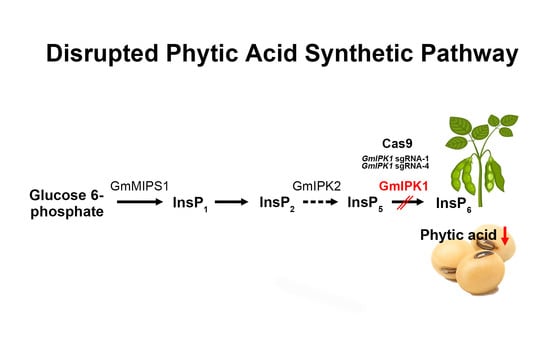
Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds
Abstract
:1. Introduction
2. Results and Discussion
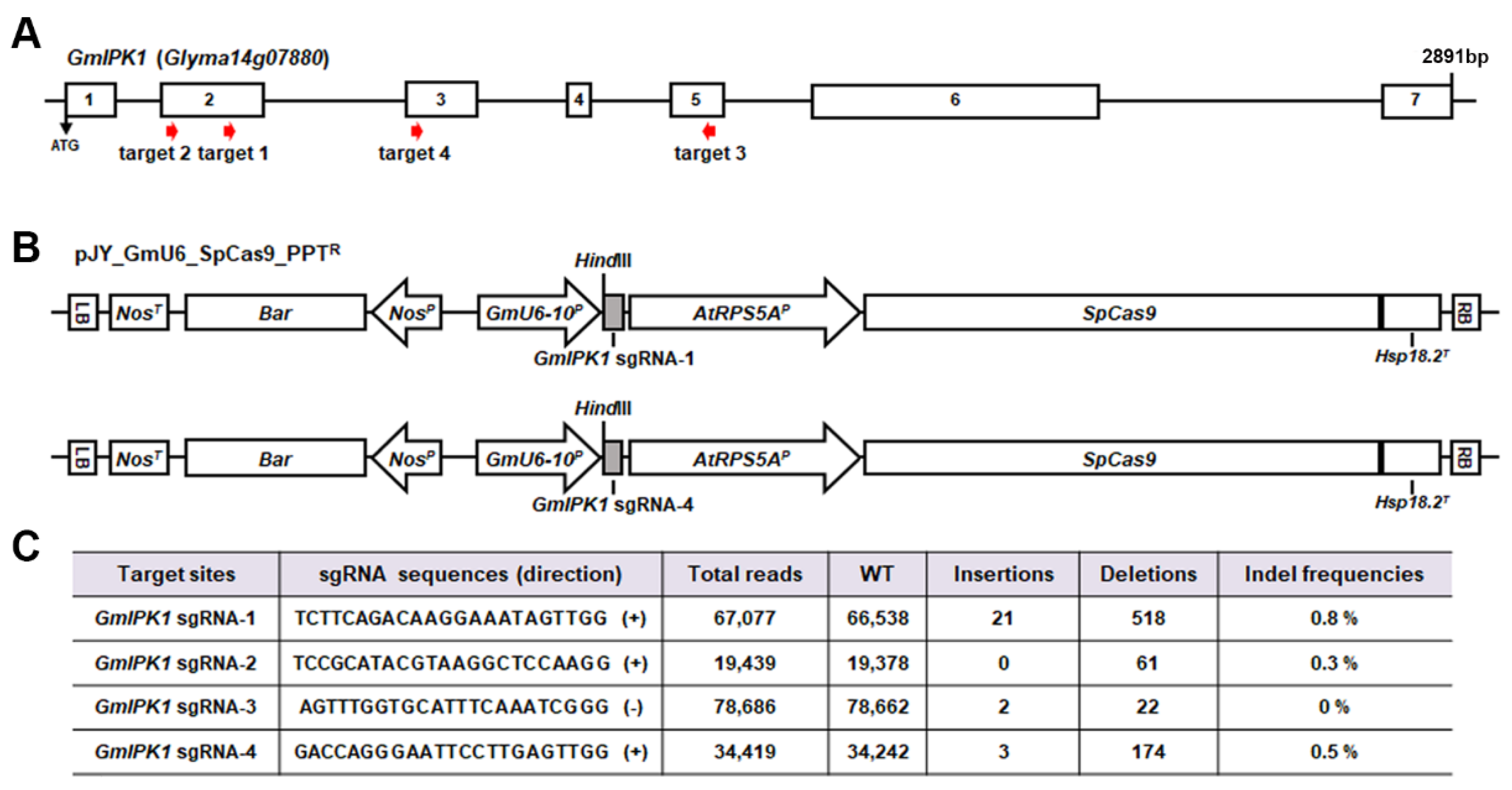
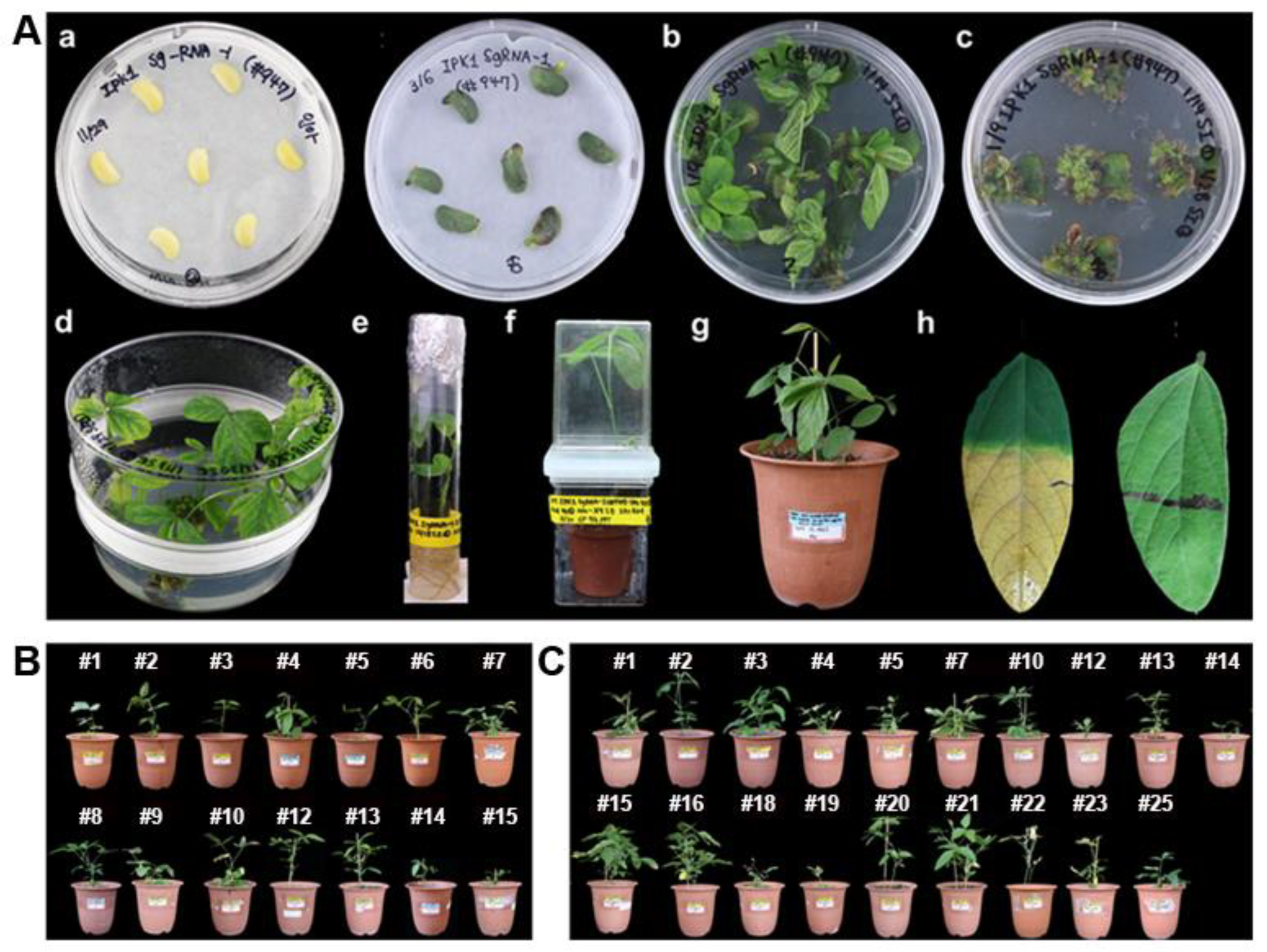
2.1. Generation of Gmipk1 Gene-Edited Soybean Plants by CRISPR/Cas9 System
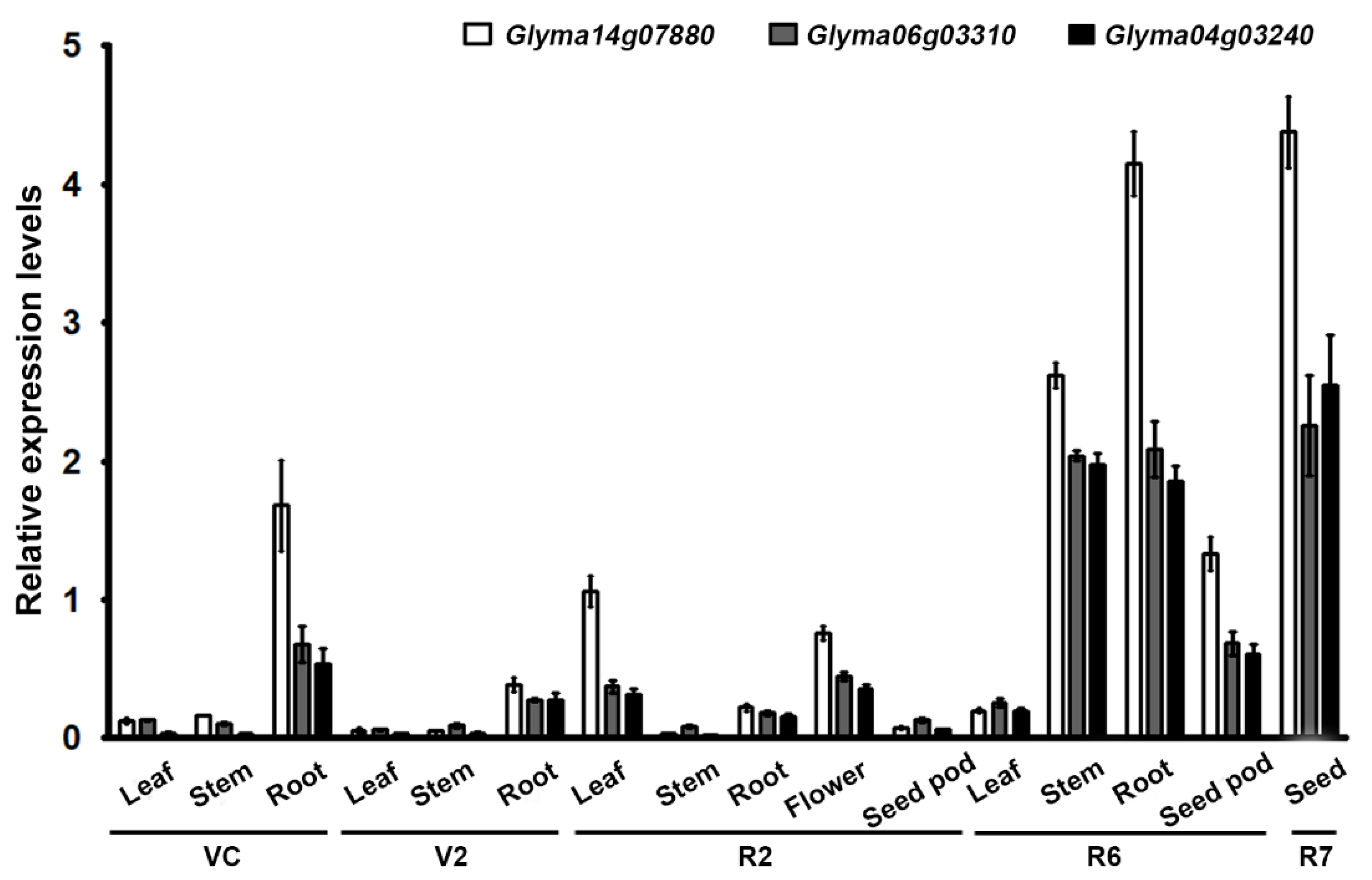
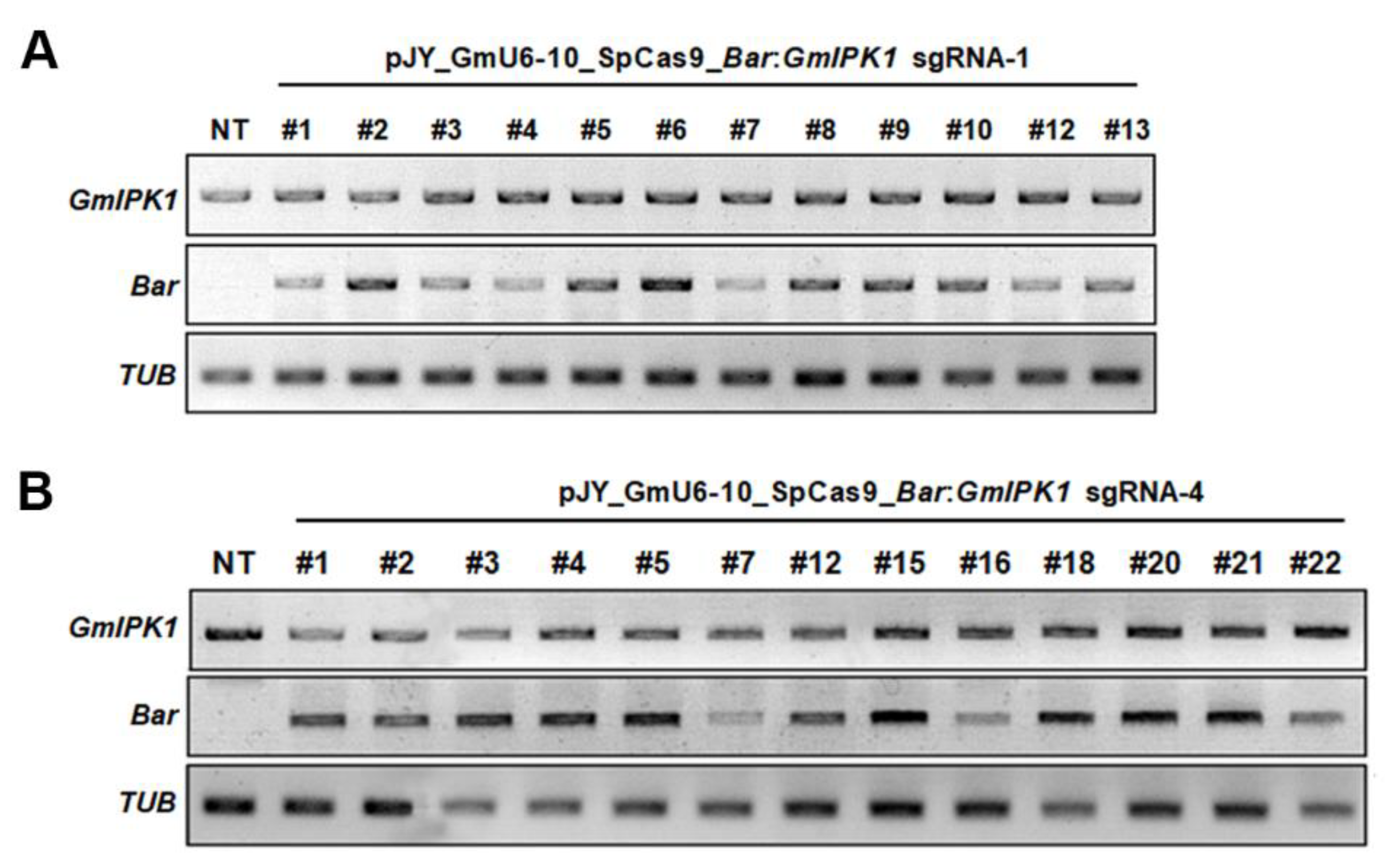
2.2. Integration and Expression of Transgenes in Transgenic Soybean Plants
2.3. Selection of CRISPR/Cas9-Induced sgRNA-4 Gmipk1 Gene-Edited Line and Measurement of PA Content
3. Materials and Methods
3.1. RNA Isolation and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
3.2. Construction of Two Gmipk1 Genome Editing Vectors for Soybean Transformation
3.3. Next-Generation Sequencing (NGS) Analysis
3.4. Agrobacterium-Mediated Soybean Transformation
3.5. Confirmation of Transgenes in Transgenic Soybean Plants (T0)
3.6. RNA Analysis of Transgenic Plants (T0)
3.7. Selection of Gmipk1 Gene-Edited Soybean Plants and Generation Advance
3.8. Determination of PA Content
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, L.; Cai, Y.; Liu, X.; Yao, W.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int. J. Mol. Sci. 2018, 19, 3039. [Google Scholar] [CrossRef] [PubMed]
- Hada, A.; Krishnan, V.; Jaabir, M.; Kumari, A.; Jolly, M.; Praveen, S.; Sachdev, A. Improved Agrobacterium tumefaciens-mediated transformation of soybean [Glycine max (L.) Merr.] following optimization of culture conditions and mechanical techniques. Vitr. Cell. Dev. Biol. Plant 2018, 54, 672–688. [Google Scholar] [CrossRef]
- Yuan, F.; Zhu, D.; Tan, D.; Dong, D.; Fu, X.; Zhu, S.; Li, B.; Shu, Q. Identification and characterization of the soybean IPK1 ortholog of a low phytic acid mutant reveals an exon-excluding splice-site mutation. Theor. Appl. Genet. 2012, 152, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Hinchee, M.A.; Connor-Ward, D.V.; Newell, C.A.; McDonnell, R.E.; Sato, S.J.; Gasser, C.S.; Fischhoff, D.A.; Re, D.B.; Fraley, R.T.; Horsch, R.B. Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat. Biotechnol. 1988, 6, 915–922. [Google Scholar] [CrossRef]
- Cho, H.S.; Lee, D.H.; Jung, H.W.; Oh, S.-W.; Kim, H.J.; Chung, Y.-S. Evaluation of yield components from transgenic soybean overexpressing chromatin architecture-controlling ATPG8 and ATPG10 genes. Plant Breed. Biotech. 2019, 7, 34–41. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, M.-J.; Pak, J.H.; Im, H.H.; Lee, D.H.; Kim, K.-H.; Lee, J.-H.; Kim, D.-H.; Choi, H.K.; Jung, H.W. RNAi-mediated Soybean mosaic virus (SMV) resistance of a Korean Soybean cultivar. Plant Biotechnol. Rep. 2016, 10, 257–267. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kim, H.J.; Pak, J.H.; Cho, H.S.; Choi, H.K.; Jung, H.W.; Lee, D.H.; Chung, Y.-S. Overexpression of AtSZF2 from Arabidopsis showed enhanced tolerance to salt stress in soybean. Plant Breed. Biotech. 2017, 5, 1–15. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, H.J.; Cho, H.S.; Jung, H.W.; Cha, J.-Y.; Yun, D.-J.; Oh, S.-W.; Chung, Y.-S. Overexpression of AtYUCCA6 in soybean crop results in reduced ROS production and increased drought tolerance. Plant Biotechnol. Rep. 2019, 13, 161–168. [Google Scholar] [CrossRef]
- Yeom, W.W.; Kim, H.J.; Lee, K.-R.; Cho, H.S.; Kim, J.-Y.; Jung, H.W.; Oh, S.-W.; Jun, S.E.; Kim, H.U.; Chung, Y.-S. Increased production of α-linolenic acid in soybean seeds by overexpression of Lesquerella FAD3-1. Front. Plant Sci. 2020, 10, 1812. [Google Scholar] [CrossRef]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Verma, K.; Saini, R.; Rani, A. Recent advances in the regeneration and genetic transformation of soybean. J. Innov. Biol. 2014, 1, 15–26. [Google Scholar]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE 2015, 10, e0136064. [Google Scholar] [CrossRef]
- Bao, A.; Chen, H.; Chen, L.; Chen, S.; Hao, Q.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef]
- Do, P.T.; Nguyen, C.X.; Bui, H.T.; Tran, L.T.N.; Stacey, G.; Gillman, J.D.; Zhang, Z.J.; Stacey, M.G. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 2019, 19, 311. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Raboy, V. Low phytic acid crops: Observations based on four decades of research. Plants 2020, 9, 140. [Google Scholar] [CrossRef]
- Zhao, H.; Frank, T.; Tan, Y.; Zhou, C.; Jabnoune, M.; Arpat, A.; Cui, H.; Huang, J.; He, Z.; Poirier, Y.; et al. Disruption of OsSULTR3;3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol. 2016, 211, 926–939. [Google Scholar] [CrossRef]
- Kumar, V.; Sinha, A.K.; Makkar, H.P.; Becker, K. Dietary roles of phytate and phytase in human nutrition: A review. Food Chem. 2010, 120, 945–959. [Google Scholar] [CrossRef]
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy, P.P.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000, 124, 355–368. [Google Scholar] [CrossRef]
- Shi, J.; Wang, H.; Schellin, K.; Li, B.; Faller, M.; Stoop, J.M.; Meeley, R.B.; Ertl, D.S.; Ranch, J.P.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937. [Google Scholar] [CrossRef]
- Adeola, O.; Lawrence, B.; Sutton, A.; Cline, T. Phytase-induced changes in mineral utilization in zinc-supplemented diets for pigs. J. Anim. Sci. 1995, 73, 3384–3391. [Google Scholar] [CrossRef] [Green Version]
- Almeida, F.F.D.; Araújo, A.P.; Alves, B.J.R. Seeds with high molybdenum concentration improved growth and nitrogen acquisition of rhizobium-inoculated and nitrogen-fertilized common bean plants. Rev. Bras. Ciência Solo 2013, 37, 367–378. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, B.; Raigond, P.; Sahu, C.; Mishra, U.N.; Sharma, S.; Lal, M.K. Phytic acid: Blessing in disguise, a prime compound required for both plant and human nutrition. Food Res. Int. 2021, 142, 110193. [Google Scholar] [CrossRef] [PubMed]
- Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009, 177, 281–296. [Google Scholar] [CrossRef]
- Yuan, F.-J.; Zhao, H.-J.; Ren, X.-L.; Zhu, S.-L.; Fu, X.-J.; Shu, Q.-Y. Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 2007, 115, 945–957. [Google Scholar] [CrossRef]
- Basak, N.; Krishnan, V.; Pandey, V.; Punjabi, M.; Hada, A.; Marathe, A.; Jolly, M.; Palaka, B.K.; Ampasala, D.R.; Sachdev, A. Expression profiling and in silico homology modeling of Inositol pentakisphosphate 2-kinase, a potential candidate gene for low phytate trait in soybean. 3 Biotech 2020, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Bhati, K.K.; Aggarwal, S.; Sharma, S.; Mantri, S.; Singh, S.P.; Bhalla, S.; Kaur, J.; Tiwari, S.; Roy, J.K.; Tuli, R. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 2014, 224, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tanaka, K.; Kuwano, M.; Yoshida, K.T. Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): Implications for the phytic acid biosynthetic pathway. Gene 2007, 405, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Di, Y.-H.; Sun, X.-J.; Hu, Z.; Jiang, Q.-Y.; Song, G.-H.; Zhang, B.; Zhao, S.-S.; Zhang, H. Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem. Biophys. Res. Commun. 2019, 519, 819–823. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, V.; Krishnan, V.; Hada, A.; Marathe, A.; Jolly, M.; Sachdev, A. Seed targeted RNAi-mediated silencing of GmMIPS1 limits phytate accumulation and improves mineral bioavailability in soybean. Sci. Rep. 2019, 9, 7744. [Google Scholar] [CrossRef] [Green Version]
- Punjabi, M.; Bharadvaja, N.; Jolly, M.; Dahuja, A.; Sachdev, A. Development and evaluation of low phytic acid soybean by siRNA triggered seed specific silencing of inositol polyphosphate 6-/3-/5-kinase gene. Front. Plant Sci. 2018, 9, 804. [Google Scholar] [CrossRef]
- Jin, H.; Yu, X.; Yang, Q.; Yuan, F.; Fu, X. Transcriptome analysis identifies differentially expressed genes involved in the metabolic regulatory network of progenies from the cross of low phytic acid GmMIPS1 and GmIPK1 soybean mutants. Sci. Rep. 2021, 11, 8740. [Google Scholar] [CrossRef]
- Yun, J.-Y.; Kim, S.-T.; Kim, S.-G.; Kim, J.-S. A zero-background CRISPR binary vector system for construction of sgRNA libraries in plant functional genomics applications. Plant Biotechnol. Rep. 2019, 13, 543–551. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Park, J.; Lim, K.; Kim, J.-S.; Bae, S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2017, 33, 286–288. [Google Scholar] [CrossRef]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.-F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.H.; Shin, G.; Kim, H.J.; Lee, S.B.; Moon, J.Y.; Jeong, J.C.; Choi, H.-K.; Kim, I.A.; Song, H.J.; Kim, C.Y.; et al. Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds. Int. J. Mol. Sci. 2022, 23, 10583. https://doi.org/10.3390/ijms231810583
Song JH, Shin G, Kim HJ, Lee SB, Moon JY, Jeong JC, Choi H-K, Kim IA, Song HJ, Kim CY, et al. Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds. International Journal of Molecular Sciences. 2022; 23(18):10583. https://doi.org/10.3390/ijms231810583
Chicago/Turabian StyleSong, Ji Hyeon, Gilok Shin, Hye Jeong Kim, Saet Buyl Lee, Ju Yeon Moon, Jae Cheol Jeong, Hong-Kyu Choi, In Ah Kim, Hyeon Jin Song, Cha Young Kim, and et al. 2022. "Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds" International Journal of Molecular Sciences 23, no. 18: 10583. https://doi.org/10.3390/ijms231810583
APA StyleSong, J. H., Shin, G., Kim, H. J., Lee, S. B., Moon, J. Y., Jeong, J. C., Choi, H.-K., Kim, I. A., Song, H. J., Kim, C. Y., & Chung, Y.-S. (2022). Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds. International Journal of Molecular Sciences, 23(18), 10583. https://doi.org/10.3390/ijms231810583