Genome-Wide Analysis of the Growth-Regulating Factor (GRF) Family in Aquatic Plants and Their Roles in the ABA-Induced Turion Formation of Spirodela polyrhiza
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of GRF Genes in 13 Aquatic Plants
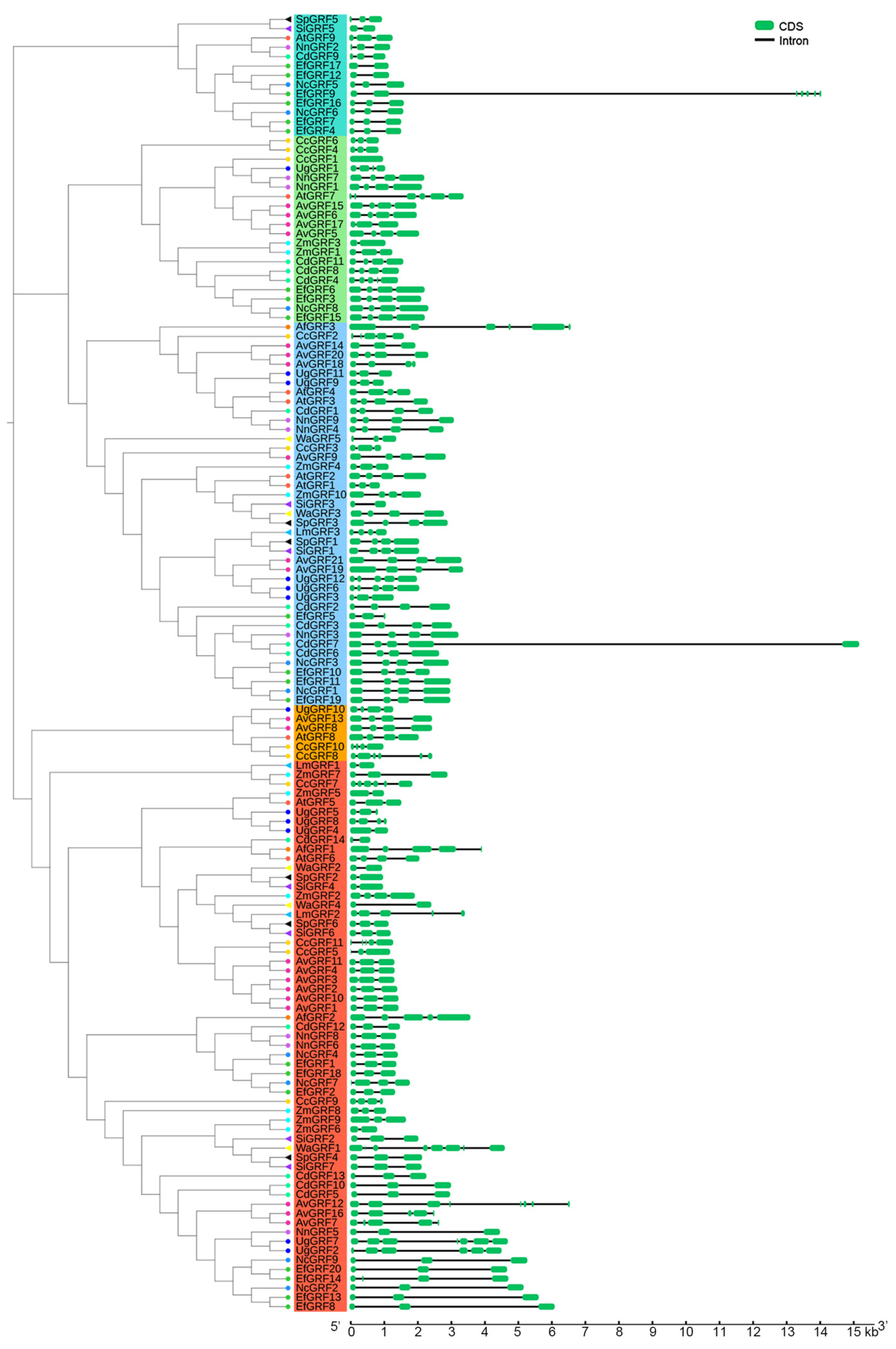
2.2. Gene Structure and Conserved Motifs of the GRF Genes
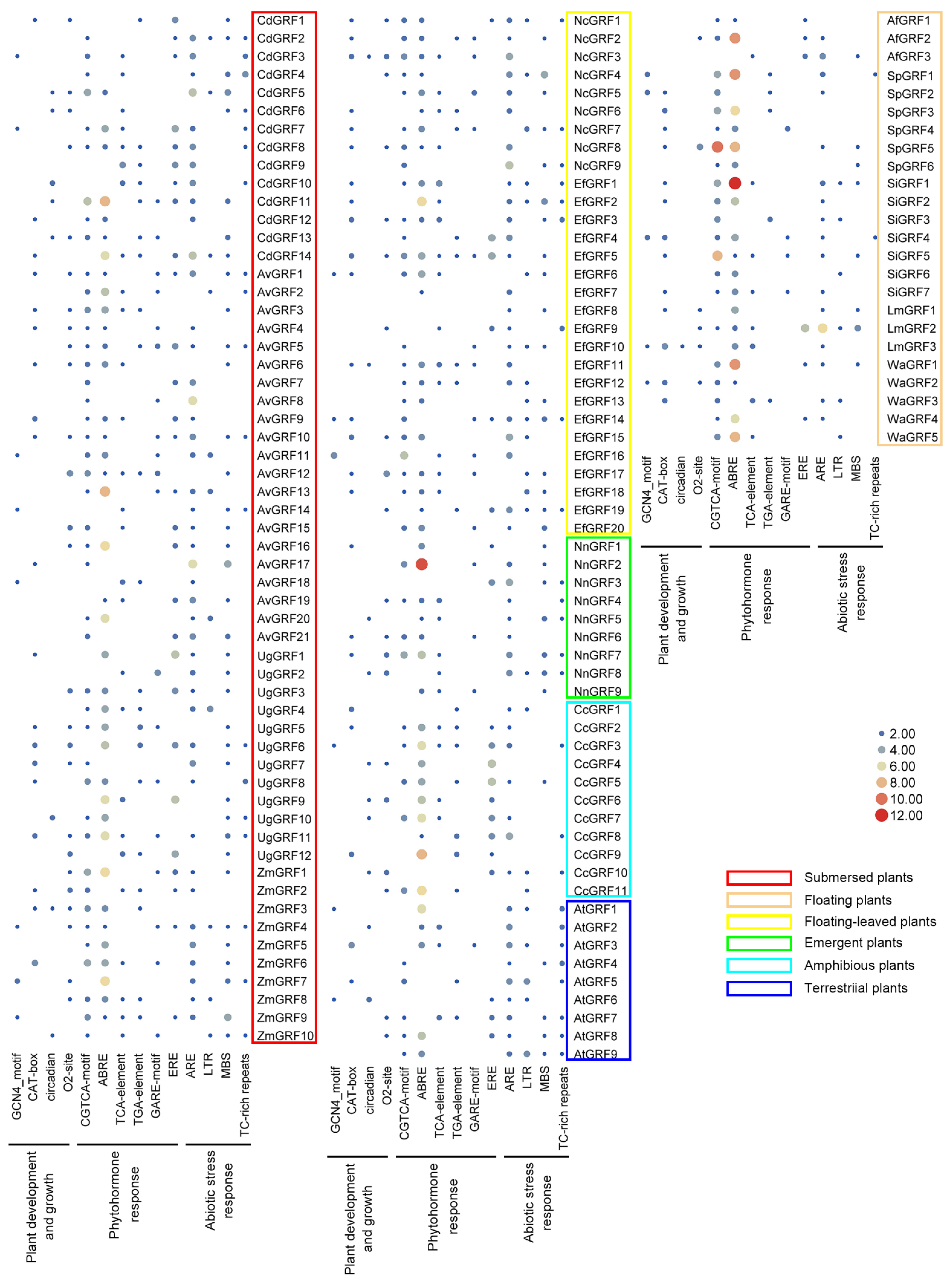
2.3. Cis-Regulatory Element Analysis of the GRF Genes
2.4. Chromosomal Localization, Gene Duplication, and Synteny Analysis of the GRFs in S. polyrhiza
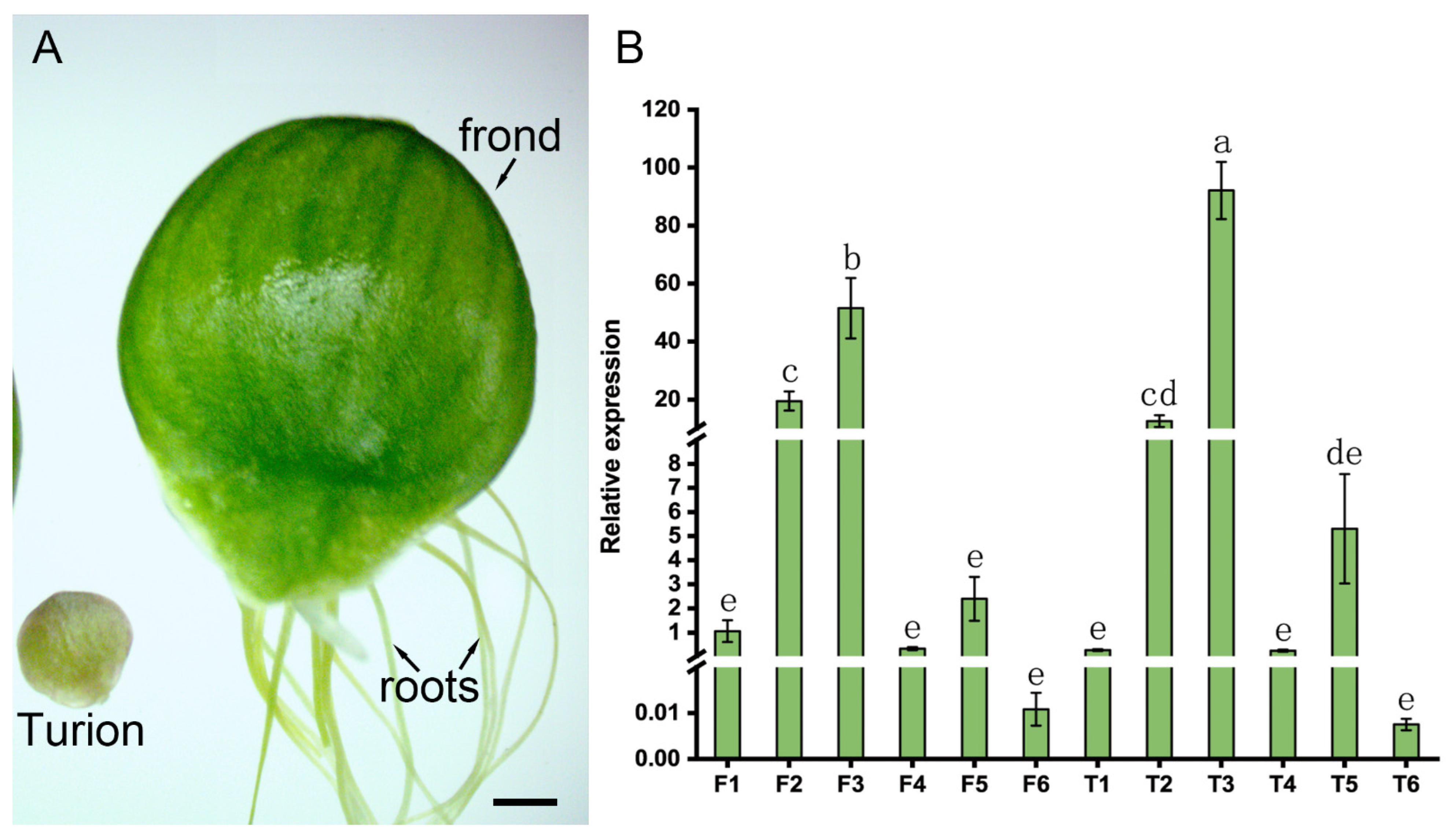
2.5. Expression Analysis of the SpGRF Genes in the Process of ABA-Induced Turion Formation
3. Discussion
4. Materials and Methods
4.1. Identification of the GRF Genes
4.2. Physiochemical Properties and Subcellular Localization
4.3. Phylogenetic Classification, Gene Structures, and Motif Analysis
4.4. Cis-Acting Elements, Chromosomal Localization, Gene Duplication, and Synteny Analysis
4.5. Expression Analysis of the SpGRF Genes by qRT-PCR (Quantitative Real-Time PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, J.H.; Tsukaya, H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 2015, 66, 6093–6107. [Google Scholar] [CrossRef]
- Li, G.; Hu, S.; Zhao, X.; Kumar, S.; Li, Y.; Yang, J.; Hou, H. Mechanisms of the morphological plasticity induced by phytohormones and the environment in plants. Int. J. Mol. Sci. 2021, 22, 765. [Google Scholar] [CrossRef]
- Tang, Y.H.; Cheng, W.; Li, S.; Li, Y.; Wang, X.; Xie, J.T.; He, Y.Y.; Wang, Y.Y.; Niu, Y.R.; Bao, X.X.; et al. Genome-wide identification and expression analysis of the growth regulating factor (GRF) family in Jatropha curcas. PLoS ONE 2021, 16, e0254711. [Google Scholar] [CrossRef] [PubMed]
- Treich, I.; Cairns, B.R.; de los Santos, T.; Brewster, E.; Carlson, M. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol. Cell. Biol. 1995, 15, 4240–4248. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, J.H.; Kende, H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef]
- van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef]
- Liu, H.H.; Guo, S.Y.; Xu, Y.Y.; Li, C.H.; Zhang, Z.Y.; Zhang, D.J.; Xu, S.J.; Zhang, C.; Chong, K. OsmiR396d-Regulated OsGRFs Function in Floral Organogenesis in Rice through Binding to Their Targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef]
- Hewezi, T.; Maier, T.R.; Nettleton, D.; Baum, T.J. The Arabidopsis MicroRNA396-GRF1/GRF3 Regulatory Module Acts as a Developmental Regulator in the Reprogramming of Root Cells during Cyst Nematode Infection. Plant Physiol. 2012, 159, 321–335. [Google Scholar] [CrossRef]
- Li, S.C.; Gao, F.Y.; Xie, K.L.; Zeng, X.H.; Cao, Y.; Zeng, J.; He, Z.S.; Ren, Y.; Li, W.B.; Deng, Q.M.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 2010, 137, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, D.S.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef]
- Zhang, J.F.; Li, Z.F.; Jin, J.J.; Xie, X.D.; Zhang, H.; Chen, Q.S.; Luo, Z.P.; Yang, J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 2018, 639, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Zan, T.; Zhang, L.; Xie, T.; Li, L. Genome-Wide Identification and Analysis of the Growth-Regulating Factor (GRF) Gene Family and GRF-Interacting Factor Family in Triticum aestivum L. Biochem. Genet. 2020, 58, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yang, Y.Z.; Luo, X.F.; Zhou, W.G.; Dai, Y.J.; Zheng, C.; Liu, W.G.; Yang, W.Y.; Shu, K. Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol. 2019, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Wang, Z.; Feng, C.; Luo, J.; Zhang, Y.; Tu, J.; Cai, X.; Gao, P. Genome-Wide Identification and Structural Characterization of Growth-Regulating Factors (GRFs) in Actinida eriantha and Actinidia chinensis. Plants 2022, 11, 1633. [Google Scholar] [CrossRef]
- Cao, J.F.; Huang, J.Q.; Liu, X.; Huang, C.C.; Zheng, Z.S.; Zhang, X.F.; Shangguan, X.X.; Wang, L.J.; Zhang, Y.G.; Wendel, J.F.; et al. Genome-wide characterization of the GRF family and their roles in response to salt stress in Gossypium. BMC Genom. 2020, 21, 575. [Google Scholar] [CrossRef]
- Li, Z.; Xie, Q.; Yan, J.; Chen, J.; Chen, Q. Genome-Wide Identification and Characterization of the Abiotic-Stress-Responsive GRF Gene Family in Diploid Woodland Strawberry (Fragaria vesca). Plants 2021, 10, 1916. [Google Scholar] [CrossRef]
- Carpenter, S.R.; Lodge, D.M. Effects of Submersed Macrophytes on Ecosystem Processes. Aquat. Bot. 1986, 26, 341–370. [Google Scholar] [CrossRef]
- Engelhardt, K.A.; Ritchie, M.E. Effects of macrophyte species richness on wetland ecosystem functioning and services. Nature 2001, 411, 687–689. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Wu, H.; Hao, B.; Huang, W.; Liu, G. Bioaccumulation of heavy metals by submerged macrophytes: Looking for hyperaccumulators in eutrophic lakes. Environ. Sci. Technol. 2013, 47, 4695–4703. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Hu, S.; Hou, H.; Kimura, S. Heterophylly: Phenotypic plasticity of leaf shape in aquatic and amphibious plants. Plants 2019, 8, 420. [Google Scholar] [CrossRef]
- He, D.; Guo, P.; Gugger, P.F.; Guo, Y.; Liu, X.; Chen, J. Investigating the molecular basis for heterophylly in the aquatic plant Potamogeton octandrus (Potamogetonaceae) with comparative transcriptomics. PeerJ 2018, 6, e4448. [Google Scholar] [CrossRef] [PubMed]
- Wanke, D. The ABA-mediated switch between submersed and emersed life-styles in aquatic macrophytes. J. Plant Res. 2011, 124, 467–475. [Google Scholar] [CrossRef]
- Brunetti, C.; Sebastiani, F.; Tattini, M. Review: ABA, flavonols, and the evolvability of land plants. Plant Sci. 2019, 280, 448–454. [Google Scholar] [CrossRef]
- Li, G.; Hu, S.; Yang, J.; Schultz, E.A.; Clarke, K.; Hou, H. Water-Wisteria as an ideal plant to study heterophylly in higher aquatic plants. Plant Cell Rep. 2017, 36, 1225–1236. [Google Scholar] [CrossRef]
- Kim, J.; Joo, Y.; Kyung, J.; Jeon, M.; Park, J.Y.; Lee, H.G.; Chung, D.S.; Lee, E.; Lee, I. A molecular basis behind heterophylly in an amphibious plant, Ranunculus trichophyllus. PLoS Genet. 2018, 14, e1007208. [Google Scholar] [CrossRef]
- Iida, S.; Ikeda, M.; Amano, M.; Sakayama, H.; Kadono, Y.; Kosuge, K. Loss of heterophylly in aquatic plants: Not ABA-mediated stress but exogenous ABA treatment induces stomatal leaves in Potamogeton perfoliatus. J. Plant Res. 2016, 129, 853–862. [Google Scholar] [CrossRef][Green Version]
- Wang, W.Q.; Messing, J. Analysis of ADP-glucose pyrophosphorylase expression during turion formation induced by abscisic acid in Spirodela polyrhiza (greater duckweed). BMC Plant Biol. 2012, 12, 5. [Google Scholar] [CrossRef]
- Smart, C.C.; Fleming, A.J. A Plant Gene with Homology to D-Myo-Inositol-3-Phosphate Synthase Is Rapidly and Spatially up-Regulated during an Abscisic-Acid-Induced Morphogenic Response in Spirodela-Polyrrhiza. Plant J. 1993, 4, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Vercruysse, J.; Baekelandt, A.; Gonzalez, N.; Inze, D. Molecular networks regulating cell division during Arabidopsis leaf growth. J. Exp. Bot. 2020, 71, 2365–2378. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar] [CrossRef] [PubMed]
- Du, W.X.; Yang, J.F.; Li, Q.; Su, Q.; Yi, D.X.; Pang, Y.Z. Genome-Wide Identification and Characterization of Growth Regulatory Factor Family Genes in Medicago. Int. J. Mol. Sci. 2022, 23, 6905. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, K.; Ning, L.; He, J.; Ma, X.; Li, Z.; Zhang, X.; Yin, D. Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2019, 20, 4120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Yang, J.J.; Li, G.J.; Sun, Z.L.; Hu, S.Q.; Chen, Y.; Guo, W.J.; Hou, H.W. Genome-wide identification and comparative analysis of the WRKY gene family in aquatic plants and their response to abiotic stresses in giant duckweed (Spirodela polyrhiza). Genomics 2021, 113, 1761–1777. [Google Scholar] [CrossRef]
- Santamaria, L. Why are most aquatic plants widely distributed? Dispersal, clonal growth and small-scale heterogeneity in a stressful environment. Acta Oecol. 2002, 23, 137–154. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y.; et al. Arabidopsis GROWTH-REGULATING FACTOR7 Functions as a Transcriptional Repressor of Abscisic Acid- and Osmotic Stress-Responsive Genes, Including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Lin, B.L.; Yang, W.J. Blue light and abscisic acid independently induce heterophyllous switch in marsilea quadrifolia. Plant Physiol. 1999, 119, 429–434. [Google Scholar] [CrossRef]
- Smart, C.C.; Trewavas, A.J. Abscisic-Acid-Induced Turion Formation in Spirodela-Polyrrhiza L.1. Production and Development of the Turion. Plant Cell Environ. 1983, 6, 507–514. [Google Scholar] [CrossRef]
- Smart, C.C.; Fleming, A.J.; Chaloupkova, K.; Hanke, D.E. The Physiological-Role of Abscisic-Acid in Eliciting Turion Morphogenesis. Plant Physiol. 1995, 108, 623–632. [Google Scholar] [CrossRef]
- Vercruyssen, L.; Tognetti, V.B.; Gonzalez, N.; Van Dingenen, J.; De Milde, L.; Bielach, A.; De Rycke, R.; Van Breusegem, F.; Inze, D. GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 2015, 167, 817–832. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Fujikura, U.; Olas, J.J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018, 14, e1007484. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, Y.; Li, G.J.; Yang, J.J.; Zhao, X.Y.; Sun, Z.L.; Hou, H.W. Role of Nramp transporter genes of Spirodela polyrhiza in cadmium accumulation. Ecotoxicol. Environ. Saf. 2021, 227, 112907. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Chen, Y.; Zhao, X.; Yang, J.; Wang, X.; Li, X.; Hu, S.; Hou, H. Genome-Wide Analysis of the Growth-Regulating Factor (GRF) Family in Aquatic Plants and Their Roles in the ABA-Induced Turion Formation of Spirodela polyrhiza. Int. J. Mol. Sci. 2022, 23, 10485. https://doi.org/10.3390/ijms231810485
Li G, Chen Y, Zhao X, Yang J, Wang X, Li X, Hu S, Hou H. Genome-Wide Analysis of the Growth-Regulating Factor (GRF) Family in Aquatic Plants and Their Roles in the ABA-Induced Turion Formation of Spirodela polyrhiza. International Journal of Molecular Sciences. 2022; 23(18):10485. https://doi.org/10.3390/ijms231810485
Chicago/Turabian StyleLi, Gaojie, Yan Chen, Xuyao Zhao, Jingjing Yang, Xiaoyu Wang, Xiaozhe Li, Shiqi Hu, and Hongwei Hou. 2022. "Genome-Wide Analysis of the Growth-Regulating Factor (GRF) Family in Aquatic Plants and Their Roles in the ABA-Induced Turion Formation of Spirodela polyrhiza" International Journal of Molecular Sciences 23, no. 18: 10485. https://doi.org/10.3390/ijms231810485
APA StyleLi, G., Chen, Y., Zhao, X., Yang, J., Wang, X., Li, X., Hu, S., & Hou, H. (2022). Genome-Wide Analysis of the Growth-Regulating Factor (GRF) Family in Aquatic Plants and Their Roles in the ABA-Induced Turion Formation of Spirodela polyrhiza. International Journal of Molecular Sciences, 23(18), 10485. https://doi.org/10.3390/ijms231810485






