Loss of Serpina1 in Mice Leads to Altered Gene Expression in Inflammatory and Metabolic Pathways
Abstract
:1. Introduction
2. Results
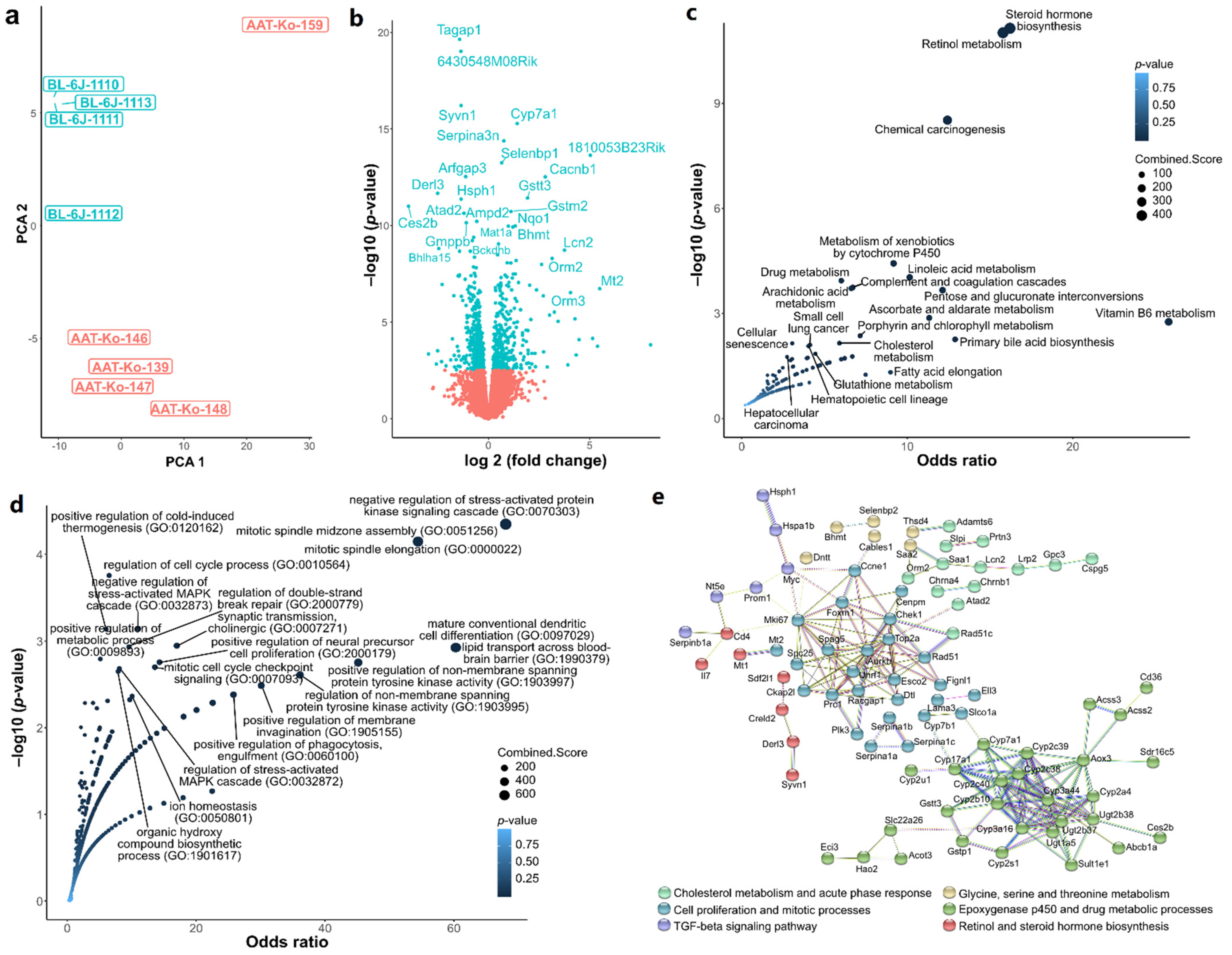
2.1. Transcriptome Quality Control and Differential Gene Expression
2.2. Hypothesis-Free Pathway Screening
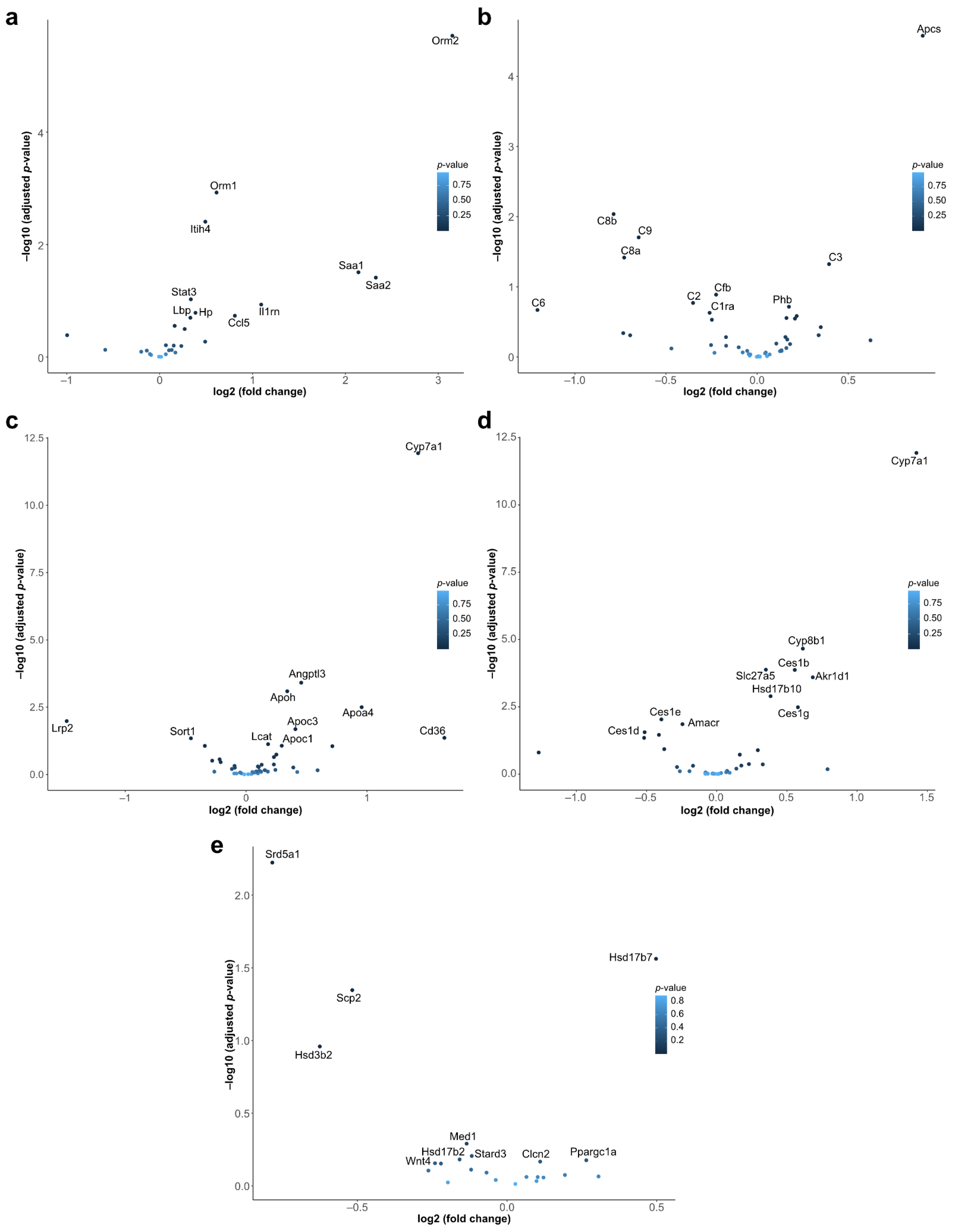
2.3. Hypothesis-Driven Pathway Analysis
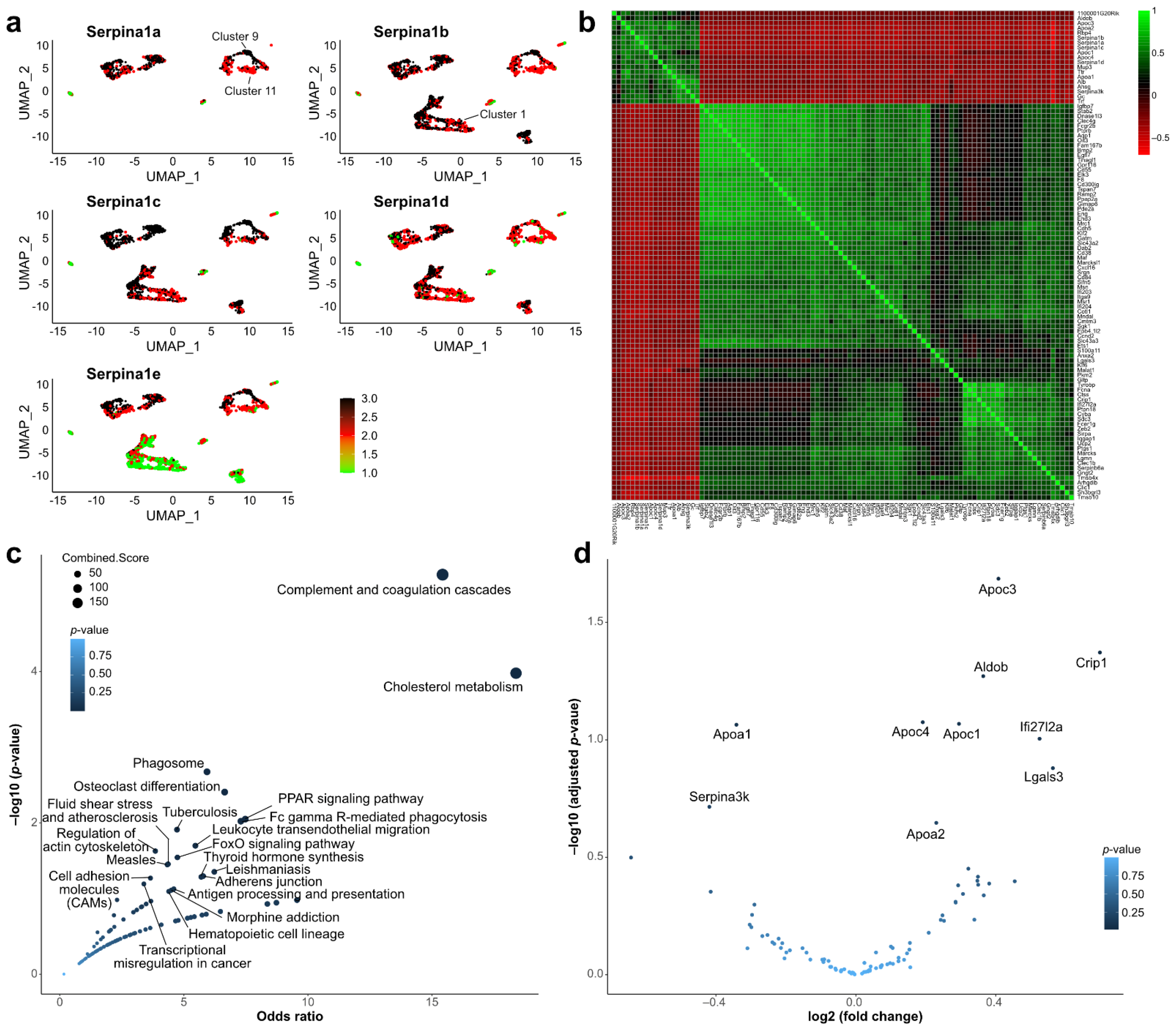
2.4. Single-Cell Sequencing Data Analysis
3. Discussion
4. Materials and Methods
4.1. Animal Model
4.2. Total RNA Isolation from the Mice Livers
4.3. Transcriptome Analysis (RNA-seq)
4.4. Data Preprocessing
4.5. Gene Expression Analysis
4.6. Gene Ontology and Pathway Analysis
4.7. Single-Cell Sequencing (scSEQ) Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hua, B.; Fan, L.; Liang, Y.; Zhao, Y.; Tuddenham, E.G. Alpha1-antitrypsin Pittsburgh in a family with bleeding tendency. Haematologica 2009, 94, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Bumcrot, D.; Bettencourt, B. Serpina1 Sirnas: Compositions of Matter and Methods of Treatment. U.S. Patent 10935917B2, 27 December 2012. [Google Scholar]
- Billingsley, G.D.; Walter, M.A.; Hammond, G.L.; Cox, D.W. Physical mapping of four serpin genes: Alpha 1-antitrypsin, alpha 1-antichymotrypsin, corticosteroid-binding globulin, and protein C inhibitor, within a 280-kb region on chromosome I4q32.1. Am. J. Hum. Genet. 1993, 52, 343–353. [Google Scholar] [PubMed]
- Perlino, E.; Cortese, R.; Ciliberto, G. The human alpha 1-antitrypsin gene is transcribed from two different promoters in macrophages and hepatocytes. EMBO J. 1987, 6, 2767–2771. [Google Scholar] [CrossRef] [PubMed]
- Kalsheker, N.; Morley, S.; Morgan, K. Gene regulation of the serine proteinase inhibitors alpha1-antitrypsin and alpha1-antichymotrypsin. Biochem. Soc. Trans. 2002, 30, 93–98. [Google Scholar] [CrossRef]
- Janciauskiene, S.; Wrenger, S.; Welte, T. Immunoregulatory Properties of Acute Phase Proteins—Specific Focus on α1-Antitrypsin. In Acute Phase Proteins; Janciauskiene, S., Ed.; Intechopen: London, UK, 2013; p. 190. [Google Scholar]
- Rotondo, J.C.; Aquila, G.; Oton-Gonzalez, L.; Selvatici, R.; Rizzo, P.; De Mattei, M.; Pavasini, R.; Tognon, M.; Campo, G.C.; Martini, F. Methylation of SERPINA1 gene promoter may predict chronic obstructive pulmonary disease in patients affected by acute coronary syndrome. Clin. Epigenet. 2021, 13, 79. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Oton-Gonzalez, L.; Selvatici, R.; Rizzo, P.; Pavasini, R.; Campo, G.C.; Lanzillotti, C.; Mazziotta, C.; De Mattei, M.; Tognon, M.; et al. SERPINA1 Gene Promoter Is Differentially Methylated in Peripheral Blood Mononuclear Cells of Pregnant Women. Front. Cell Dev. Biol. 2020, 8, 550543. [Google Scholar] [CrossRef]
- Barbour, K.W.; Wei, F.; Brannan, C.; Flotte, T.R.; Baumann, H.; Berger, F.G. The murine alpha(1)-proteinase inhibitor gene family: Polymorphism, chromosomal location, and structure. Genomics 2002, 80, 515–522. [Google Scholar] [CrossRef]
- Ostermann, L.; Maus, R.; Stolper, J.; Schutte, L.; Katsarou, K.; Tumpara, S.; Pich, A.; Mueller, C.; Janciauskiene, S.; Welte, T.; et al. Alpha-1 antitrypsin deficiency impairs lung antibacterial immunity in mice. JCI Insight 2021, 6, e140816. [Google Scholar] [CrossRef]
- Borel, F.; Sun, H.; Zieger, M.; Cox, A.; Cardozo, B.; Li, W.; Oliveira, G.; Davis, A.; Gruntman, A.; Flotte, T.R.; et al. Editing out five Serpina1 paralogs to create a mouse model of genetic emphysema. Proc. Natl. Acad. Sci. USA 2018, 115, 2788–2793. [Google Scholar] [CrossRef]
- Yuan, Y.; DiCiaccio, B.; Li, Y.; Elshikha, A.S.; Titov, D.; Brenner, B.; Seifer, L.; Pan, H.; Karic, N.; Akbar, M.A.; et al. Anti-inflammaging effects of human alpha-1 antitrypsin. Aging Cell 2018, 17, e12694. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Paciencia, S.; Saint-Germain, E.; Rowell, M.C.; Ruiz, A.F.; Kalegari, P.; Ferbeyre, G. The senescence-associated secretory phenotype and its regulation. Cytokine 2019, 117, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Pott, G.B.; Chan, E.D.; Dinarello, C.A.; Shapiro, L. Alpha-1-antitrypsin is an endogenous inhibitor of proinflammatory cytokine production in whole blood. J. Leukoc. Biol. 2009, 85, 886–895. [Google Scholar] [CrossRef]
- Gottlieb, P.A.; Alkanani, A.K.; Michels, A.W.; Lewis, E.C.; Shapiro, L.; Dinarello, C.A.; Zipris, D. Alpha1-Antitrypsin therapy downregulates toll-like receptor-induced IL-1beta responses in monocytes and myeloid dendritic cells and may improve islet function in recently diagnosed patients with type 1 diabetes. J. Clin. Endocrinol. Metab. 2014, 99, E1418–E1426. [Google Scholar] [CrossRef]
- Shapiro, L.; Pott, G.B.; Ralston, A.H. Alpha-1-antitrypsin inhibits human immunodeficiency virus type 1. FASEB J. 2001, 15, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Tabula Muris, C. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 2020, 583, 590–595. [Google Scholar] [CrossRef]
- Meek, R.L.; Benditt, E.P. Amyloid A gene family expression in different mouse tissues. J. Exp. Med. 1986, 164, 2006–2017. [Google Scholar] [CrossRef] [Green Version]
- Sack, G.H., Jr. Serum amyloid A—A review. Mol. Med. 2018, 24, 46. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.J.; Sorci-Thomas, M.G. SAA: A link between cholesterol efflux capacity and inflammation? J. Lipid Res. 2015, 56, 1383–1385. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; de Beer, M.C.; de Beer, F.C.; van der Westhuyzen, D.R. Serum amyloid A is a ligand for scavenger receptor class B type I and inhibits high density lipoprotein binding and selective lipid uptake. J. Biol. Chem. 2005, 280, 2954–2961. [Google Scholar] [CrossRef]
- Porez, G.; Gross, B.; Prawitt, J.; Gheeraert, C.; Berrabah, W.; Alexandre, J.; Staels, B.; Lefebvre, P. The hepatic orosomucoid/alpha1-acid glycoprotein gene cluster is regulated by the nuclear bile acid receptor FXR. Endocrinology 2013, 154, 3690–3701. [Google Scholar] [CrossRef]
- Cole, G.B.; Keum, G.; Liu, J.; Small, G.W.; Satyamurthy, N.; Kepe, V.; Barrio, J.R. Specific estrogen sulfotransferase (SULT1E1) substrates and molecular imaging probe candidates. Proc. Natl. Acad. Sci. USA 2010, 107, 6222–6227. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Up to date on cholesterol 7 alpha-hydroxylase (CYP7A1) in bile acid synthesis. Liver Res. 2020, 4, 47–63. [Google Scholar] [CrossRef]
- Lopez-Serra, P.; Marcilla, M.; Villanueva, A.; Ramos-Fernandez, A.; Palau, A.; Leal, L.; Wahi, J.E.; Setien-Baranda, F.; Szczesna, K.; Moutinho, C.; et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 2014, 5, 3608. [Google Scholar] [CrossRef]
- Kedishvili, N.Y. Enzymology of retinoic acid biosynthesis and degradation. J. Lipid Res. 2013, 54, 1744–1760. [Google Scholar] [CrossRef] [PubMed]
- Strnad, P.; Mandorfer, M.; Choudhury, G.; Griffiths, W.; Trautwein, C.; Loomba, R.; Schluep, T.; Chang, T.; Yi, M.; Given, B.D.; et al. Fazirsiran for Liver Disease Associated with Alpha1-Antitrypsin Deficiency. N. Engl. J. Med. 2022, 387, 514–524. [Google Scholar] [CrossRef]
- Paterson, T.; Moore, S. The expression and characterization of five recombinant murine alpha 1-protease inhibitor proteins. Biochem. Biophys. Res. Commun. 1996, 219, 64–69. [Google Scholar] [CrossRef]
- Frenzel, E.; Wrenger, S.; Immenschuh, S.; Koczulla, R.; Mahadeva, R.; Deeg, H.J.; Dinarello, C.A.; Welte, T.; Marcondes, A.M.; Janciauskiene, S. Acute-phase protein alpha1-antitrypsin-a novel regulator of angiopoietin-like protein 4 transcription and secretion. J. Immunol. 2014, 192, 5354–5362. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meghadri, S.H.; Martinez-Delgado, B.; Ostermann, L.; Gomez-Mariano, G.; Perez-Luz, S.; Tumpara, S.; Wrenger, S.; DeLuca, D.S.; Maus, U.A.; Welte, T.; et al. Loss of Serpina1 in Mice Leads to Altered Gene Expression in Inflammatory and Metabolic Pathways. Int. J. Mol. Sci. 2022, 23, 10425. https://doi.org/10.3390/ijms231810425
Meghadri SH, Martinez-Delgado B, Ostermann L, Gomez-Mariano G, Perez-Luz S, Tumpara S, Wrenger S, DeLuca DS, Maus UA, Welte T, et al. Loss of Serpina1 in Mice Leads to Altered Gene Expression in Inflammatory and Metabolic Pathways. International Journal of Molecular Sciences. 2022; 23(18):10425. https://doi.org/10.3390/ijms231810425
Chicago/Turabian StyleMeghadri, Sri Harsha, Beatriz Martinez-Delgado, Lena Ostermann, Gema Gomez-Mariano, Sara Perez-Luz, Srinu Tumpara, Sabine Wrenger, David S. DeLuca, Ulrich A. Maus, Tobias Welte, and et al. 2022. "Loss of Serpina1 in Mice Leads to Altered Gene Expression in Inflammatory and Metabolic Pathways" International Journal of Molecular Sciences 23, no. 18: 10425. https://doi.org/10.3390/ijms231810425






