Clomifene and Assisted Reproductive Technology in Humans Are Associated with Sex-Specific Offspring Epigenetic Alterations in Imprinted Control Regions
Abstract
1. Introduction
2. Results
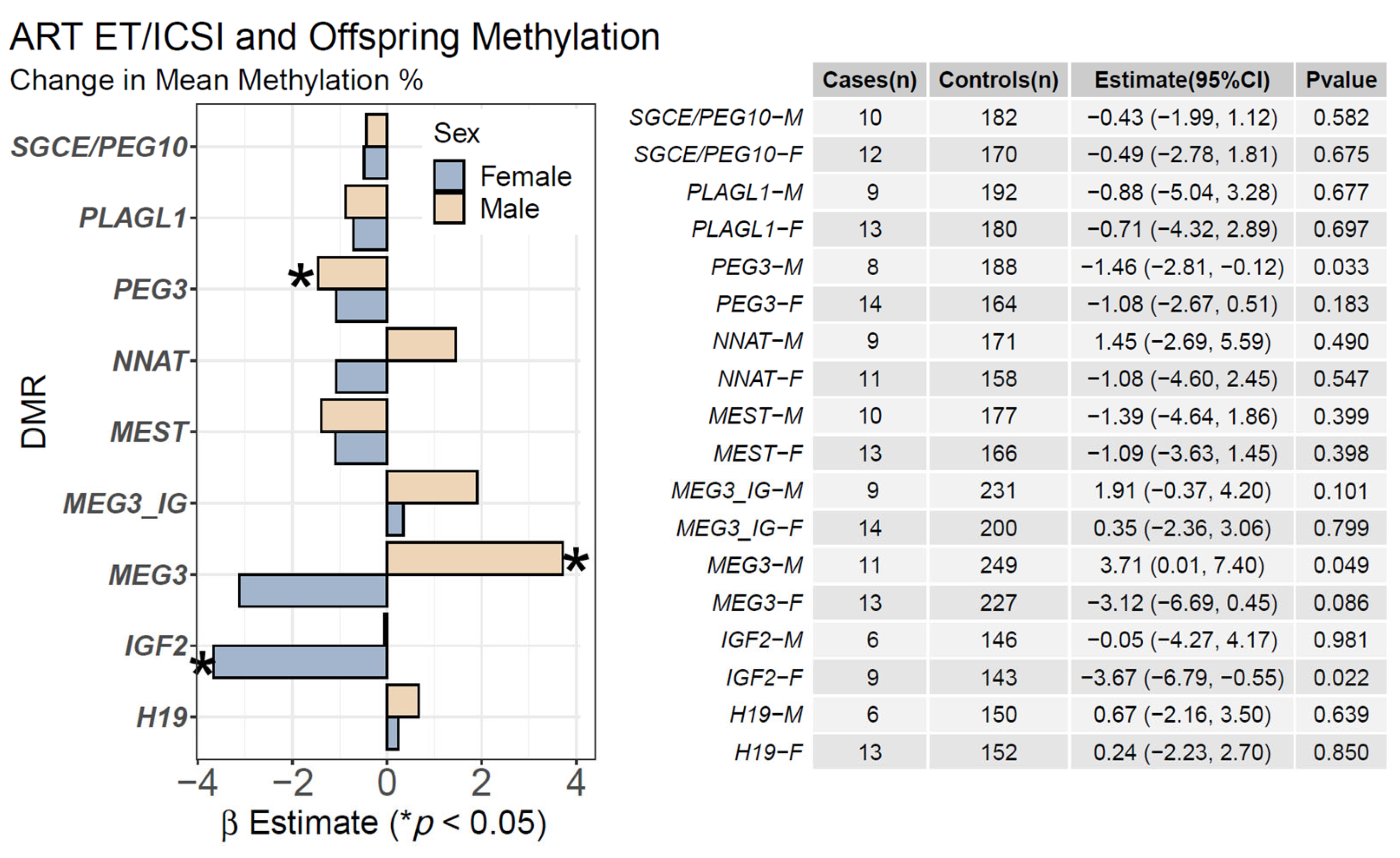
2.1. ART ET/ICSI and Mean Offspring CpG Methylation
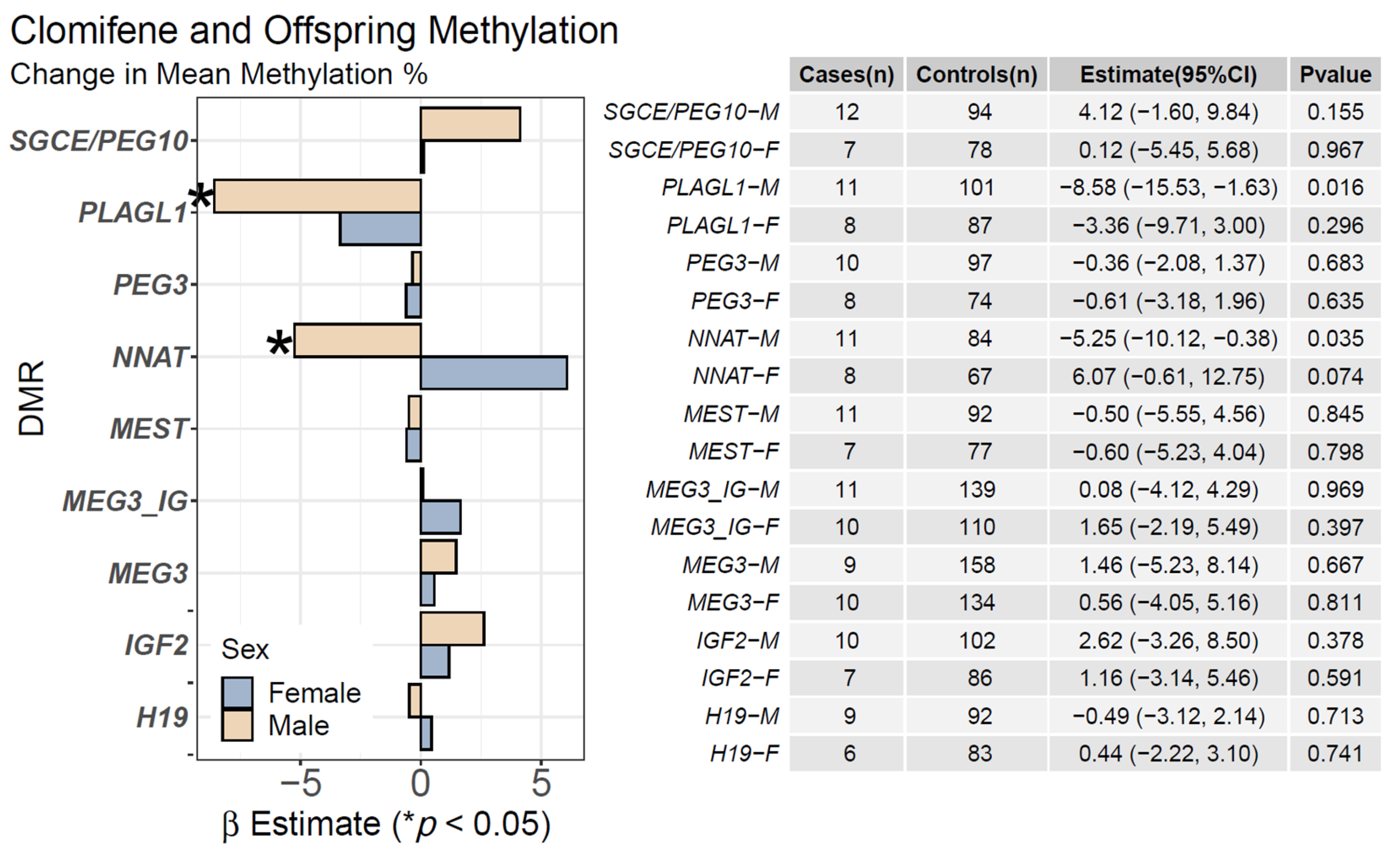
2.2. Clomifene Only and Mean Offspring CpG Methylation
3. Discussion
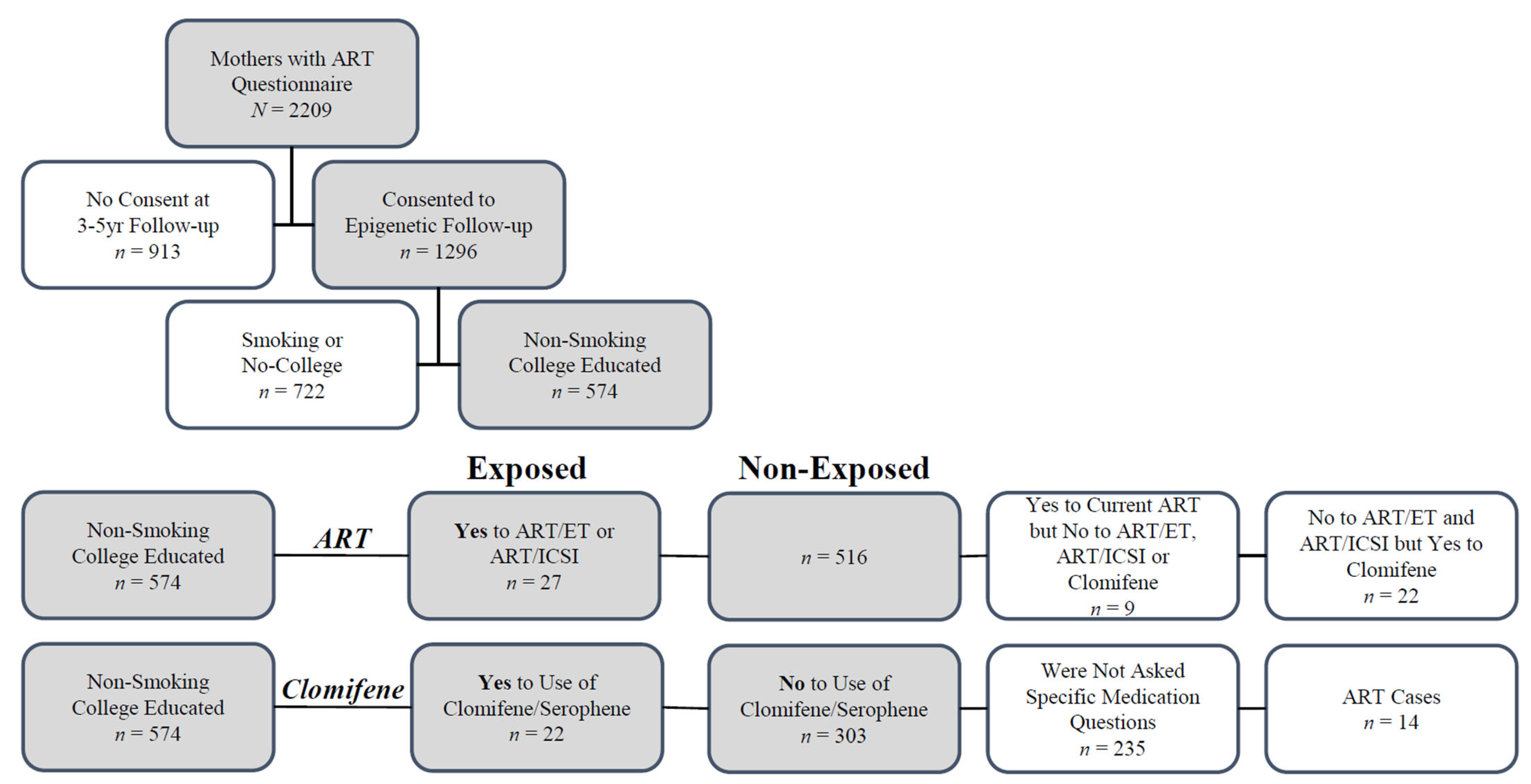
4. Materials and Methods
4.1. Study Participants
4.2. Data Collection
4.2.1. Outcome Assessment
4.2.2. Exposure Assessment
4.3. Statistical Analyses
4.4. Nomenclature
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sunderam, S.; Kissin, D.M.; Zhang, Y.; Folger, S.G.; Boulet, S.L.; Warner, L.; Callaghan, W.M.; Barfield, W.D. Assisted Reproductive Technology Surveillance—United States, 2016. MMWR Surveill. Summ. 2019, 68, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ely, D.M.; Hamilton, B.E. Trends in Fertility and Mother’s Age at First Birth Among Rural and Metropolitan Counties: United States, 2007–2017. NCHS Data Brief 2018, 323, 1–8. [Google Scholar]
- Qin, J.; Liu, X.; Sheng, X.; Wang, H.; Gao, S. Assisted reproductive technology and the risk of pregnancy-related complications and adverse pregnancy outcomes in singleton pregnancies: A meta-analysis of cohort studies. Fertil. Steril. 2016, 105, 73–85.e6. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Shetty, A.; Hamilton, M.; Bhattacharya, S.; Maheshwari, A. Obstetric and perinatal outcomes in singleton pregnancies resulting from IVF/ICSI: A systematic review and meta-analysis. Hum. Reprod. Update 2012, 18, 485–503. [Google Scholar] [CrossRef]
- Hansen, M.; Kurinczuk, J.J.; Milne, E.; de Klerk, N.; Bower, C. Assisted reproductive technology and birth defects: A systematic review and meta-analysis. Hum. Reprod. Update 2013, 19, 330–353. [Google Scholar] [CrossRef]
- Hansen, M.; Bower, C. The impact of assisted reproductive technologies on intra-uterine growth and birth defects in singletons. Semin. Fetal Neonatal Med. 2014, 19, 228–233. [Google Scholar] [CrossRef]
- McDonald, S.D.; Murphy, K.; Beyene, J.; Ohlsson, A. Perinatel outcomes of singleton pregnancies achieved by in vitro fertilization: A systematic review and meta-analysis. J. Obstet. Gynaecol. Can. 2005, 27, 449–459. [Google Scholar] [CrossRef]
- Rimm, A.A.; Katayama, A.C.; Diaz, M.; Katayama, K.P. A meta-analysis of controlled studies comparing major malformation rates in IVF and ICSI infants with naturally conceived children. J. Assist. Reprod. Genet. 2004, 21, 437–443. [Google Scholar] [CrossRef]
- Wen, J.; Jiang, J.; Ding, C.; Dai, J.; Liu, Y.; Xia, Y.; Liu, J.; Hu, Z. Birth defects in children conceived by in vitro fertilization and intracytoplasmic sperm injection: A meta-analysis. Fertil. Steril. 2012, 97, 1331–1337.e4. [Google Scholar] [CrossRef]
- Rhon-Calderon, E.A.; Vrooman, L.A.; Riesche, L.; Bartolomei, M.S. The effects of Assisted Reproductive Technologies on genomic imprinting in the placenta. Placenta 2019, 84, 37–43. [Google Scholar] [CrossRef]
- Murphy, S.K.; Huang, Z.; Hoyo, C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS ONE 2012, 7, e40924. [Google Scholar] [CrossRef] [PubMed]
- Ollikainen, M.; Craig, J.M. Epigenetic discordance at imprinting control regions in twins. Epigenomics 2011, 3, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Skaar, D.A.; Li, Y.; Bernal, A.J.; Hoyo, C.; Murphy, S.K.; Jirtle, R.L. The human imprintome: Regulatory mechanisms, methods of ascertainment, and roles in disease susceptibility. ILAR J. 2012, 53, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Woodfine, K.; Huddleston, J.E.; Murrell, A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin 2011, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, T.; Perez de Nanclares, G.; Maher, E.R.; Temple, I.K.; Tumer, Z.; Monk, D.; Mackay, D.J.; Gronskov, K.; Riccio, A.; Linglart, A.; et al. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin. Epigenetics 2015, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Market-Velker, B.A.; Zhang, L.; Magri, L.S.; Bonvissuto, A.C.; Mann, M.R. Dual effects of superovulation: Loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum. Mol. Genet. 2010, 19, 36–51. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef]
- Barlow, D.P.; Stoger, R.; Herrmann, B.G.; Saito, K.; Schweifer, N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 1991, 349, 84–87. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Kuroiwa, Y.; Miyoshi, N.; Kohda, T.; Suzuki, R.; Yokoyama, M.; Viville, S.; Barton, S.C.; Ishino, F.; Surani, M.A. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 1995, 11, 52–59. [Google Scholar] [CrossRef]
- Gomes, M.V.; Huber, J.; Ferriani, R.A.; Amaral Neto, A.M.; Ramos, E.S. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol. Hum. Reprod. 2009, 15, 471–477. [Google Scholar] [CrossRef]
- Katagiri, Y.; Aoki, C.; Tamaki-Ishihara, Y.; Fukuda, Y.; Kitamura, M.; Matsue, Y.; So, A.; Morita, M. Effects of assisted reproduction technology on placental imprinted gene expression. Obstet. Gynecol. Int. 2010, 2010, 437528. [Google Scholar] [CrossRef] [PubMed]
- Katari, S.; Turan, N.; Bibikova, M.; Erinle, O.; Chalian, R.; Foster, M.; Gaughan, J.P.; Coutifaris, C.; Sapienza, C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum. Mol. Genet. 2009, 18, 3769–3778. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.; Hyslop, L.; Stojkovic, P.; Leary, C.; Murdoch, A.; Reik, W.; Stojkovic, M.; Herbert, M.; Dean, W. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum. Reprod. 2010, 25, 2387–2395. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Esmaeili, M.; Taheri, M. H19 lncRNA: Roles in tumorigenesis. Biomed. Pharm. 2020, 123, 109774. [Google Scholar] [CrossRef]
- Fontana, L.; Tabano, S.; Maitz, S.; Colapietro, P.; Garzia, E.; Gerli, A.G.; Sirchia, S.M.; Miozzo, M. Clinical and Molecular Diagnosis of Beckwith-Wiedemann Syndrome with Single- or Multi-Locus Imprinting Disturbance. Int. J. Mol. Sci. 2021, 22, 3445. [Google Scholar] [CrossRef] [PubMed]
- Bergman, D.; Halje, M.; Nordin, M.; Engstrom, W. Insulin-like growth factor 2 in development and disease: A mini-review. Gerontology 2013, 59, 240–249. [Google Scholar] [CrossRef]
- House, J.S.; Hall, J.; Park, S.S.; Planchart, A.; Money, E.; Maguire, R.L.; Huang, Z.; Mattingly, C.J.; Skaar, D.; Tzeng, J.Y.; et al. Cadmium exposure and MEG3 methylation differences between Whites and African Americans in the NEST Cohort. Environ. Epigenet. 2019, 5, dvz014. [Google Scholar] [CrossRef]
- He, H.; Kim, J. Regulation and function of the peg3 imprinted domain. Genom. Inform. 2014, 12, 105–113. [Google Scholar] [CrossRef]
- He, H.; Perera, B.P.; Ye, A.; Kim, J. Parental and sexual conflicts over the Peg3 imprinted domain. Sci. Rep. 2016, 6, 38136. [Google Scholar] [CrossRef]
- Kamiya, M.; Judson, H.; Okazaki, Y.; Kusakabe, M.; Muramatsu, M.; Takada, S.; Takagi, N.; Arima, T.; Wake, N.; Kamimura, K.; et al. The cell cycle control gene ZAC/PLAGL1 is imprinted—A strong candidate gene for transient neonatal diabetes. Hum. Mol. Genet. 2000, 9, 453–460. [Google Scholar] [CrossRef]
- Xie, T.; Pan, S.; Zheng, H.; Luo, Z.; Tembo, K.M.; Jamal, M.; Yu, Z.; Yu, Y.; Xia, J.; Yin, Q.; et al. PEG10 as an oncogene: Expression regulatory mechanisms and role in tumor progression. Cancer Cell Int. 2018, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Caramaschi, D.; Jungius, J.; Page, C.M.; Novakovic, B.; Saffery, R.; Halliday, J.; Lewis, S.; Magnus, M.C.; London, S.J.; Haberg, S.E.; et al. Association of medically assisted reproduction with offspring cord blood DNA methylation across cohorts. Hum. Reprod. 2021, 36, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.H.; Mendola, P.; Sundaram, R.; Zeng, X.; Guan, W.; Tsai, M.Y.; Robinson, S.L.; Stern, J.E.; Ghassabian, A.; Lawrence, D.; et al. Conception by fertility treatment and offspring deoxyribonucleic acid methylation. Fertil. Steril. 2021, 116, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Anckaert, E.; De Rycke, M.; Smitz, J. Culture of oocytes and risk of imprinting defects. Hum. Reprod. Update 2013, 19, 52–66. [Google Scholar] [CrossRef]
- Denomme, M.M.; Mann, M.R. Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction 2012, 144, 393–409. [Google Scholar] [CrossRef]
- Fauque, P. Ovulation induction and epigenetic anomalies. Fertil. Steril. 2013, 99, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Cortessis, V.K.; Azadian, M.; Buxbaum, J.; Sanogo, F.; Song, A.Y.; Sriprasert, I.; Wei, P.C.; Yu, J.; Chung, K.; Siegmund, K.D. Comprehensive meta-analysis reveals association between multiple imprinting disorders and conception by assisted reproductive technology. J. Assist. Reprod. Genet. 2018, 35, 943–952. [Google Scholar] [CrossRef]
- Provenzi, L.; Carli, P.; Fumagalli, M.; Giorda, R.; Casavant, S.; Beri, S.; Citterio, A.; D’Agata, A.; Morandi, F.; Mosca, F.; et al. Very preterm birth is associated with PLAGL1 gene hypomethylation at birth and discharge. Epigenomics 2018, 10, 1121–1130. [Google Scholar] [CrossRef]
- Harrison, K.; Hoad, G.; Scott, P.; Simpson, L.; Horgan, G.W.; Smyth, E.; Heys, S.D.; Haggarty, P. Breast cancer risk and imprinting methylation in blood. Clin. Epigenetics 2015, 7, 92. [Google Scholar] [CrossRef]
- Hoyo, C.; Murtha, A.P.; Schildkraut, J.M.; Jirtle, R.L.; Demark-Wahnefried, W.; Forman, M.R.; Iversen, E.S.; Kurtzberg, J.; Overcash, F.; Huang, Z.; et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics 2011, 6, 928–936. [Google Scholar] [CrossRef] [PubMed]
- King, K.E.; Darrah, T.H.; Money, E.; Meentemeyer, R.; Maguire, R.L.; Nye, M.D.; Michener, L.; Murtha, A.P.; Jirtle, R.; Murphy, S.K.; et al. Geographic clustering of elevated blood heavy metal levels in pregnant women. BMC Public Health 2015, 15, 1035. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.C.; Semenova, V.; Darrah, T.; Vengosh, A.; Huang, Z.; King, K.; Nye, M.D.; Fry, R.; Skaar, D.; Maguire, R.; et al. Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharm. Toxicol. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Belva, F.; Painter, R.; Bonduelle, M.; Roelants, M.; Devroey, P.; De Schepper, J. Are ICSI adolescents at risk for increased adiposity? Hum. Reprod. 2012, 27, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Gonzalez, R.; Moreira, P.N.; Perez-Crespo, M.; Sanchez-Martin, M.; Ramirez, M.A.; Pericuesta, E.; Bilbao, A.; Bermejo-Alvarez, P.; de Dios Hourcade, J.; de Fonseca, F.R.; et al. Long-term effects of mouse intracytoplasmic sperm injection with DNA-fragmented sperm on health and behavior of adult offspring. Biol. Reprod. 2008, 78, 761–772. [Google Scholar] [CrossRef]
- Ulaner, G.A.; Vu, T.H.; Li, T.; Hu, J.F.; Yao, X.M.; Yang, Y.; Gorlick, R.; Meyers, P.; Healey, J.; Ladanyi, M.; et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 2003, 12, 535–549. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Taniguchi, T.; Jhee, A.; Kerr, N.; Reeve, A.E. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene 1999, 18, 7527–7534. [Google Scholar] [CrossRef][Green Version]
- Cui, H.; Cruz-Correa, M.; Giardiello, F.M.; Hutcheon, D.F.; Kafonek, D.R.; Brandenburg, S.; Wu, Y.; He, X.; Powe, N.R.; Feinberg, A.P. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science 2003, 299, 1753–1755. [Google Scholar] [CrossRef]
- Nakagawa, H.; Chadwick, R.B.; Peltomaki, P.; Plass, C.; Nakamura, Y.; de La Chapelle, A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 591–596. [Google Scholar] [CrossRef]
- Choufani, S.; Turinsky, A.L.; Melamed, N.; Greenblatt, E.; Brudno, M.; Berard, A.; Fraser, W.D.; Weksberg, R.; Trasler, J.; Monnier, P.; et al. Impact of assisted reproduction, infertility, sex and paternal factors on the placental DNA methylome. Hum. Mol. Genet. 2019, 28, 372–385. [Google Scholar] [CrossRef]
- Poplinski, A.; Tuttelmann, F.; Kanber, D.; Horsthemke, B.; Gromoll, J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int. J. Androl. 2010, 33, 642–649. [Google Scholar] [CrossRef]
- Huntriss, J.D.; Hemmings, K.E.; Hinkins, M.; Rutherford, A.J.; Sturmey, R.G.; Elder, K.; Picton, H.M. Variable imprinting of the MEST gene in human preimplantation embryos. Eur. J. Hum. Genet. 2013, 21, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Velker, B.A.M.; Denomme, M.M.; Krafty, R.T.; Mann, M.R.W. Maintenance of Mest imprinted methylation in blastocyst-stage mouse embryos is less stable than other imprinted loci following superovulation or embryo culture. Environ. Epigenet. 2017, 3, dvx015. [Google Scholar] [CrossRef] [PubMed]
- Cannarella, R.; Crafa, A.; Condorelli, R.A.; Mongioi, L.M.; La Vignera, S.; Calogero, A.E. Relevance of sperm imprinted gene methylation on assisted reproductive technique outcomes and pregnancy loss: A systematic review. Syst. Biol. Reprod. Med. 2021, 67, 251–259. [Google Scholar] [CrossRef]
- Barberet, J.; Binquet, C.; Guilleman, M.; Doukani, A.; Choux, C.; Bruno, C.; Bourredjem, A.; Chapusot, C.; Bourc’his, D.; Duffourd, Y.; et al. Do assisted reproductive technologies and in vitro embryo culture influence the epigenetic control of imprinted genes and transposable elements in children? Hum. Reprod. 2021, 36, 479–492. [Google Scholar] [CrossRef]
- Cowley, M.; Skaar, D.A.; Jima, D.D.; Maguire, R.L.; Hudson, K.M.; Park, S.S.; Sorrow, P.; Hoyo, C. Effects of cadmium exposure on DNA methylation at imprinting control regions and genome-wide in mothers and newborn children. Environ. Health Perspect. 2018, 126, 037003. [Google Scholar] [CrossRef] [PubMed]
- Tobi, E.W.; Almqvist, C.; Hedman, A.; Andolf, E.; Holte, J.; Olofsson, J.I.; Wramsby, H.; Wramsby, M.; Pershagen, G.; Heijmans, B.T.; et al. DNA methylation differences at birth after conception through ART. Hum. Reprod. 2021, 36, 248–259. [Google Scholar] [CrossRef]
- Chen, M.; Heilbronn, L.K. The health outcomes of human offspring conceived by assisted reproductive technologies (ART). J. Dev. Orig. Health Dis. 2017, 8, 388–402. [Google Scholar] [CrossRef]
- Kawwass, J.F.; Badell, M.L. Maternal and Fetal Risk Associated With Assisted Reproductive Technology. Obstet. Gynecol. 2018, 132, 763–772. [Google Scholar] [CrossRef]
- Grondahl, M.L.; Christiansen, S.L.; Kesmodel, U.S.; Agerholm, I.E.; Lemmen, J.G.; Lundstrom, P.; Bogstad, J.; Raaschou-Jensen, M.; Ladelund, S. Effect of women’s age on embryo morphology, cleavage rate and competence—A multicenter cohort study. PLoS ONE 2017, 12, e0172456. [Google Scholar] [CrossRef]
- Grotegut, C.A.; Chisholm, C.A.; Johnson, L.N.; Brown, H.L.; Heine, R.P.; James, A.H. Medical and obstetric complications among pregnant women aged 45 and older. PLoS ONE 2014, 9, e96237. [Google Scholar] [CrossRef]
- Denomme, M.M.; Zhang, L.; Mann, M.R. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil. Steril. 2011, 96, 734–738.e732. [Google Scholar] [CrossRef] [PubMed]
- Stouder, C.; Deutsch, S.; Paoloni-Giacobino, A. Superovulation in mice alters the methylation pattern of imprinted genes in the sperm of the offspring. Reprod. Toxicol. 2009, 28, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Fauque, P.; Jouannet, P.; Lesaffre, C.; Ripoche, M.A.; Dandolo, L.; Vaiman, D.; Jammes, H. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev. Biol. 2007, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Menezo, Y.; Clement, P.; Dale, B. DNA Methylation Patterns in the Early Human Embryo and the Epigenetic/Imprinting Problems: A Plea for a More Careful Approach to Human Assisted Reproductive Technology (ART). Int. J. Mol. Sci. 2019, 20, 1342. [Google Scholar] [CrossRef]
- Rahimi, S.; Martel, J.; Karahan, G.; Angle, C.; Behan, N.A.; Chan, D.; MacFarlane, A.J.; Trasler, J.M. Moderate maternal folic acid supplementation ameliorates adverse embryonic and epigenetic outcomes associated with assisted reproduction in a mouse model. Hum. Reprod. 2019, 34, 851–862. [Google Scholar] [CrossRef]
- Morbeck, D.E.; Krisher, R.L.; Herrick, J.R.; Baumann, N.A.; Matern, D.; Moyer, T. Composition of commercial media used for human embryo culture. Fertil. Steril. 2014, 102, 759–766.e759. [Google Scholar] [CrossRef]
- Nakamura, A.; Pryor, L.; Ballon, M.; Lioret, S.; Heude, B.; Charles, M.-A.; Melchior, M.; El-Khoury Lesueur, F. Maternal education and offspring birth weight for gestational age: The mediating effect of smoking during pregnancy. Eur. J. Public Health 2020, 30, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Ntani, G.; Inskip, H.; Barker, M.; Cummins, S.; Cooper, C.; Moon, G.; Baird, J. Education and the Relationship Between Supermarket Environment and Diet. Am. J. Prev. Med. 2016, 51, e27–e34. [Google Scholar] [CrossRef]
- Song, S.; Ghosh, J.; Mainigi, M.; Turan, N.; Weinerman, R.; Truongcao, M.; Coutifaris, C.; Sapienza, C. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin. Epigenetics 2015, 7, 41. [Google Scholar] [CrossRef]
- Kindsfather, A.J.; Czekalski, M.A.; Pressimone, C.A.; Erisman, M.P.; Mann, M.R.W. Perturbations in imprinted methylation from assisted reproductive technologies but not advanced maternal age in mouse preimplantation embryos. Clin. Epigenetics 2019, 11, 162. [Google Scholar] [CrossRef]
- Liu, Y.; Murphy, S.K.; Murtha, A.P.; Fuemmeler, B.F.; Schildkraut, J.; Huang, Z.; Overcash, F.; Kurtzberg, J.; Jirtle, R.; Iversen, E.S.; et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics 2012, 7, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Nye, M.D.; Hoyo, C.; Huang, Z.; Vidal, A.C.; Wang, F.; Overcash, F.; Smith, J.S.; Vasquez, B.; Hernandez, B.; Swai, B.; et al. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS ONE 2013, 8, e56325. [Google Scholar] [CrossRef]
- Joubert, B.R.; Felix, J.F.; Yousefi, P.; Bakulski, K.M.; Just, A.C.; Breton, C.; Reese, S.E.; Markunas, C.A.; Richmond, R.C.; Xu, C.J.; et al. DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am. J. Hum. Genet. 2016, 98, 680–696. [Google Scholar] [CrossRef] [PubMed]
- Rotroff, D.M.; Joubert, B.R.; Marvel, S.W.; Haberg, S.E.; Wu, M.C.; Nilsen, R.M.; Ueland, P.M.; Nystad, W.; London, S.J.; Motsinger-Reif, A. Maternal smoking impacts key biological pathways in newborns through epigenetic modification in Utero. BMC Genom. 2016, 17, 976. [Google Scholar] [CrossRef]
- Joubert, B.R.; Haberg, S.E.; Nilsen, R.M.; Wang, X.; Vollset, S.E.; Murphy, S.K.; Huang, Z.; Hoyo, C.; Midttun, O.; Cupul-Uicab, L.A.; et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012, 120, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Szyf, M. The epigenetics of social adversity in early life: Implications for mental health outcomes. Neurobiol. Dis. 2010, 39, 66–72. [Google Scholar] [CrossRef]
- Szyf, M. The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life. Epigenetics 2011, 6, 971–978. [Google Scholar] [CrossRef]
- Levesque, M.L.; Casey, K.F.; Szyf, M.; Ismaylova, E.; Ly, V.; Verner, M.P.; Suderman, M.; Brendgen, M.; Vitaro, F.; Dionne, G.; et al. Genome-wide DNA methylation variability in adolescent monozygotic twins followed since birth. Epigenetics 2014, 9, 1410–1421. [Google Scholar] [CrossRef]
- House, J.S.; Mendez, M.; Maguire, R.L.; Gonzalez-Nahm, S.; Huang, Z.; Daniels, J.; Murphy, S.K.; Fuemmeler, B.F.; Wright, F.A.; Hoyo, C. Periconceptional maternal mediterranean diet is associated with favorable offspring behaviors and altered CpG methylation of imprinted genes. Front. Cell Dev. Biol. 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Canovas, S.; Ross, P.J.; Kelsey, G.; Coy, P. DNA Methylation in Embryo Development: Epigenetic Impact of ART (Assisted Reproductive Technologies). Bioessays 2017, 39, 1700106. [Google Scholar] [CrossRef]
| Demographic | ART ET/ICSI | |||
|---|---|---|---|---|
| Mean (SD) or Frequency (% of Total) | ||||
| Exposed | Non-Exposed | All | ||
| n = 27 (4.7%) | n = 516 (89.9%) | n = 543 (100%) | ||
| Maternal age # | 36.3 (3.6) | 30.1 (5.5) | 30.4 (5.5) | |
| Maternal pre-pregnancy BMI # | 24.5 (6.1) | 27.7 (8.8) | 27.5 (8.7) | |
| Race / Ethnicity # | White | 21 (77.8%) | 242 (46.9%) | 263 (48.4%) |
| Non-white | 6 (22.2%) | 274 (53.1%) | 280 (51.6%) | |
| Parity * | Nulliparous | 17 (63%) | 213 (41.4%) | 230 (42.5%) |
| One or more | 10 (37%) | 301 (58.6%) * | 311 (57.5%) * | |
| Diabetes † | None | 25 (96.2%) | 444 (88.1%) | 469 (88.5%) |
| Any type | 1 (3.8%) | 60 (11.9%) | 61 (11.5%) | |
| Sex | Male | 11 (40.7%) | 272 (52.7%) | 283 (52.1%) |
| Female | 16 (59.3%) | 244 (47.3%) | 260 (47.9%) | |
| Demographic | Clomifene Only | |||
| n = 22 (6.8%) | n = 303 (93.2%) | n = 325 (100%) | ||
| Maternal age # | 33.6 (4.9) | 30.0 (5.1) | 30.2 (5.2) | |
| Maternal pre-pregnancy BMI | 25.6 (5.8) | 27.4 (8.1) | 27.2 (7.9) | |
| Race/Ethnicity # | White | 18 (81.8%) | 143 (47.2%) | 161 (49.5%) |
| Non-white | 4 (18.2%) | 160 (52.8%) | 164 (50.5%) | |
| Parity | Nulliparous | 7 (31.8%) | 136 (45.0%) | 143 (44.0%) |
| One or more | 15 (68.2%) | 167 (55.0%) | 182 (55.0%) | |
| Diabetes ‡ | No | 20 (90.9%) | 269 (90.0%) | 289 (90.0%) |
| Any type | 2 (9.1%) | 30 (10.0%) | 32 (10.0%) | |
| Sex | Male | 12 (54.5%) | 165 (54.5%) | 177 (54.5%) |
| Female | 10 (45.5%) | 138 (45.5%) | 148 (45.5%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lloyd, D.T.; Skinner, H.G.; Maguire, R.; Murphy, S.K.; Motsinger-Reif, A.A.; Hoyo, C.; House, J.S. Clomifene and Assisted Reproductive Technology in Humans Are Associated with Sex-Specific Offspring Epigenetic Alterations in Imprinted Control Regions. Int. J. Mol. Sci. 2022, 23, 10450. https://doi.org/10.3390/ijms231810450
Lloyd DT, Skinner HG, Maguire R, Murphy SK, Motsinger-Reif AA, Hoyo C, House JS. Clomifene and Assisted Reproductive Technology in Humans Are Associated with Sex-Specific Offspring Epigenetic Alterations in Imprinted Control Regions. International Journal of Molecular Sciences. 2022; 23(18):10450. https://doi.org/10.3390/ijms231810450
Chicago/Turabian StyleLloyd, Dillon T., Harlyn G. Skinner, Rachel Maguire, Susan K. Murphy, Alison A. Motsinger-Reif, Cathrine Hoyo, and John S. House. 2022. "Clomifene and Assisted Reproductive Technology in Humans Are Associated with Sex-Specific Offspring Epigenetic Alterations in Imprinted Control Regions" International Journal of Molecular Sciences 23, no. 18: 10450. https://doi.org/10.3390/ijms231810450
APA StyleLloyd, D. T., Skinner, H. G., Maguire, R., Murphy, S. K., Motsinger-Reif, A. A., Hoyo, C., & House, J. S. (2022). Clomifene and Assisted Reproductive Technology in Humans Are Associated with Sex-Specific Offspring Epigenetic Alterations in Imprinted Control Regions. International Journal of Molecular Sciences, 23(18), 10450. https://doi.org/10.3390/ijms231810450





