Structural Significance of His73 in F-Actin Dynamics: Insights from Ab Initio Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Structural Significance of His73 Methylation in F-Actin Dynamics
2.2. His73/Gly158 H-Bond Breaking Is Not Obligatory for Material Exchange in F-Actin Dynamics
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pollard, T.D.; Cooper, J.A. Actin, a Central Player in Cell Shape and Movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Carlier, M.-F. Actin polymerization and ATP hydrolysis. Adv. Biophys. 1990, 26, 51–73. [Google Scholar] [CrossRef]
- Kudryashov, D.S.; Grintsevich, E.E.; Rubenstein, P.A.; Reisler, E. A Nucleotide State-sensing Region on Actin. J. Biol. Chem. 2010, 285, 25591–25601. [Google Scholar] [CrossRef]
- Porta, J.C.; Borgstahl, G.E.O. Structural Basis for Profilin-Mediated Actin Nucleotide Exchange. J. Mol. Biol. 2012, 418, 103–116. [Google Scholar] [CrossRef]
- Otterbein, L.; Graceffa, P.; Dominguez, R. The Crystal Structure of Uncomplexed Actin in the ADP State. Science 2001, 293, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Graceffa, P.; Dominguez, R. Crystal Structure of Monomeric Actin in the ATP State: Structural Basis of Nucleotide-Dependent Actin Dynamics. J. Biol. Chem. 2003, 278, 34172–34180. [Google Scholar] [CrossRef]
- Rould, M.; Wan, Q.; Joel, P.; Lowey, S.; Trybus, K. Crystal Structures of Expressed Non-polymerizable Monomeric Actin in the ADP and ATP States. J. Biol. Chem. 2006, 281, 31909–31919. [Google Scholar] [CrossRef]
- Murakami, K.; Yasunaga, T.; Noguchi, T.Q.P.; Gomibuchi, Y.; Ngo, K.X.; Uyeda, T.Q.P.; Wakabayashi, T. Structural Basis for Actin Assembly, Activation of ATP Hydrolysis, and Delayed Phosphate Release. Cell 2010, 143, 275–287. [Google Scholar] [CrossRef]
- Yao, X.; Grade, S.; Wriggers, W.; Rubenstein, P.A. His73, Often Methylated, Is an Important Structural Determinant for Actin: A Mutagenic Analysis of His73 of Yeast Actin. J. Biol. Chem. 1999, 274, 37443–37449. [Google Scholar] [CrossRef]
- Belmont Lisa, D.; Orlova, A.; Drubin David, G.; Egelman Edward, H. A change in actin conformation associated with filament instability after Pi release. Proc. Natl. Acad. Sci. USA 1999, 96, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Nyman, T.; Schüler, H.; Korenbaum, E.; Schutt, C.E.; Karlsson, R.; Lindberg, U. The role of MeH73 in actin polymerization and ATP hydrolysis11Edited by R. Huber. J. Mol. Biol. 2002, 317, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Parr, R.G.; Craig, D.P.; Ross, I.G. Molecular Orbital Calculations of the Lower Excited Electronic Levels of Benzene, Configuration Interaction Included. J. Chem. Phys. 1950, 18, 1561–1563. [Google Scholar] [CrossRef]
- Freedman, H.; Laino, T.; Curioni, A. Reaction Dynamics of ATP Hydrolysis in Actin Determined by ab Initio Molecular Dynamics Simulations. J. Chem. Theory Comput. 2012, 8, 3373–3383. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef]
- Curtiss, L.A.; Raghavachari, K.; Trucks, G.W.; Pople, J.A. Gaussian-2 theory for molecular energies of first and second row compounds. J. Chem. Phys. 1991, 94, 7221–7230. [Google Scholar] [CrossRef]
- Šponer, J.; Jurečka, P.; Hobza, P. Accurate Interaction Energies of Hydrogen-Bonded Nucleic Acid Base Pairs. J. Am. Chem. Soc. 2004, 126, 10142–10151. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Hanwell, M.; Curtis, D.; Lonie, D.; Vandermeersch, T.; Zurek, E.; Hutchison, G. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin: DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef]
- Otomo, T.; Tomchick, D.R.; Otomo, C.; Panchal, S.C.; Machius, M.; Rosen, M.K. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 2005, 433, 488–494. [Google Scholar] [CrossRef]
- Chereau, D.; Kerff, F.; Graceffa, P.; Grabarek, Z.; Langsetmo, K.; Dominguez, R. Actin-bound structures of Wiskott–Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA 2005, 102, 16644–16649. [Google Scholar] [CrossRef] [PubMed]
- Aguda, A.H.; Xue, B.; Irobi, E.; Préat, T.; Robinson, R.C. The Structural Basis of Actin Interaction with Multiple WH2/β-Thymosin Motif-Containing Proteins. Structure 2006, 14, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Mouilleron, S.; Guettler, S.; Langer, C.A.; Treisman, R.; McDonald, N.Q. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 2008, 27, 3198–3208. [Google Scholar] [CrossRef]
- Tsuge, H.; Nagahama, M.; Oda, M.; Iwamoto, S.; Utsunomiya, H.; Marquez, V.E.; Katunuma, N.; Nishizawa, M.; Sakurai, J. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens ι-toxin. Proc. Natl. Acad. Sci. USA 2008, 105, 7399–7404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
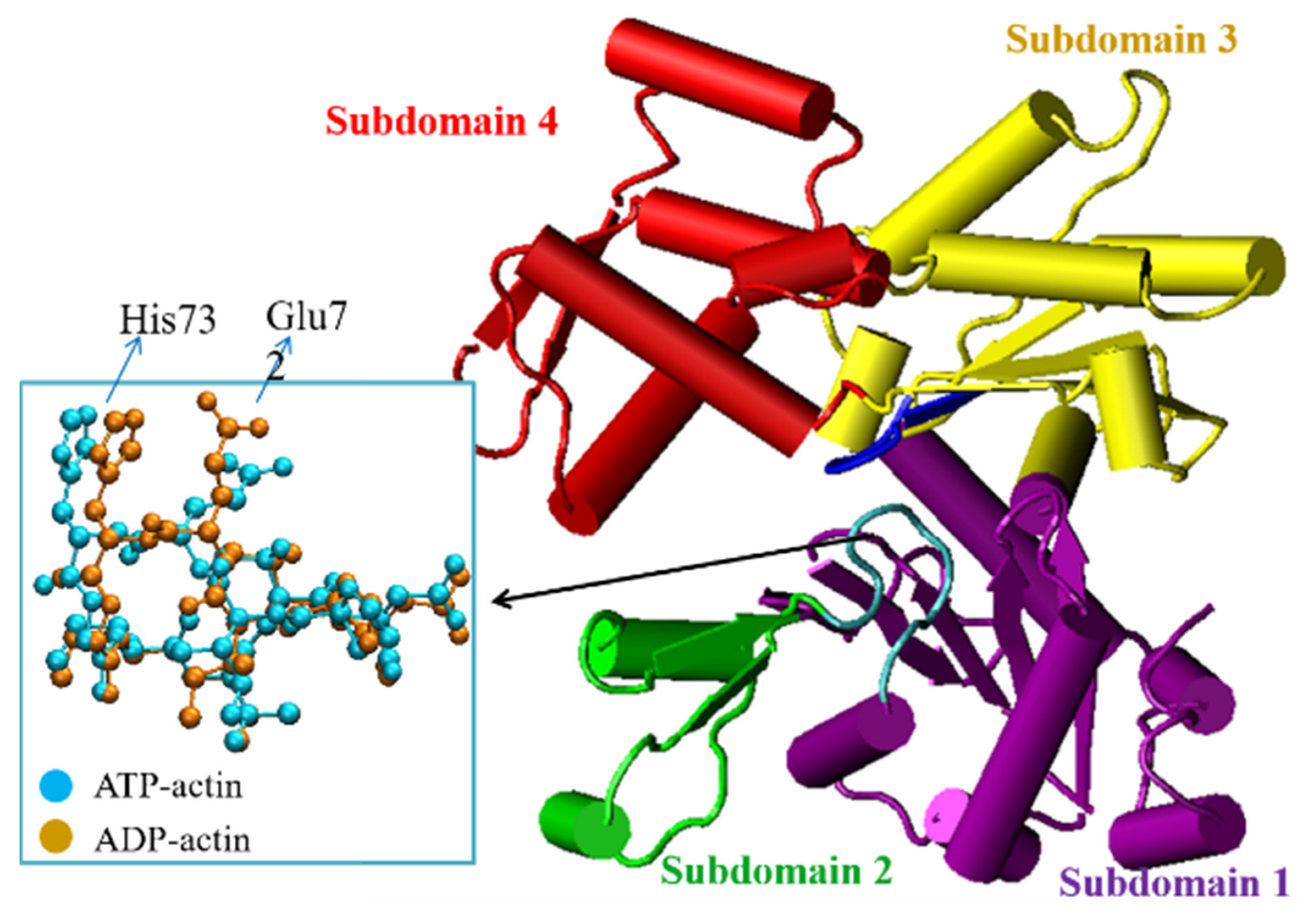

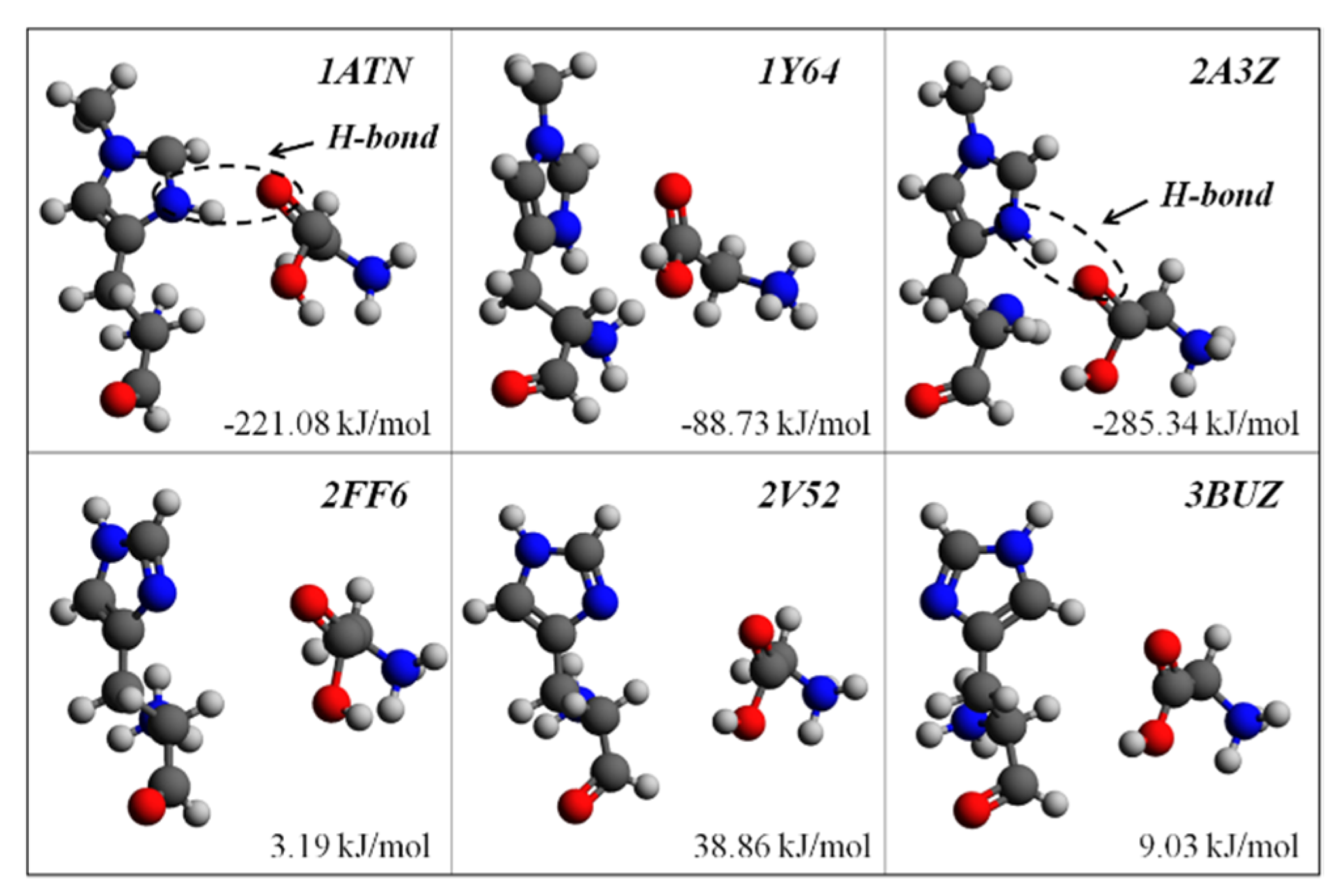
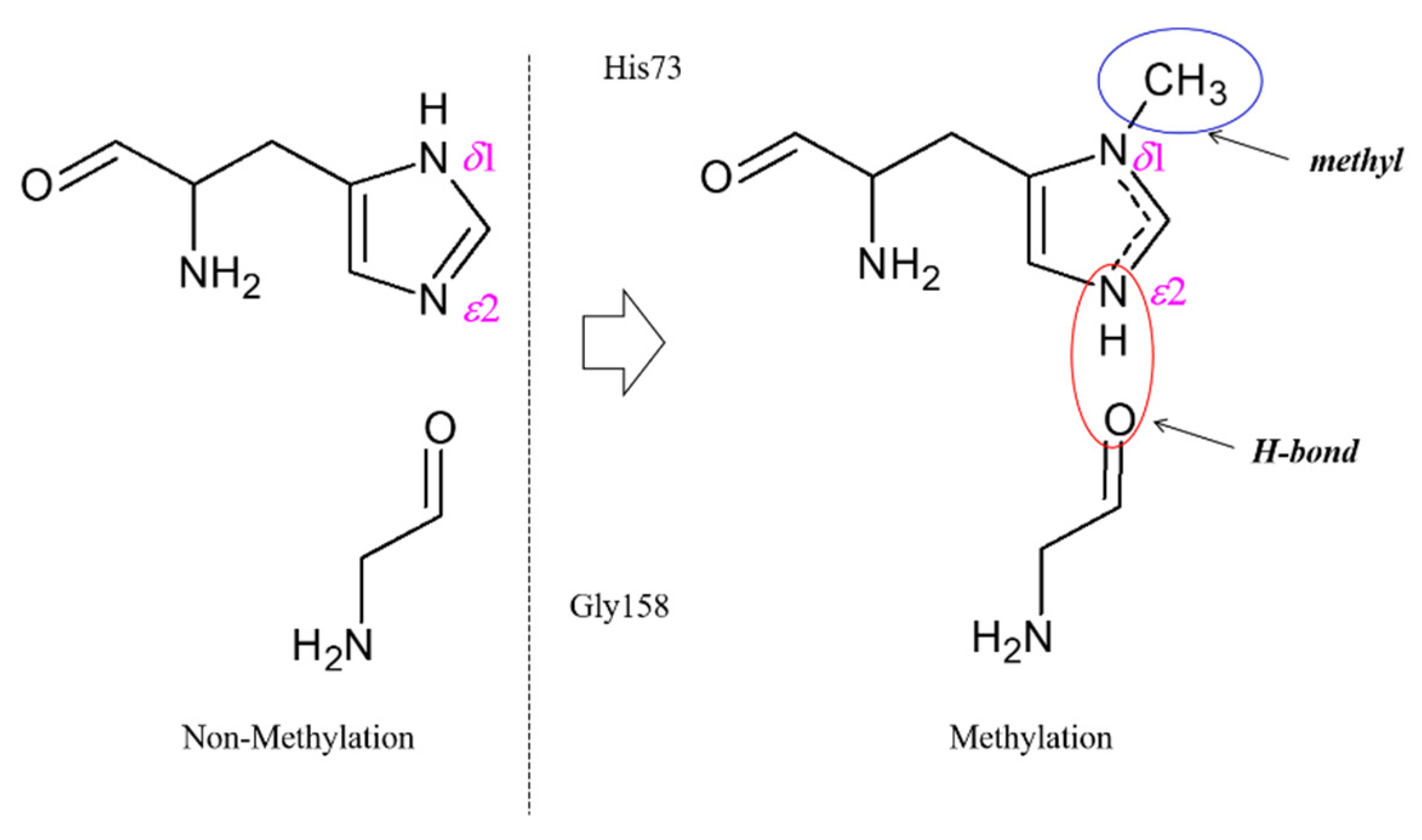
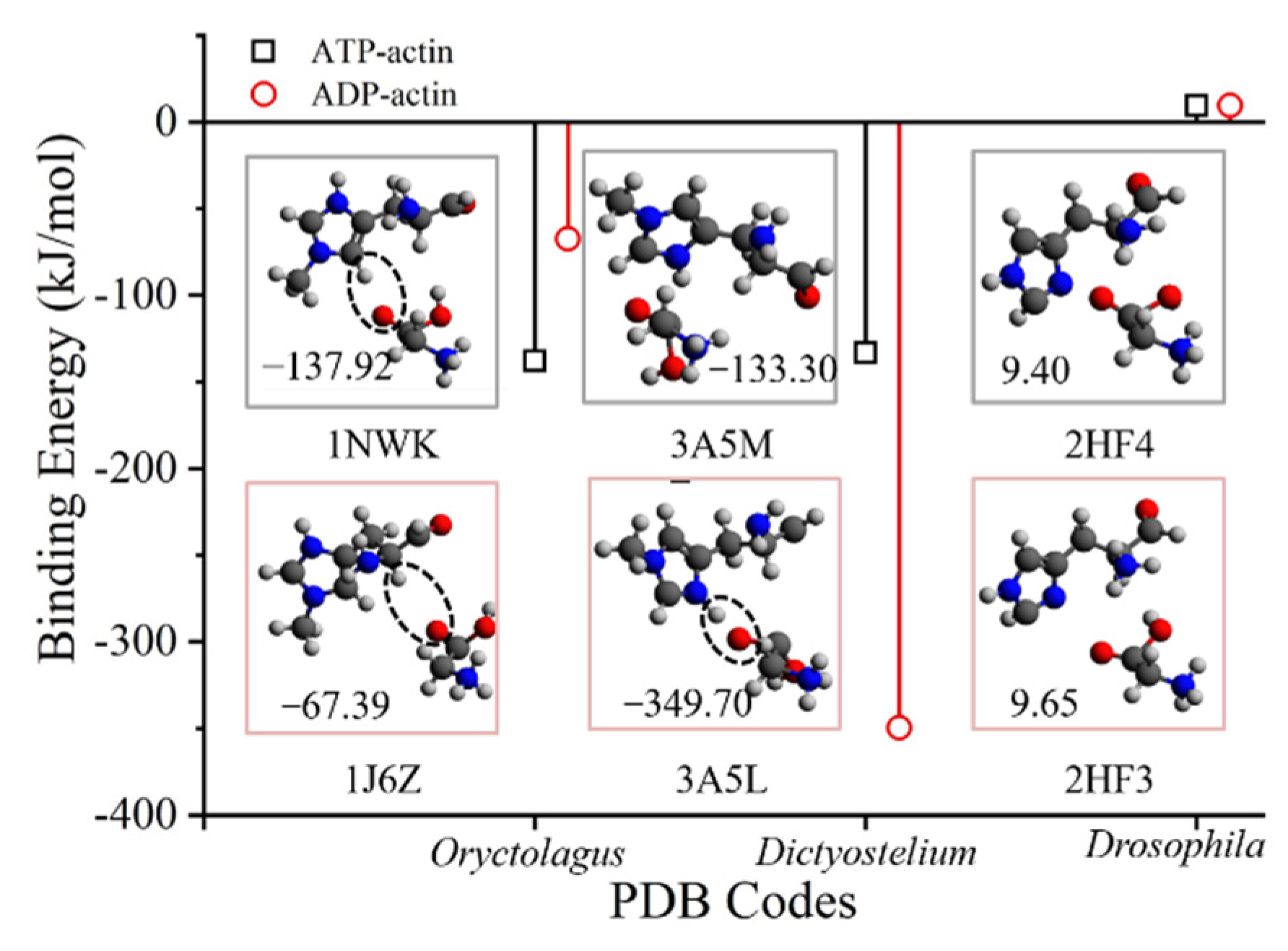
| PDB ID | States 1 | Ecomb 2 | EHis73 | EGly158 | Ebsse | Eb |
|---|---|---|---|---|---|---|
| 1ATN | ATP/Y | −2,111,763.69 | −1,357,798.71 | −753,724.23 | 17.43 | −221.08 |
| 1Y64 | ATP/Y | −2,111,713.21 | −1,357,771.33 | −753,839.34 | 12.91 | −88.73 |
| 2A3Z | ATP/Y | −2,111,797.72 | −1,357,715.98 | −753,774.68 | 18.84 | −285.34 |
| 2FF6 | ATP/N | −2,007,737.54 | −1,253,918.49 | −753,814.92 | 7.35 | 3.19 |
| 2V52 | ATP/N | −2,007,744.46 | −1,253,947.10 | −753,829.20 | 7.41 | 38.86 |
| 3BUZ | ATP/N | −2,007,754.82 | −1,253,882.65 | −753,872.34 | 8.95 | 9.03 |
| PDB ID | States 1 | Ecomb 2 | EHis73 | EGly158 | Ebsse | Eb |
|---|---|---|---|---|---|---|
| 1NWK | ATP/Y | −2,111,727.89 | −1,357,765.61 | −753,810.78 | 12.18 | −137.92 |
| 1J6Z | ADP/Y | −2,111,661.74 | −1,357,758.88 | −753,828.18 | 6.61 | −67.39 |
| 3A5M | ATP/Y | −2,111,746.51 | −1,357,783.93 | −753,814.66 | 13.26 | −133.30 |
| 3A5L | ADP/Y | −2,111,751.50 | −1,357,775.95 | −753,597.98 | 24.34 | −349.71 |
| 2HF4 | ATP/N | −2,007,709.88 | −1,253,871.19 | −753,841.34 | 6.85 | 9.40 |
| 2HF3 | ADP/N | −2007699.82 | −1253870.48 | −753830.77 | 8.32 | 9.65 |
| PDB ID | Nucleotide State | Methylation | Sources | Reference |
|---|---|---|---|---|
| 1NWK | ATP | Yes | Oryctolagus | Dominguez, et al. 2001, 2003 [5,6] |
| 1J6Z | ADP | Yes | ||
| 3A5M | ATP | Yes | Dictyostelium | Murakami, et al. 2010 [8] |
| 3A5L | ADP | Yes | ||
| 2HF4 | ATP | No | Drosophila | Rould, et al. 2006 [7] |
| 2HF3 | ADP | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Du, J.; Ren, M. Structural Significance of His73 in F-Actin Dynamics: Insights from Ab Initio Study. Int. J. Mol. Sci. 2022, 23, 10447. https://doi.org/10.3390/ijms231810447
Li T, Du J, Ren M. Structural Significance of His73 in F-Actin Dynamics: Insights from Ab Initio Study. International Journal of Molecular Sciences. 2022; 23(18):10447. https://doi.org/10.3390/ijms231810447
Chicago/Turabian StyleLi, Tong, Juan Du, and Mingfa Ren. 2022. "Structural Significance of His73 in F-Actin Dynamics: Insights from Ab Initio Study" International Journal of Molecular Sciences 23, no. 18: 10447. https://doi.org/10.3390/ijms231810447






