MAF | MM | Mm | mm | p Value | Beta | SE | p * Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10788994 | 1 | 55,500,976 | C/T | intergenic | BSND;PCSK9 | 0.7616 | 0.3475 | 121.80 ± 31.26 (31,728) | 120.41 ± 30.84 (33,743) | 119.23 ± 30.86 (9015) | 6.44 × 10−13 | −0.0047 | 0.0006 | 1.99 × 10−14 |
| rs151193009 | 1 | 55,509,585 | C/T | exon2:c.C277T:p.R93C | PCSK9 | 0.4419 | 0.0027 | 120.98 ± 30.98 (73,449) | 106.76 ± 28.53 (405) | -- | 1.76 × 10−18 | −0.0579 | 0.0057 | 1.19 × 10−24 |
| rs557211 | 1 | 55,514,215 | T/G | Intron Variant | PCSK9 | 0.8433 | 0.1916 | 121.42 ± 31.179 (49,044) | 119.81 ± 30.701 (23,300) | 119.30 ± 30.654 (2740) | 1.69 × 10−11 | −0.0052 | 0.0007 | 3.75 × 10−12 |
| rs768846693 | 1 | 55,518,412 | C/A | exon5:c.C747A:p.S249R | PCSK9 | 0.9078 | 0.0004 | 120.88 ± 31.02 (75,324) | 91.73 ± 28.18 (63) | -- | 1.18 × 10−9 | −0.1301 | 0.0143 | 1.12 × 10−19 |
| rs757143429 | 1 | 55,523,828 | C/T | exon8:c.C1300T:p.R434W | PCSK9 | 0.7454 | 0.0011 | 120.89 ± 31.02 (75,239) | 107.81 ± 30.25 (175) | -- | 1.47 × 10−7 | −0.0509 | 0.0086 | 3.46 × 10−9 |
| rs565436 | 1 | 55,524,601 | G/A | Intron Variant | PCSK9 | 0.3577 | 0.1033 | 121.22 ± 31.11 (60,123) | 119.41 ±30.65 (13,901) | 118.19 ± 31.05 (780) | 8.08 × 10−10 | −0.0067 | 0.0010 | 3.81 × 10−12 |
| rs505151 | 1 | 55529187 | G/A | exon12:c.G2009A:p.G670E | PCSK9 | 0.9202 | 0.0534 | 119.0 ± 31.00 (67,530) | 120.00 ± 31.21 (7603) | 121.00 ± 32.37 (215) | 6.00 × 10−6 | 0.0065 | 0.0013 | 5.89 × 10−7 |
| Serum Total Cholesterol Level | Serum LDL-C Level | |||||
|---|---|---|---|---|---|---|
| Beta | r2 | p Value | Beta | r2 | p Value | |
| Age (years) | 0.0014 | 0.0362 | <10−307 | 0.0015 | 0.0186 | <10−307 |
| Sex (male vs. female) | 0.0154 | 0.0049 | 1.15 × 10−131 | -- | -- | -- |
| Body mass index (kg/m2) | 0.0017 | 0.0066 | 1.09 × 10−108 | 0.0051 | 0.0268 | <10−307 |
| Current smoking status (%) | 0.0060 | 0.0004 | 8.49 × 10−9 | -- | -- | -- |
| rs10788994 (TT vs.TC vs. CC) | −0.0022 | 0.0007 | 9.21 × 10−6 | −0.0039 | 0.0009 | 6.80 × 10−8 |
| rs151193009 (CC vs. CT) | −0.0388 | 0.0013 | 1.47 × 10−24 | −0.0615 | 0.0014 | 3.03 × 10−27 |
| rs557211 (TT vs.TG vs. GG) | −0.0018 | 0.0001 | 0.0024 | −0.0024 | 0.0001 | 0.0052 |
| rs768846693 (CC vs. CA) | −0.0662 | 0.0006 | 8.4 × 10−12 | −0.1261 | 0.0011 | 3.87 × 10−18 |
| rs757143429 (CC vs. CT) | −0.0310 | 0.0004 | 7.69 × 10−8 | −0.0511 | 0.0005 | 3.43 × 10−9 |
| rs565436 (AA vs. AG vs. GG) | −0.0038 | 0.0005 | 7.55 × 10−9 | −0.0060 | 0.0005 | 1.19 × 10−9 |
| rs505151 (AA vs. AG vs. GG) | 0.0036 | 0.0002 | 0.0001 | 0.0065 | 0.0003 | 1.23 × 10−6 |
| TA | TB | GA | TA-TB | GA-TA | GA-TB | IVA-TB | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p a | Beta | SE | p a | Beta | SE | p a | Beta | SE | p a (p b) | |||
| LDL-C | DM | WGRS_PCSK9_7SNVs | −1.9993 | 0.1185 | 6.76 × 10−64 | 0.5599 | 0.0301 | 4.66 × 10−77 | −2.3685 | 0.8918 | 0.0079 | −4.2294 | 1.5926 | 0.0079 (0.0098 c) |
| WGRS_LDL-C_41SNVs | −1.9993 | 0.1185 | 6.76 × 10−64 | 0.9823 | 0.0202 | <10−307 | −1.9743 | 0.6972 | 0.0046 | −1.9710 | 0.6961 | 0.0046 (5.02 × 10−7 c) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, L.-A.; Teng, M.-S.; Wu, S.; Chou, H.-H.; Ko, Y.-L. Common and Rare PCSK9 Variants Associated with Low-Density Lipoprotein Cholesterol Levels and the Risk of Diabetes Mellitus: A Mendelian Randomization Study. Int. J. Mol. Sci. 2022, 23, 10418. https://doi.org/10.3390/ijms231810418
Hsu L-A, Teng M-S, Wu S, Chou H-H, Ko Y-L. Common and Rare PCSK9 Variants Associated with Low-Density Lipoprotein Cholesterol Levels and the Risk of Diabetes Mellitus: A Mendelian Randomization Study. International Journal of Molecular Sciences. 2022; 23(18):10418. https://doi.org/10.3390/ijms231810418
Chicago/Turabian StyleHsu, Lung-An, Ming-Sheng Teng, Semon Wu, Hsin-Hua Chou, and Yu-Lin Ko. 2022. "Common and Rare PCSK9 Variants Associated with Low-Density Lipoprotein Cholesterol Levels and the Risk of Diabetes Mellitus: A Mendelian Randomization Study" International Journal of Molecular Sciences 23, no. 18: 10418. https://doi.org/10.3390/ijms231810418
APA StyleHsu, L.-A., Teng, M.-S., Wu, S., Chou, H.-H., & Ko, Y.-L. (2022). Common and Rare PCSK9 Variants Associated with Low-Density Lipoprotein Cholesterol Levels and the Risk of Diabetes Mellitus: A Mendelian Randomization Study. International Journal of Molecular Sciences, 23(18), 10418. https://doi.org/10.3390/ijms231810418





