The Multifaceted Role of Signal Peptide-CUB-EGF Domain-Containing Protein (SCUBE) in Cancer
Abstract
:1. Introduction
2. Defects in SCUBE Family in Cancer
3. SCUBE Association with Other Proteins
4. Emerging Role of SCUBE Family in Human Cancers
4.1. Role of SCUBE as a Tumor Suppressor
4.2. SCUBE in Cell Proliferation and Cancer Progression
4.3. SCUBE in Regulating Angiogenesis
4.4. SCUBE in Invasion and Metastasis
4.5. SCUBE in the Maintenance of Cancer Stemness and Drug Resistance
4.6. SCUBE and Immune Response
4.7. SCUBE as a Biomarker
5. Role of SCUBE in Patient Survival
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SCUBE | Signal peptide CUB and EGF-like domain-containing protein |
| BMP | Bone morphogenetic protein |
| NSCLC | Nonsmall-cell lung cancer |
| VEGF | Vascular endothelial growth factor |
| FOXO1 | Forkhead box protein 1 |
| TGF-β | Transforming growth factor-beta |
| BC | Breast cancer |
| TNBC | Triple-negative breast cancer |
| lncRNA | Long noncoding RNA |
References
- Tsai, M.-T.; Cheng, C.-J.; Lin, Y.-C.; Chen, C.-C.; Wu, A.-R.; Wu, M.-T.; Hsu, C.-C.; Yang, R.-B. Isolation and characterization of a secreted, cell-surface glycoprotein SCUBE2 from humans. Biochem. J. 2009, 422, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Grimmond, S.; Larder, R.; Van Hateren, N.; Siggers, P.; Hulsebos, T.J.; Arkell, R.; Greenfield, A. Cloning, Mapping, and Expression Analysis of a Gene Encoding a Novel Mammalian EGF-Related Protein (SCUBE1). Genomics 2000, 70, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Grimmond, S.; Larder, R.; Van Hateren, N.; Siggers, P.; Morse, S.; Hacker, T.; Arkell, R.; Greenfield, A. Expression of a novel mammalian epidermal growth factor-related gene during mouse neural development. Mech. Dev. 2001, 102, 209–211. [Google Scholar] [CrossRef]
- Tu, C.-F.; Su, Y.-H.; Huang, Y.-N.; Tsai, M.-T.; Li, L.-T.; Chen, Y.-L.; Cheng, C.-J.; Dai, D.-F.; Yang, R.-B. Localization and characterization of a novel secreted protein SCUBE1 in human platelets. Cardiovasc. Res. 2006, 71, 486–495. [Google Scholar] [CrossRef]
- Yang, R.-B.; Ng, C.K.D.; Wasserman, S.M.; Colman, S.D.; Shenoy, S.; Mehraban, F.; Kömüves, L.G.; Tomlinson, J.E.; Topper, J.N. Identification of a Novel Family of Cell-surface Proteins Expressed in Human Vascular Endothelium. J. Biol. Chem. 2002, 277, 46364–46373. [Google Scholar] [CrossRef]
- Jakobs, P.; Exner, S.; Schürmann, S.; Pickhinke, U.; Bandari, S.; Ortmann, C.; Kupich, S.; Schulz, P.; Hansen, U.; Seidler, D.G.; et al. Scube2 enhances proteolytic Shh processing from the surface of Shh-producing cells. J. Cell Sci. 2014, 127, 1726–1737. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Chao, T.-Y.; Yeh, C.-T.; Roffler, S.R.; Kannagi, R.; Yang, R.-B. Endothelial SCUBE2 Interacts with VEGFR2 and Regulates VEGF-Induced Angiogenesis. Arter. Thromb. Vasc. Biol. 2017, 37, 144–155. [Google Scholar] [CrossRef]
- Wu, B.-T.; Su, Y.-H.; Tsai, M.-T.; Wasserman, S.M.; Topper, J.N.; Yang, R.-B. A Novel Secreted, Cell-surface Glycoprotein Containing Multiple Epidermal Growth Factor-like Repeats and One CUB Domain Is Highly Expressed in Primary Osteoblasts and Bones. J. Biol. Chem. 2004, 279, 37485–37490. [Google Scholar] [CrossRef]
- Karagüzel, E.; Menteşe, A.; Kazaz, I.O.; Demir, S.; Örem, A.; Okatan, A.E.; Altay, D.U.; Yaman, S. SCUBE1: A promising biomarker in renal cell cancer. Int. Braz J. Urol 2017, 43, 638–643. [Google Scholar] [CrossRef]
- Icel, E.; Icel, A.; Mertoglu, C.; Tasli, N.G.; Karakurt, Y.; Ucak, T.; Gunay, M. Serum SCUBE-1 levels in patients with diabetic retinopathy. Int. Ophthalmol. 2019, 40, 859–865. [Google Scholar] [CrossRef]
- International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN); Harley, J.B.; Alarcón-Riquelme, M.E.; Criswell, L.A.; Jacob, C.O.; Kimberly, R.P.; Moser, K.L.; Tsao, B.P.; Vyse, T.J.; Langefeld, C.D.; et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008, 40, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Tachmazidou, I.; Hatzikotoulas, K.; Southam, L.; Esparza-Gordillo, J.; Haberland, V.; Zheng, J.; Johnson, T.; Koprulu, M.; Zengini, E.; Steinberg, J.; et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat. Genet. 2019, 51, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Capkin, A.A.; Demir, S.; Mentese, A.; Bulut, Ç.; Ayar, A. Can signal peptide-CUB-EGF domain-containing protein (SCUBE) levels be a marker of angiogenesis in patients with psoriasis? Arch. Dermatol. Res. 2017, 309, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Tekin, Y.B.; Erin, K.B.; Yilmaz, A. Evaluation of SCUBE-1 levels as a placental dysfunction marker at gestational diabetes mellitus. Gynecol. Endocrinol. 2019, 36, 417–420. [Google Scholar] [CrossRef]
- Bolayır, H.A. The role of SCUBE1 in the pathogenesis of no-reflow phenomenon presenting with ST segment elevation myocardial infarction. Anatol. J. Cardiol. 2017, 18, 122–127. [Google Scholar] [CrossRef]
- Heit, J.A.; Cunningham, J.M.; Petterson, T.M.; Armasu, S.M.; Rider, D.N.; DE Andrade, M. Genetic variation within the anticoagulant, procoagulant, fibrinolytic and innate immunity pathways as risk factors for venous thromboembolism. J. Thromb. Haemost. 2011, 9, 1133–1142. [Google Scholar] [CrossRef]
- Kühnisch, J.; The GINI-10 plus study group; Thiering, E.; Heitmüller, D.; Tiesler, C.M.T.; Grallert, H.; Heinrich-Weltzien, R.; Hickel, R.; Heinrich, J.; The LISA-10 plus study group. Genome-wide association study (GWAS) for molar–incisor hypomineralization (MIH). Clin. Oral Investig. 2013, 18, 677–682. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Chen, C.-C.; Cheng, C.-J.; Yang, R.-B. Domain and Functional Analysis of a Novel Breast Tumor Suppressor Protein, SCUBE2. J. Biol. Chem. 2011, 286, 27039–27047. [Google Scholar] [CrossRef]
- Chen, J.-H.; Kuo, K.-T.; Bamodu, O.A.; Lin, Y.-C.; Yang, R.-B.; Yeh, C.-T.; Chao, T.-Y. Upregulated SCUBE2 expression in breast cancer stem cells enhances triple negative breast cancer aggression through modulation of notch signaling and epithelial-to-mesenchymal transition. Exp. Cell Res. 2018, 370, 444–453. [Google Scholar] [CrossRef]
- Parris, T.Z.; Aziz, L.; Kovács, A.; Hajizadeh, S.; Nemes, S.; Semaan, M.; Chen, C.Y.; Karlsson, P.; Helou, K. Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer 2014, 14, 324. [Google Scholar] [CrossRef] [Green Version]
- Skrzypczak, M.; Lattrich, C.; Häring, J.; Schüler, S.; Ortmann, O.; Treeck, O. Expression of SCUBE2 gene declines in high grade endometrial cancer and associates with expression of steroid hormone receptors and tumor suppressor PTEN. Gynecol. Endocrinol. 2013, 29, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Miao, S.; Li, Y. SCUBE2 inhibits the proliferation, migration and invasion of human non-small cell lung cancer cells through regulation of the sonic hedgehog signaling pathway. Gene 2018, 672, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Li, C.; Feng, X.; Yu, A.; Tang, H.; Peng, Z.; Wang, X. Decreased expression of SCUBE2 is associated with progression and prognosis in colorectal cancer. Oncol. Rep. 2015, 33, 1956–1964. [Google Scholar] [CrossRef] [PubMed]
- Guo, E.; Liu, H.; Liu, X. Overexpression of SCUBE2 Inhibits Proliferation, Migration, and Invasion in Glioma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 437–444. [Google Scholar] [CrossRef]
- Han, N.; Lu, H.; Zhang, Z.; Ruan, M.; Yang, W.; Zhang, C. Comprehensive and in-depth analysis of microRNA and mRNA expression profile in salivary adenoid cystic carcinoma. Gene 2018, 678, 349–360. [Google Scholar] [CrossRef]
- Liang, W.; Yang, C.; Peng, J.; Qian, Y.; Wang, Z. The Expression of HSPD1, SCUBE3, CXCL14 and Its Relations with the Prognosis in Osteosarcoma. Cell Biophys. 2015, 73, 763–768. [Google Scholar] [CrossRef]
- Morris, M.R.; Ricketts, C.J.; Gentle, D.; McRonald, F.; Carli, N.; Khalili, H.; Brown, M.; Kishida, T.; Yao, M.; Banks, R.E.; et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 2010, 30, 1390–1401. [Google Scholar] [CrossRef]
- Yang, M.; Guo, M.; Hu, Y.; Jiang, Y. Scube regulates synovial angiogenesis-related signaling. Med. Hypotheses 2013, 81, 948–953. [Google Scholar] [CrossRef]
- Tai, T.-S.; Pai, S.-Y.; Ho, I.-C. Itm2a, a Target Gene of GATA-3, Plays a Minimal Role in Regulating the Development and Function of T Cells. PLoS ONE 2014, 9, e96535. [Google Scholar] [CrossRef]
- Kulus, M.; Sujka-Kordowska, P.; Konwerska, A.; Celichowski, P.; Kranc, W.; Kulus, J.; Piotrowska-Kempisty, H.; Antosik, P.; Bukowska, D.; Iżycki, D.; et al. New Molecular Markers Involved in Regulation of Ovarian Granulosa Cell Morphogenesis, Development and Differentiation during Short-Term Primary In Vitro Culture—Transcriptomic and Histochemical Study Based on Ovaries and Individual Separated Follicles. Int. J. Mol. Sci. 2019, 20, 3966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gal, P. C1s, the protease messenger of C1. Structure, function, and physiological significance. Immunobiology 2002, 205, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Arlaud, G.J.; Rossi, V.; Thielens, N.; Gaboriaud, C.; Bersch, B.; Hernandez, J.-F. Structural and Functional Studies on C1r and C1s: New Insights into the Mechanisms Involved in C1 Activity and Assembly. Immunobiology 1998, 199, 303–316. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Roffler, S.R.; Yan, Y.-T.; Yang, R.-B. Disruption of Scube2 Impairs Endochondral Bone Formation. J. Bone Miner. Res. 2015, 30, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Malik, M.F.A.; Saeed, M.; Haq, F. Copy number profiling of Oncotype DX genes reveals association with survival of breast cancer patients. Mol. Biol. Rep. 2018, 45, 2185–2192. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Y.; Peck, K.; Chang, Y.-L.; Pan, S.-H.; Cheng, Y.-F.; Lin, J.-C.; Yang, R.-B.; Hong, T.-M.; Yang, P.-C. SCUBE3 is an endogenous TGF-β receptor ligand and regulates the epithelial-mesenchymal transition in lung cancer. Oncogene 2011, 30, 3682–3693. [Google Scholar] [CrossRef]
- Cheng, C.-J.; Lin, Y.-C.; Tsai, M.-T.; Chen, C.-S.; Hsieh, M.-C.; Chen, C.-L.; Yang, R.-B. SCUBE2 Suppresses Breast Tumor Cell Proliferation and Confers a Favorable Prognosis in Invasive Breast Cancer. Cancer Res. 2009, 69, 3634–3641. [Google Scholar] [CrossRef]
- Andres, S.A.; Wittliff, J.L. Co-expression of genes with estrogen receptor-α and progesterone receptor in human breast carcinoma tissue. Horm. Mol. Biol. Clin. Investig. 2012, 12, 377–390. [Google Scholar] [CrossRef]
- Fei, H.; Chen, S.; Xu, C. RNA-sequencing and microarray data mining revealing: The aberrantly expressed mRNAs were related with a poor outcome in the triple negative breast cancer patients. Ann. Transl. Med. 2020, 8, 363. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Flores, E.R.; Brohl, A.S. Chromosome 19 miRNA cluster and CEBPB expression specifically mark and potentially drive triple negative breast cancers. PLoS ONE 2018, 13, e0206008. [Google Scholar] [CrossRef]
- Kadio, B.; Yaya, S.; Basak, A.; Djè, K.; Gomes, J.; Mesenge, C. Calcium role in human carcinogenesis: A comprehensive analysis and critical review of literature. Cancer Metastasis Rev. 2016, 35, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Heng, L.; Jia, Z.; Sun, J.; Zhao, Y.; Zhang, K.; Zhu, Y.; Lu, S. Integrated Analysis of Competing Endogenous RNAs Network Reveals Potential Signatures in Osteosarcoma Development. Technol. Cancer Res. Treat. 2020, 19, 1533033820957025. [Google Scholar] [CrossRef] [PubMed]
- Huo, Q.; He, X.; Li, Z.; Yang, F.; He, S.; Shao, L.; Hu, Y.; Chen, S.; Xie, N. SCUBE3 serves as an independent poor prognostic factor in breast cancer. Cancer Cell Int. 2021, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Orr, B.; Grace, O.C.; Brown, P.; Riddick, A.C.P.; Stewart, G.D.; Franco, O.E.; Hayward, S.; Thomson, A.A. Reduction of pro-tumorigenic activity of human prostate cancer-associated fibroblasts using Dlk1 or SCUBE1. Dis. Model. Mech. 2012, 6, 530–536. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Liu, C.-Y.; Kannagi, R.; Yang, R.-B. Inhibition of Endothelial SCUBE2 (Signal Peptide-CUB-EGF Domain-Containing Protein 2), a Novel VEGFR2 (Vascular Endothelial Growth Factor Receptor 2) Coreceptor, Suppresses Tumor Angiogenesis. Arter. Thromb. Vasc. Biol. 2018, 38, 1202–1215. [Google Scholar] [CrossRef]
- Jiang, W.; Han, X.; Wang, J.; Wang, L.; Xu, Z.; Wei, Q.; Zhang, W.; Wang, H. miR-22 enhances the radiosensitivity of small-cell lung cancer by targeting the WRNIP1. J. Cell. Biochem. 2019, 120, 17650–17661. [Google Scholar] [CrossRef]
- Kim, C.; Paik, S. Gene-expression-based prognostic assays for breast cancer. Nat. Rev. Clin. Oncol. 2010, 7, 340–347. [Google Scholar] [CrossRef]
- Sparano, J.A.; Paik, S. Development of the 21-Gene Assay and Its Application in Clinical Practice and Clinical Trials. J. Clin. Oncol. 2008, 26, 721–728. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Lee, Y.-C.; Li, L.-H.; Cheng, C.-J.; Yang, R.-B. Tumor suppressor SCUBE2 inhibits breast-cancer cell migration and invasion through the reversal of epithelial-mesenchymal transition. J. Cell Sci. 2013, 127, 85–100. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, M.; Da, L.; Huang, W.; Zhang, C. Circular RNA circ_SETD2 represses breast cancer progression via modulating the miR-155-5p/SCUBE2 axis. Open Med. 2020, 15, 940–953. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, R.-Y.; Xiang, X.-J. Reduced expression of SCUBE2 predicts poor prognosis in gastric cancer patients. Int. J. Clin. Exp. Pathol. 2018, 11, 972–980. [Google Scholar] [PubMed]
- Islam, S.; Mokhtari, R.; Noman, A.; Uddin, M.; Rahman, M.; Azadi, M.; Zlotta, A.; van der Kwast, T.; Yeger, H.; Farhat, W. Sonic hedgehog (Shh) signaling promotes tumorigenicity and stemness via activation of epithelial-to-mesenchymal transition (EMT) in bladder cancer. Mol. Carcinog. 2015, 55, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Shi, W.; Guo, H.; Long, W.; Wang, Y.; Qi, J.; Liu, J.; Xu, Y. The Inhibitory Effect of (−)-Epigallocatechin-3-Gallate on Breast Cancer Progression via Reducing SCUBE2 Methylation and DNMT Activity. Molecules 2019, 24, 2899. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Cheng, Y.-F.; Siow, T.Y.; Kumar, A.; Peck, K.; Chang, C. SCUBE3 regulation of early lung cancer angiogenesis and metastatic progression. Clin. Exp. Metastasis 2013, 30, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Qin, Q.; Wang, Q.; Zhang, J.; Xu, Y.; Li, W.; Gu, M.; Chen, S.; Deng, A. SCUBE3 overexpression predicts poor prognosis in non-small cell lung cancer. Biosci. Trends 2013, 7, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Kallarackal, J.; Burger, F.; Bianco, S.; Romualdi, A.; Schad, M. A 3-gene biomarker signature to predict response to taxane-based neoadjuvant chemotherapy in breast cancer. PLoS ONE 2020, 15, e0230313. [Google Scholar] [CrossRef]
- Shuai, C.; Yuan, F.; Liu, Y.; Wang, C.; Wang, J.; He, H. Estrogen receptor—positive breast cancer survival prediction and analysis of resistance–related genes introduction. PeerJ 2021, 9, e12202. [Google Scholar] [CrossRef]
- Erol, O.; Ellidağ, H.Y.; Özel, M.K.; Derbent, A.U.; Eren, E.; Yılmaz, N. Circulating SCUBE1 levels in women with polycystic ovary syndrome. J. Turk. Soc. Obstet. Gynecol. 2018, 15, 152–158. [Google Scholar] [CrossRef]
- Mentese, A.; Fidan, E.; Sumer, A.U.; Karahan, S.C.; Sonmez, M.; Altay, D.U.; Kavgaci, H.; Alver, A. Is SCUBE 1 a new biomarker for gastric cancer? Cancer Biomark. 2012, 11, 191–195. [Google Scholar] [CrossRef]
- Topcu, T.O.; Kavgaci, H.; Ozdemir, F.; Aksoy, A.; Erdem, D.; Mentese, A.; Yaman, H.; Tufan, G.; Orem, A.; Aydin, F. Elevated Serum Levels of SCUBE1, a Marker for Coagulation, in Patients with Breast Cancer. Tohoku J. Exp. Med. 2015, 237, 127–132. [Google Scholar] [CrossRef]
- Diamanti-Kandarakis, E.; Paterakis, T.; Kandarakis, H.A. Indices of Low-Grade Inflammation in Polycystic Ovary Syndrome. Ann. N. Y. Acad. Sci. 2006, 1092, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Akdoğan, E.; Ayaz, T.; Kırbaş, A.; Rakıcı, H. Is SCUBE1 helpful to predict the arterial thrombotic risk in patients with multi-ple myeloma: A preliminary study. Hippokratia 2019, 23, 21–24. [Google Scholar] [PubMed]
- Rickles, F.R.; Patierno, S.; Fernandez, P.M. Tissue Factor, Thrombin, and Cancer. Chest 2003, 124, 58S–68S. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.-F.; Thajeb, P.; Tu, C.-F.; Chiang, F.-T.; Chen, C.-H.; Yang, R.-B.; Chen, J.-J. Plasma Concentration of SCUBE1, a Novel Platelet Protein, Is Elevated in Patients with Acute Coronary Syndrome and Ischemic Stroke. J. Am. Coll. Cardiol. 2008, 51, 2173–2180. [Google Scholar] [CrossRef]
- Koleck, T.A.; Conley, Y.P. Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer Targets Ther. 2016, 8, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, C.; Li, H.; Li, H.; Li, Y. Identifying Key Genes for Nasopharyngeal Carcinoma by Prioritized Consensus Differentially Expressed Genes Caused by Aberrant Methylation. J. Cancer 2021, 12, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Kumbrink, J.; Mayr, D.; Seiler, A.; Hagemann, F.; Degenhardt, T.; Sagebiel, S.; Würstlein, R.; Kates, R.; Harbeck, N.; et al. Molecular Prognostic Factors for Distant Metastases in Premenopausal Patients with HR+/HER2− Early Breast Cancer. J. Pers. Med. 2021, 11, 835. [Google Scholar] [CrossRef]
- Joosten, S.C.; Deckers, I.A.; Aarts, M.J.; Hoeben, A.; Van Roermund, J.G.; Smits, K.M.; Melotte, V.; Van Engeland, M.; Tjan-Heijnen, V.C. Prognostic DNA methylation markers for renal cell carcinoma: A systematic review. Epigenomics 2017, 9, 1243–1257. [Google Scholar] [CrossRef]
- Fatima, A.; Tariq, F.; Malik, M.F.A.; Qasim, M.; Haq, F. Copy Number Profiling of MammaPrint™ Genes Reveals Association with the Prognosis of Breast Cancer Patients. J. Breast Cancer 2017, 20, 246–253. [Google Scholar] [CrossRef]
- Zhen, L.; Ning, G.; Wu, L.; Zheng, Y.; Yang, F.; Chen, T.; Xu, W.; Liu, Y.; Xie, C.; Peng, L. Prognostic value of aberrantly expressed methylation genes in human hepatocellular carcinoma. Biosci. Rep. 2020, 40, BSR20192593. [Google Scholar] [CrossRef]
- Esmaeili, R.; Mohammadi, S.; Jafarbeik-Iravani, N.; Yadegari, F.; Olfatbakhsh, A.; Mazaheri, M.; Kaviani, A.; Rezaee, M.; Majidzadeh-A, K. Expression of SCUBE2 and BCL2 Predicts Favorable Response in ERα Positive Breast Cancer. Arch. Iran. Med. 2021, 24, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Poorhosseini, S.M.; Hashemi, M.; Olyaei, N.A.; Izadi, A.; Moslemi, E.; Ravesh, Z.; Hashemi-Gorji, F.; Kheiri, H.R.; Yassaee, V.R. New Gene Profiling in Determination of Breast Cancer Recurrence and Prognosis in Iranian Women. Asian Pac. J. Cancer Prev. 2016, 17, 155–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| SCUBE | Disease Class | Semantic Type | Association Type | References |
|---|---|---|---|---|
| SCUBE1 | ||||
| RCC | Neoplasms, female urogenital diseases, pregnancy complications, male urogenital diseases. | Neoplastic process | Biomarker | [9] |
| Diabetic retinopathy | Eye diseases, endocrine system diseases, cardiovascular diseases. | Disease or syndrome | Altered expression | [10] |
| SLE | Skin and connective tissue diseases, immune system diseases. | Disease or syndrome | Genetic variation | [11] |
| Degenerative polyarthritis | Musculoskeletal diseases. | Disease or syndrome | Genetic variation | [12] |
| Psoriasis | Skin and connective tissue diseases. | Disease or syndrome | Altered expression | [13] |
| Thrombosis | Cardiovascular diseases. | Pathologic function | Altered expression | [5] |
| Gestational diabetes | Nutritional and metabolic diseases, female urogenital diseases and pregnancy complications, endocrine system diseases. | Disease or syndrome | Altered expression | [14] |
| ST segment elevation myocardial infarction | Pathological conditions, signs and symptoms, cardiovascular diseases. | Disease or syndrome | Altered expression | [15] |
| Venous thromboembolism | Cardiovascular diseases. | Disease or syndrome | Genetic variation | [16] |
| Molar incisor hypo-mineralization | Congenital, hereditary, and neonatal diseases and abnormalities, stomatognathic diseases. | Disease or syndrome | Genetic variation | [17] |
| SCUBE2 | ||||
| Mammary neoplasms | Neoplasms, skin and connective tissue diseases. | Neoplastic process | Biomarker | [18] |
| Triple negative breast neoplasms | Neoplasms, skin and connective tissue diseases. | Neoplastic process | Altered expression | [19] |
| Tumor angiogenesis | Pathological conditions, signs, and symptoms. | Neoplastic process | Biomarker | [7] |
| Squamous cell carcinoma of mouth | Digestive system diseases, neoplasms, stomatognathic diseases. | Neoplastic process | Altered expression | [20] |
| Malignant neoplasm of endometrium | Neoplasms, female urogenital diseases and pregnancy complications. | Neoplastic process | Biomarker | [21] |
| NSCLC | Neoplasms, respiratory tract diseases. | Neoplastic process | Altered expression | [22] |
| CRC | Digestive system diseases, neoplasms. | Neoplastic process | Biomarker | [23] |
| Glioma | Neoplasms. | Neoplastic process | Biomarker | [24] |
| Thrombosis | Cardiovascular diseases. | Pathologic function | Altered expression | [5] |
| SCUBE3 | ||||
| SACC | Pathological conditions, signs and symptoms, neoplasms. | Neoplastic process | Biomarker | [25] |
| Osteosarcoma of bone | Neoplasms. | Neoplastic process | Altered expression | [26] |
| RCC | Neoplasms, female urogenital diseases and pregnancy complications, male urogenital diseases. | Neoplastic process | Genetic variation | [9,27] |
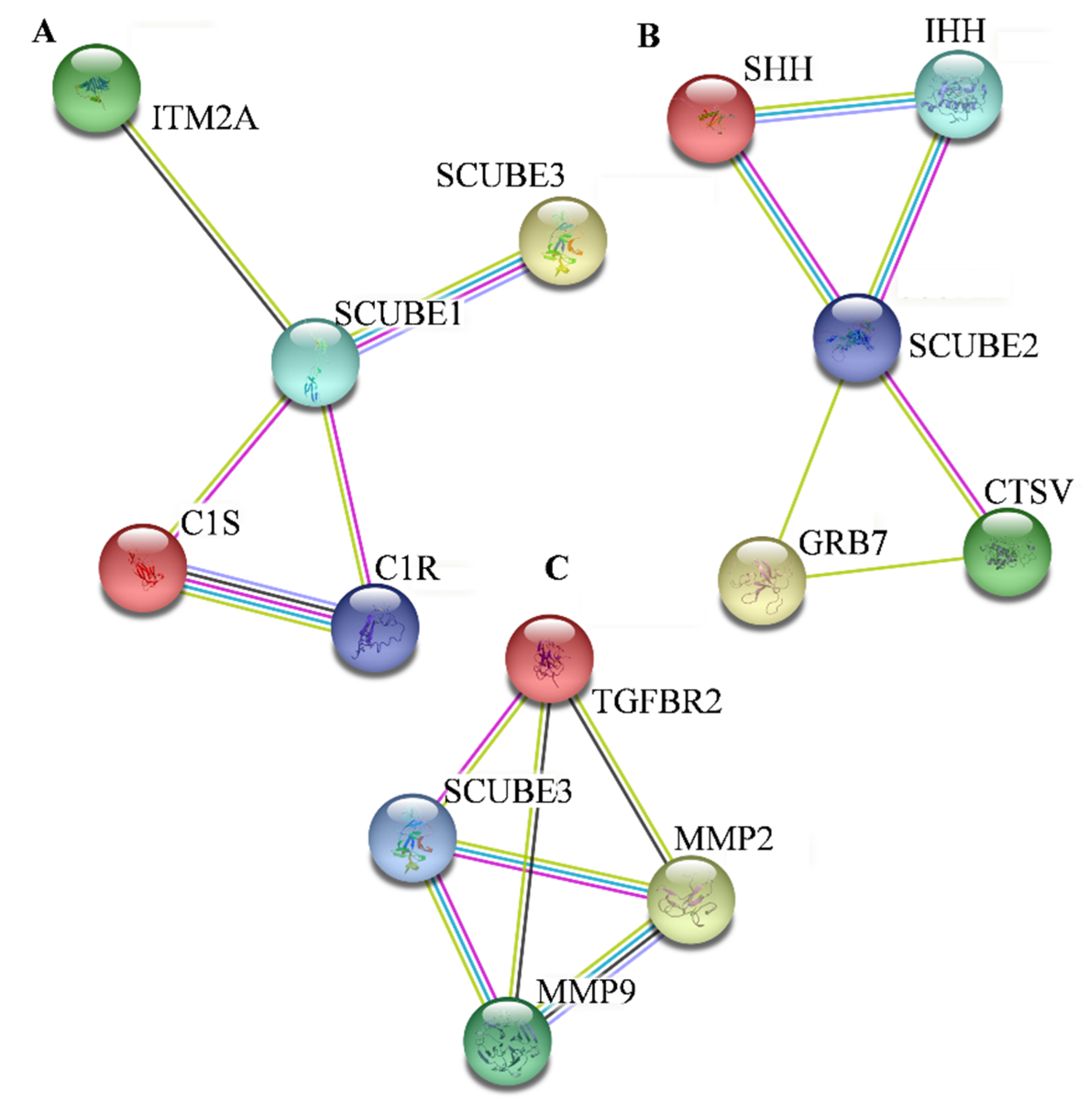
| SCUBE Family Protein | Associated Protein and Its Function | Study on Association between SCUBE and Proteins | Mechanism | References |
|---|---|---|---|---|
| SCUBE1 | SCUBE3 | Inverse relation between SCUBE1 and SCUBE3 protein in synovial angiogenesis driven RA | SCUBE1 antagonizes BMP2 signaling, suppressing BMP2-induced phospho-Smad1/5/8 expression level, whereas Scube3 functions as a ligand for endogenous TGFβ receptor that increases Smad2/3 phosphorylation, and thus upexpresses target genes involved in angiogenesis. | [28] |
| SCUBE1 | ITM2A is the BRICHOS (consisting of variety of proteins, each of 100 amino acids) resembling proteins, and structurally related to ITM2B and ITM2C. It is generally expressed in T-lineage among hematopoietic cells and is induced by MHC-mediated positive selection. It is involved in osteo- and chondrogenic differentiation. The downregulation of this gene causes ankylosing spondylitis, grave disease pathogenesis, and ovarian cancer. | SCUBE1 and ITM2A both downregulated, related to the proliferation of porcine follicular granulosa cells in an in vitro model. | [29,30] | |
| SCUBE1 | C1s is a single-chain glycoprotein and a serine protease that performs the catalytic function of the C1 complex. | C1s is a part of SCUBE1 that helps in executing the functions of SCUBE1. | [31] | |
| SCUBE1 | C1r is the serine protease enzyme that activates the proteolytic activity of complement complex C1. | C1r is a part of SCUBE1 that helps in executing the functions of SCUBE1. | [32] | |
| SCUBE2 | IHH is a member of the hedgehog signaling family that regulates embryonic morphogenesis and bone development. | SCUBE 2 is the most potent modulator of IIH signaling. | SCUBE 2 exerts an osteogenic function by increasing the IHH-mediated osteoblast differentiation in an in vitro mouse model. The deregulation of SCUBE2 impedes IHH signaling-mediated chondrocyte differentiation and proliferation, and thus bone formation. | [33] |
| SCUBE2 | SHH is the secreted protein plays an essential role during embryo development, and aberrant expression leads to the generation of diseases, including atherosclerosis and cancer. | Overexpression of SCUBE2 inhibited the expression of Shh signaling pathway, thereby inhibiting proliferation, migration, and invasion in glioma and NSCLC cells. | Overexpression of SCUBE2 decreases the expression of Smo, Gli1, and Ptch1 (components of Shh signaling pathway), which ultimately decreases Shh signaling expression in glioma and NSCLC. | [22,24] |
| SCUBE2 | GRB7 is an adapter protein that interacts with various receptor tyrosine kinases and modulates downstream signaling molecules such as AKT1, MAPK, and STAT3. This protein plays an important role in cell migration by binding with FAK. | SCUBE2 and GRB7 are both signature genes involved in the poor survival of breast-cancer patients in CNV-based assay of oncotype DX genes. | - | [34] |
| SCUBE2 | CTSV is a lysosomal cysteine protease particularly found in human testes, thymus, and corneal epithelium. Overexpression of CTSV causes colorectal and breast cancers, and squamous cell carcinoma. | SCUBE2 and CTSV are also both signature genes involved in the poor survival of breast-cancer patients in CNV-based assay of Oncotye DX genes | - | [34] |
| SCUBE3 | MMP2/9 is an enzyme that breaks ECM during normal developmental processes (embryo development, reproduction, etc.). In cancer, it allows for the primary tumor cells to migrate to form metastatic cancer cells. TGFBR2 is a transmembrane ser/thr kinase receptor expressed in all cell types such as fibroblasts. It is a tumor-suppressor gene. Loss of TGFBR2 expression is associated with many pathological conditions, including cancer. | SCUBE3, MMP2/9, and TGFBR2 are associated with the invasiveness of lung cancer. | The secreted form of SCUBE3 cleaved by MMP2/9 into N-and C-terminal. C fragments of the SCUBE3 bound to TGFBR2 activate TGF-β signaling and increase the transcription of Smad2/3, and upregulate the expression of genes involved in lung-cancer cell invasion. | [35] |
| Cancer | SEL | Effect | Targets | Pathway | References |
|---|---|---|---|---|---|
| SCUBE2 | |||||
| NSCLC | D | Promotes proliferation, migration, and invasion. | Ptch1, Shh, Smo, Gli1 | Hedgehog | [22] |
| Glioma | D | Promotes proliferation, migration, and invasion. | Ptch, Shh, Smo, Gli1 | Hedgehog | [24] |
| BCa | D | Promotes proliferation, migration, and invasion, and induces apoptosis. | MiR-155-5p, BMP, TGF-β, FOXA1 | MiR-155-5p/SCUBE2 axis, β-catenin | [18,36] |
| BCaSC | U | Enhances stemness phenotype, self-renewal, aggressiveness, and EMT. | Sox2, Oct4, Nanog, Vimentin | Notch | [19] |
| TA | U | Increases tube formation, proliferation, and migration. | VEGF, VEGFR2 | PI3K/Akt, MAPK | [44] |
| CRC | D | Promotes proliferation, migration, invasion, and apoptosis inhibition. | - | - | [23] |
| GC | D | Increases lymph-node metastasis, high-grade tumors, T3 or T4 lesions, distant metastasis, and vascular invasion | - | - | [50] |
| SCUBE1 | |||||
| PCOS | U | - | TNF-α and IL-1β | - | [57] |
| RCC | U | - | - | - | [9] |
| GC | U | - | - | - | [58] |
| BC | U | - | - | - | [59] |
| SCUBE3 | |||||
| LC | U | Increases metastasis. | MMP2/9, TGF-β, Cadherin, Vimentin | EMT | [35] |
| BlCa | U | Increases proliferation and metastasis. | E-Cadherin, MMP2/9 | - | [42] |
| SACC | U | Increases SACC proliferation. | miR-885-5p | - | [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Prajapati, K.S.; Gupta, S. The Multifaceted Role of Signal Peptide-CUB-EGF Domain-Containing Protein (SCUBE) in Cancer. Int. J. Mol. Sci. 2022, 23, 10577. https://doi.org/10.3390/ijms231810577
Kumar S, Prajapati KS, Gupta S. The Multifaceted Role of Signal Peptide-CUB-EGF Domain-Containing Protein (SCUBE) in Cancer. International Journal of Molecular Sciences. 2022; 23(18):10577. https://doi.org/10.3390/ijms231810577
Chicago/Turabian StyleKumar, Shashank, Kumari Sunita Prajapati, and Sanjay Gupta. 2022. "The Multifaceted Role of Signal Peptide-CUB-EGF Domain-Containing Protein (SCUBE) in Cancer" International Journal of Molecular Sciences 23, no. 18: 10577. https://doi.org/10.3390/ijms231810577





