Unveiling the Essential Role of Arkadia’s Non-RING Elements in the Ubiquitination Process
Abstract
:1. Introduction
2. Results and Discussion
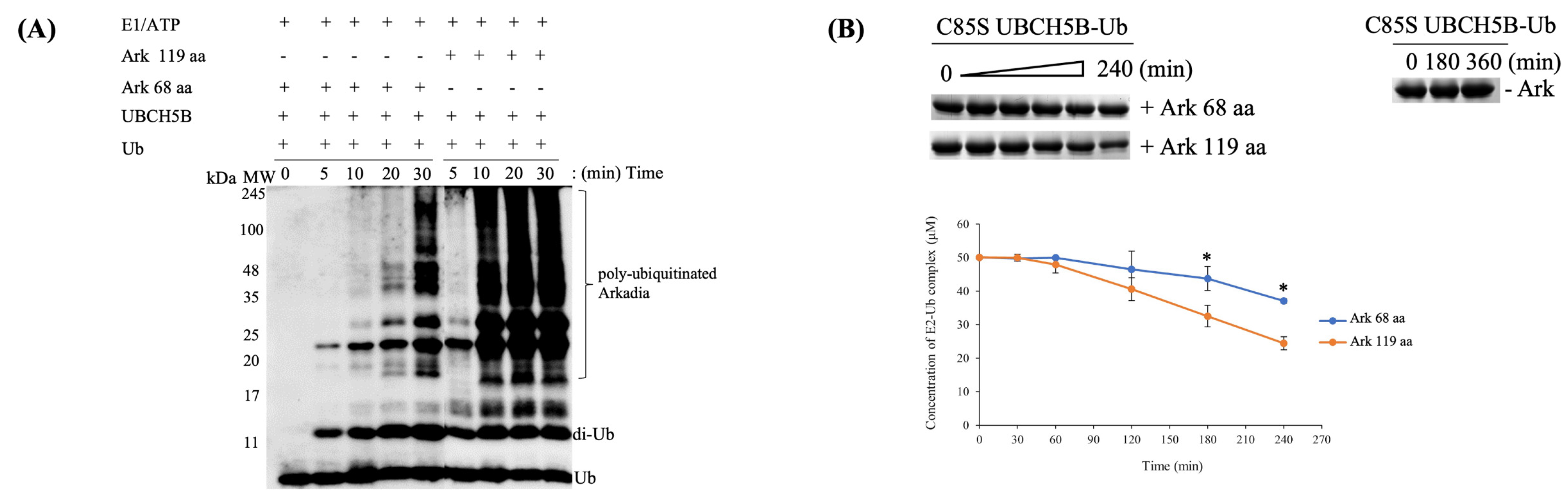
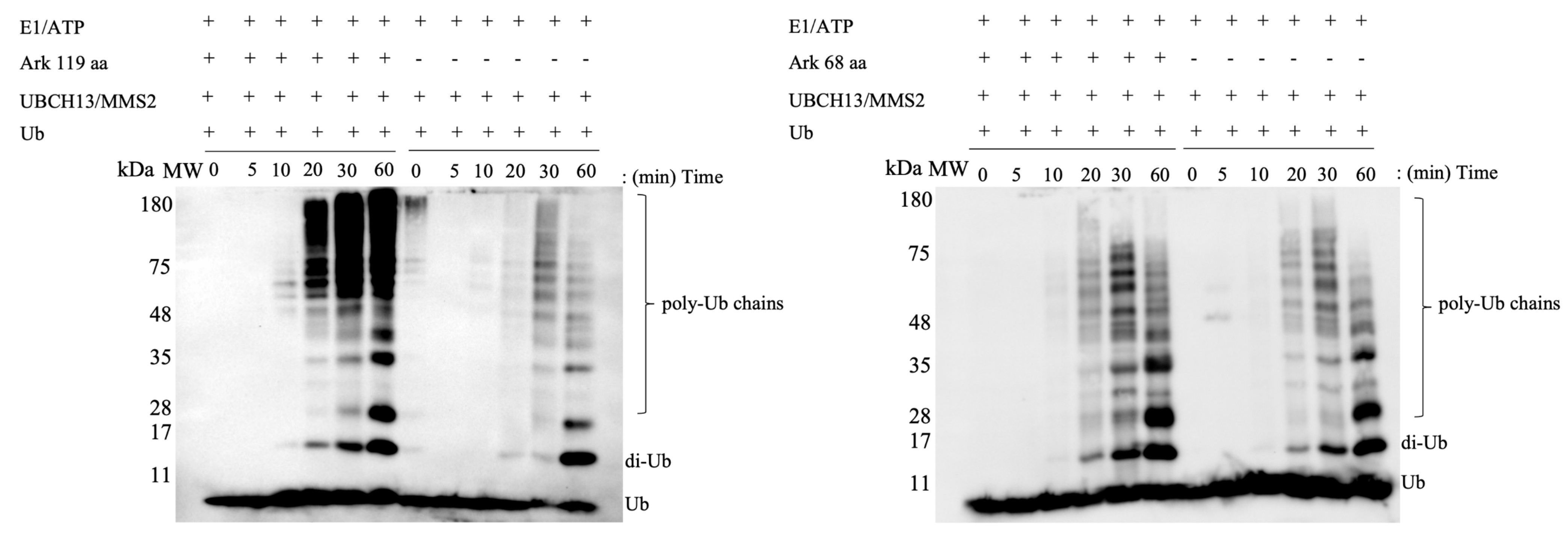
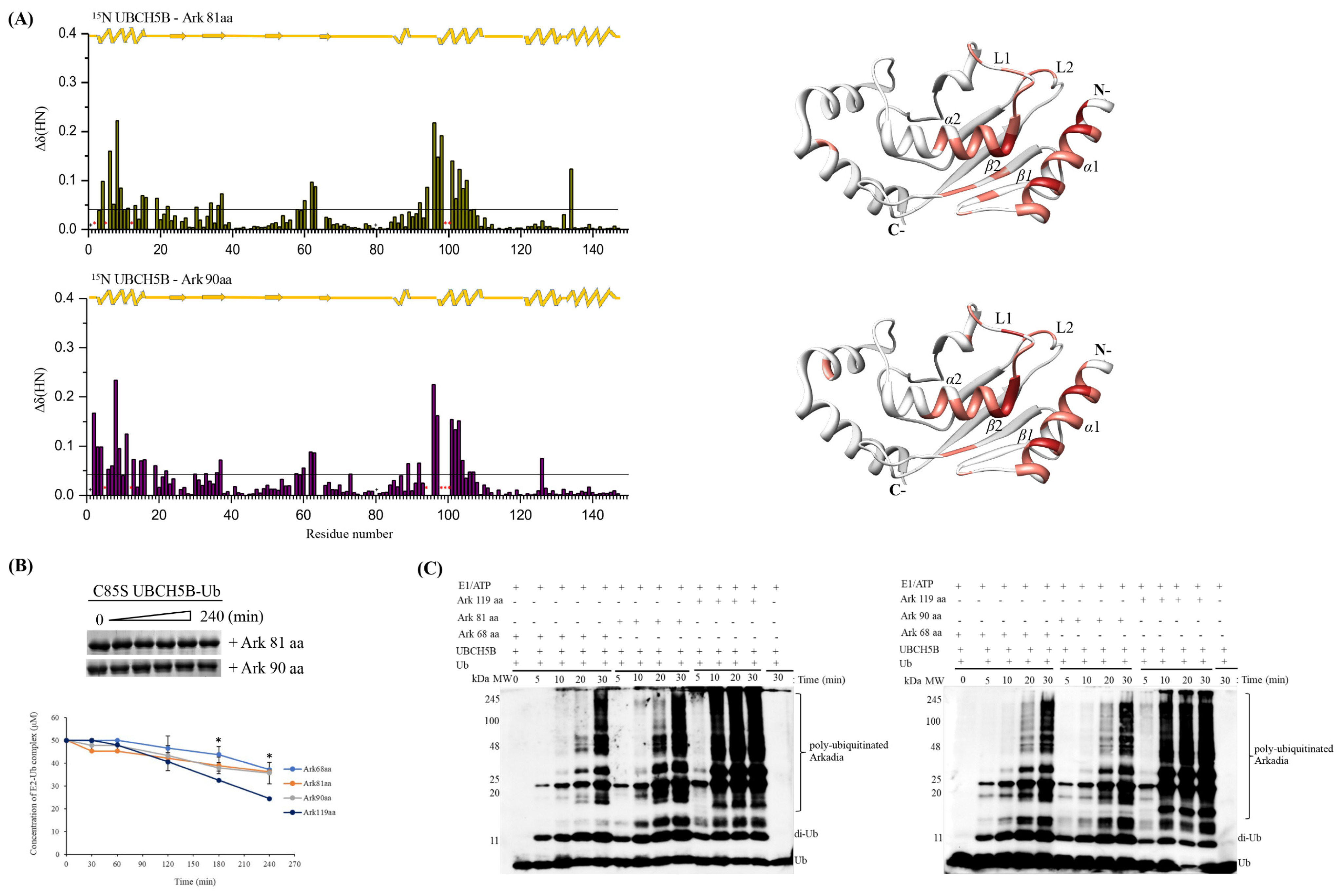
2.1. The RING Domain of Arkadia Is Not Sufficient for Enzymatic Activity
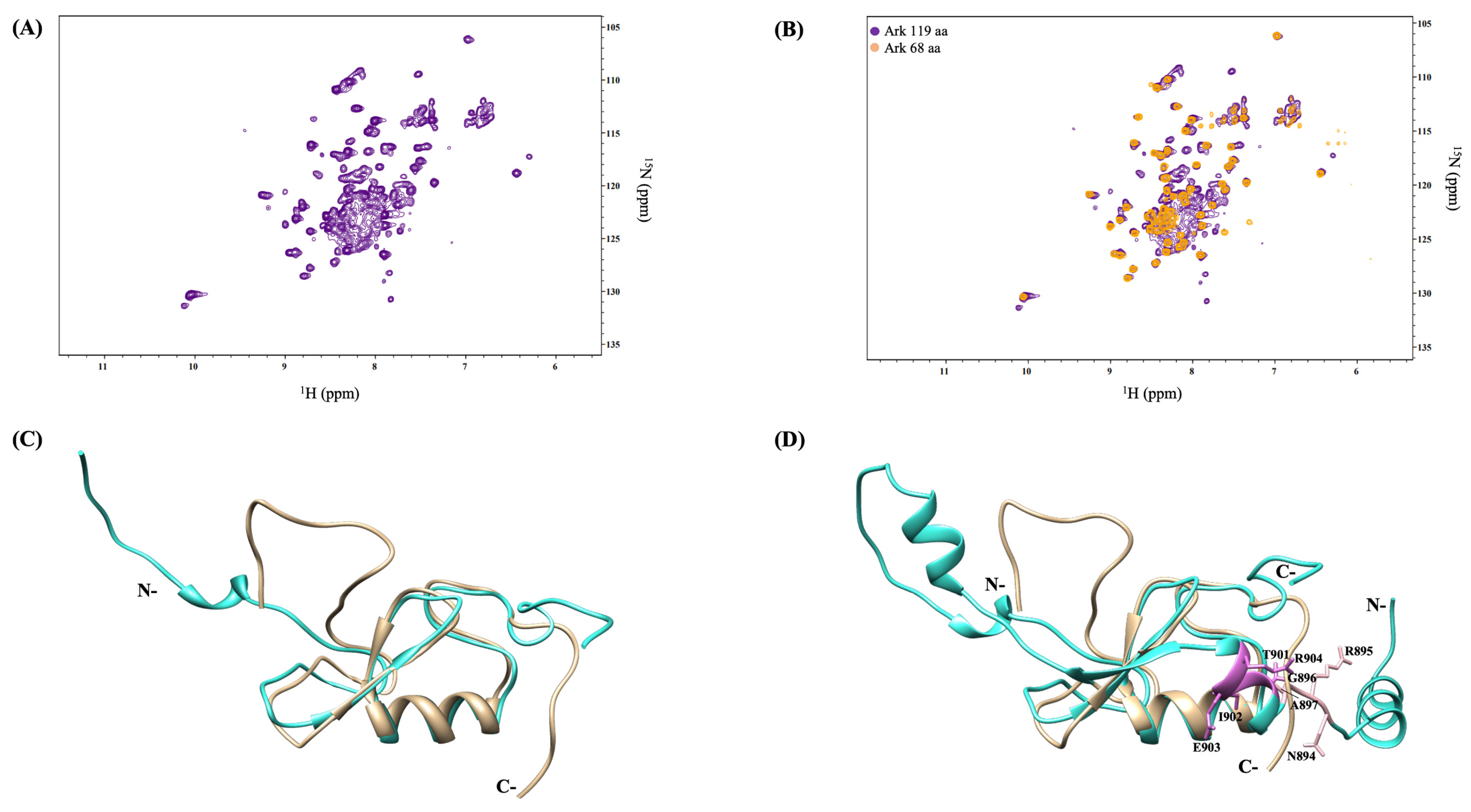
2.2. Non-RING Elements Present in Ark 119 aa Polypeptide Enhance E2-E3 Interaction
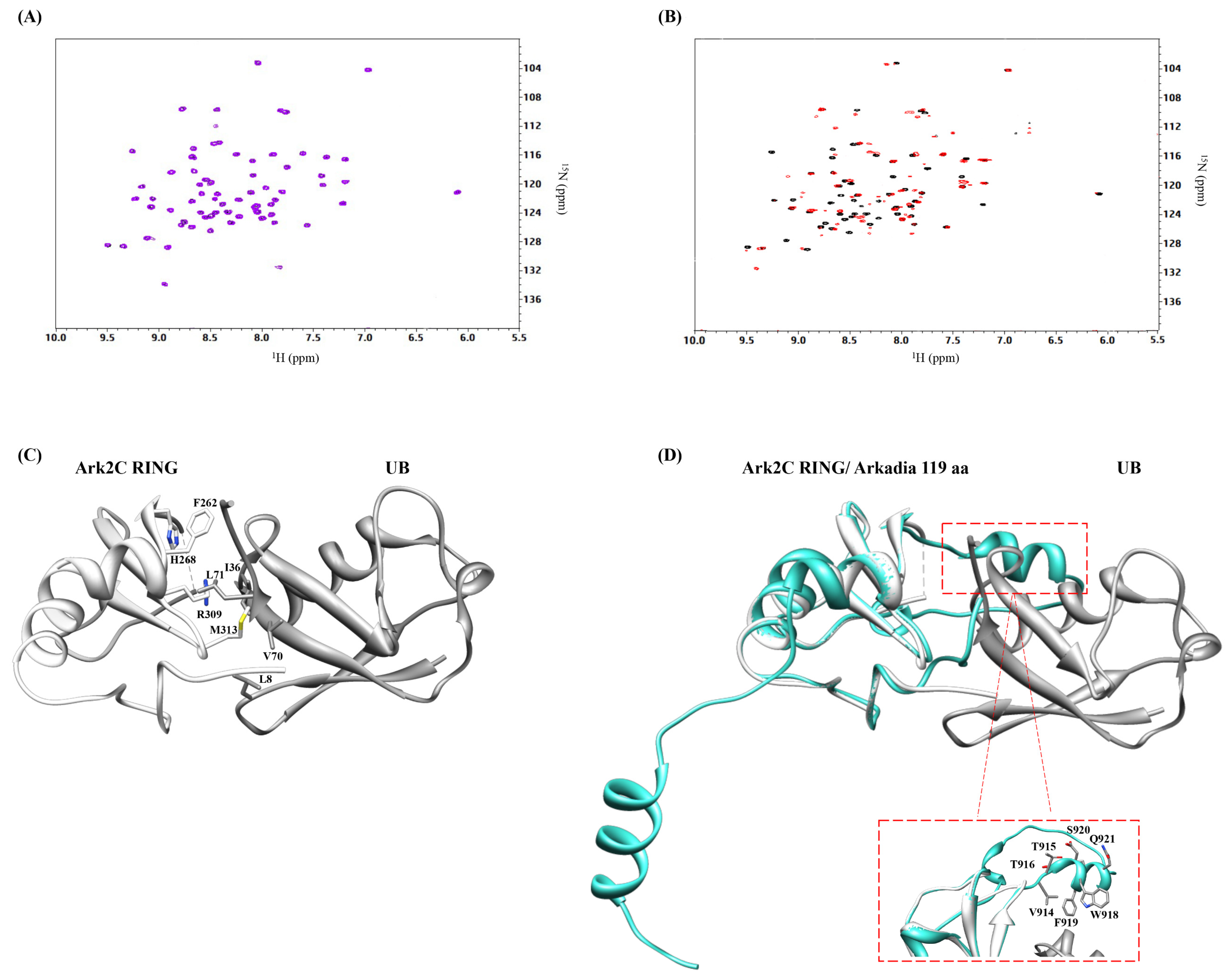
2.3. Arkadia Isoform 1 Does Not Bind Ubiquitin
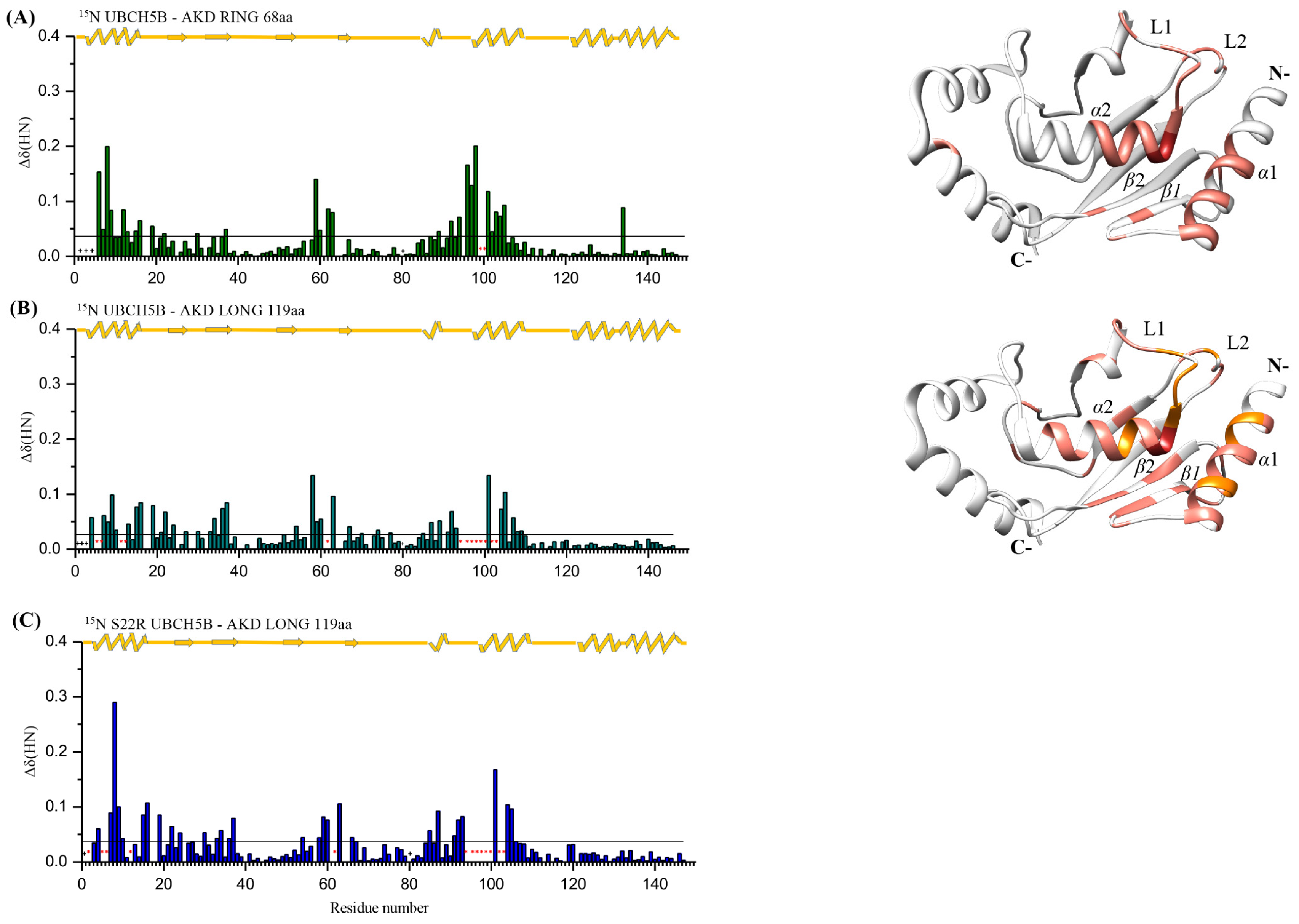
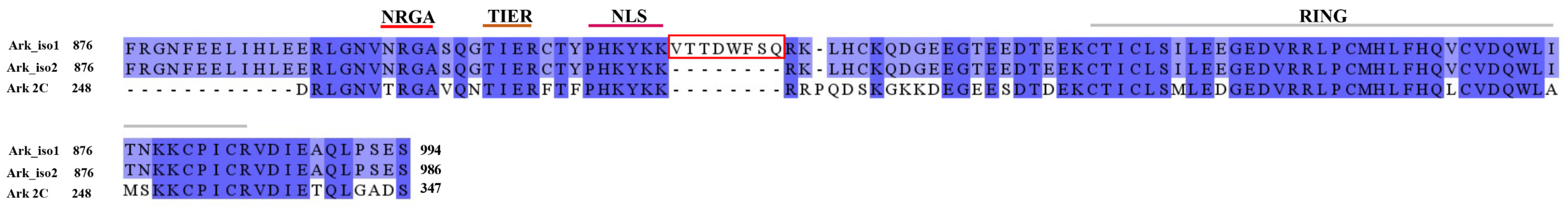
2.4. Screening Arkadia’s Non-RING Sequence That Is Crucial for E2 Interaction and E3 Ligation Activity
2.5. NRGA and TIER Segments Implication in E2 Interaction and Ubiquitination
3. Materials and Methods
3.1. Construct Design
3.2. Constructs, Protein Expression and Purification
3.3. Isothermal Titration Calorimetry (ITC)
3.4. Auto-Ubiquitination Assays
3.5. Synthesis of E2-Ub Conjugate
3.6. Oxyester Hydrolysis Assays
3.7. NMR Titration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.D.; Hicke, L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003, 278, 35857–35860. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2014, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Schwartz, A.L. The ubiquitin-proteasome pathway: The complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 1998, 95, 2727–2730. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Smit, J.J.; Sixma, T.K. RBR E3-ligases at work. EMBO Rep. 2014, 15, 142–154. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A.P. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Lissounov, A.; Christensen, D.E.; Hoyt, D.W.; Klevit, R.E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 2006, 21, 873–880. [Google Scholar] [CrossRef]
- Buetow, L.; Gabrielsen, M.; Anthony, N.G.; Dou, H.; Patel, A.; Aitkenhead, H.; Sibbet, G.J.; Smith, B.O.; Huang, D.T. Activation of a Primed RING E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin. Mol. Cell 2015, 58, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.; Sibbet, G.J.; Huang, D.T. Structural insights into non-covalent ubiquitin activation of the cIAP1-UbcH5B ubiquitin complex. J. Biol. Chem. 2019, 294, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Mariano, J.; Tsai, Y.C.; Kalathur, R.C.; Kostova, Z.; Li, J.; Tarasov, S.G.; McFeeters, R.L.; Altieri, A.S.; Ji, X.; et al. Allosteric Activation of E2-RING Finger-Mediated Ubiquitylation by a Structurally Defined Specific E2-Binding Region of gp78. Mol. Cell 2009, 34, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Hibbert, R.G.; Huang, A.; Boelens, R.; Sixma, T.K. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. USA 2011, 108, 5590–5595. [Google Scholar] [CrossRef]
- Dou, H.; Buetow, L.; Sibbet, G.J.; Cameron, K.; Huang, D.T. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 2013, 20, 982–986. [Google Scholar] [CrossRef]
- Metzger, M.B.; Liang, Y.-H.; Das, R.; Mariano, J.; Li, S.; Li, J.; Kostova, Z.; Byrd, R.A.; Ji, X.; Weissman, A.M. A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms. Mol. Cell 2013, 50, 516–527. [Google Scholar] [CrossRef]
- Li, S.; Liang, Y.-H.; Mariano, J.; Metzger, M.B.; Stringer, D.K.; Hristova, V.A.; Li, J.; Randazzo, P.A.; Tsai, Y.C.; Ji, X.; et al. Insights into ubiquitination from the unique clamp-like binding of the RING E3 AO7 to the E2 UbcH5B. J. Biol. Chem. 2015, 290, 30225–30239. [Google Scholar] [CrossRef]
- Nagano, Y.; Mavrakis, K.J.; Lee, K.L.; Fujii, T.; Koinuma, D.; sase, H.; Yuki, K.; Isogaya, K.; Saitoh, M.; Imamura, T.; et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J. Biol. Chem. 2007, 282, 20492–20501. [Google Scholar] [CrossRef] [PubMed]
- Koinuma, D.; Shinozaki, M.; Komuro, A.; Goto, K.; Saitoh, M.; Hanyu, A.; Ebina, M.; Nukiwa, T.; Miyazawa, K.; Imamura, T.; et al. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. EMBO J. 2003, 22, 6458–6470. [Google Scholar] [CrossRef]
- Mavrakis, K.J.; Andrew, R.L.; Lee, K.L.; Petropoulou, C.; Dixon, J.E.; Navaratnam, N.; Norris, D.P.; Episkopou, V. Arkadia enhances Nodal/TGF-β signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 2007, 5, 0586–0603. [Google Scholar] [CrossRef]
- le Scolan, E.; Zhu, Q.; Wang, L.; Bandyopadhyay, A.; Javelaud, D.; Mauviel, A.; Sun, L.; Luo, K. Transforming growth factor-β suppresses the ability of ski to inhibit tumor metastasis by inducing its degradation. Cancer Res. 2008, 68, 3277–3285. [Google Scholar] [CrossRef] [Green Version]
- Briones-Orta, M.A.; Levy, L.; Madsen, C.D.; Das, D.; Erker, Y.; Sahai, E.; Hill, C.S. Arkadia regulates tumor metastasis by modulation of the TGF-β pathway. Cancer Res. 2013, 73, 1800–1810. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Antonacopoulou, A.G.; Tanaka, S.; Panoutsopoulos, A.A.; Bravou, V.; Kalofonos, H.P.; Episkopou, V. Enhancement of TGF-β signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res. 2011, 71, 6438–6449. [Google Scholar] [CrossRef] [PubMed]
- Chasapis, C.T.; Kandias, N.G.; Episkopou, V.; Bentrop, D.; Spyroulias, G.A. NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase. Proteins Struct. Funct. Bioinform. 2012, 80, 1484–1489. [Google Scholar] [CrossRef]
- Kandias, N.G.; Chasapis, C.T.; Bentrop, D.; Episkopou, V.; Spyroulias, G.A. High yield expression and NMR characterization of Arkadia E3 ubiquitin ligase RING-H2 finger domain. Biochem. Biophys. Res. Commun. 2009, 378, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, S.L.; Hansen, R.K.; Wagner, S.A.; van Cuijk, L.; van Belle, G.J.; Streicher, W.; Wikström, M.; Choudhary, C.; Houtsmuller, A.B.; Marteijin, J.A.; et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J. Cell Biol. 2013, 201, 787–807. [Google Scholar] [CrossRef]
- van Cuijk, L.; van Belle, G.J.; Turkyilmaz, Y.; Poulsen, S.L.; Janssens, R.C.; Theil, A.F.; Sabatella, M.; Lans, H.; Mailand, N.; Houtsmuller, A.B.; et al. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat. Commun. 2015, 6, 7499. [Google Scholar] [CrossRef]
- Stewart, G.S.; Panier, S.; Townsend, K.; Al-Hakim, A.K.; Kolas, N.K.; Miller, E.S.; Nakada, S.; Ylanko, J.; Olivarious, S.; Mendez, M.; et al. The RIDDLE Syndrome Protein Mediates a Ubiquitin-Dependent Signaling Cascade at Sites of DNA Damage. Cell 2009, 136, 420–434. [Google Scholar] [CrossRef]
- Wright, J.D.; Mace, P.D.; Day, C.L. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat. Struct. Mol. Biol. 2016, 23, 45–52. [Google Scholar] [CrossRef]
- Bijlmakers, M.J.; Teixeira, J.M.C.; Boer, R.; Mayzel, M.; Puig-Sàrries, P.; Karlsson, G.; Coll, M.; Pons, M.; Crosas, B. A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Chaugule, V.K.; Arkinson, C.; Rennie, M.L.; Kämäräinen, O.; Toth, R.; Walden, H. Allosteric mechanism for site-specific ubiquitination of FANCD2. Nat. Chem. Biol. 2020, 16, 291–301. [Google Scholar] [CrossRef]
- Pruneda, J.N.; Stoll, K.E.; Bolton, L.J.; Brzovic, P.S.; Klevit, R.E. Ubiquitin in motion: Structural studies of the ubiquitin-conjugating enzyme~ubiquitin conjugate. Biochemistry 2011, 50, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Branigan, E.; Penedo, J.C.; Hay, R.T. Ubiquitin transfer by a RING E3 ligase occurs from a closed E2~ubiquitin conformation. Nat. Commun. 2020, 11, 2846. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Houben, K.; Dominguez, C.; van Schaik, F.; Timmers, M.; Bonvin, A.; Boelens, R. Solution structure of the ubiquitin-conjugating enzyme UbcH5B. J. Mol. Biol. 2004, 344, 513–526. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kar, G.; Keskin, O.; Nussinov, R.; Gursoy, A. Human proteome-scale structural modeling of E2-E3 interactions exploiting interface motifs. J. Proteome Res. 2012, 11, 1196–1207. [Google Scholar] [CrossRef]
- Pierce, M.M.; Raman, C.S.; Nall, B.T. Isothermal titration calorimetry of protei-n-protein interactions. Methods: A Companion Methods Enzymol. 1999, 19, 213–221. [Google Scholar] [CrossRef]
- Birkou, M.; Raptis, V.; Marousis, K.D.; Tsevis, A.; Bourikas, K.; Bentrop, D.; Episkopou, V.; Spyroulias, G.A. Impact of a Single Nucleotide Polymorphism on the 3D Protein Structure and Ubiquitination Activity of E3 Ubiquitin Ligase Arkadia. Front. Mol. Biosci. 2022, 9, 844129. [Google Scholar] [CrossRef]
- Plechanovov, A.; Jaffray, E.G.; Tatham, M.H.; Naismith, J.H.; Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 2012, 489, 115–120. [Google Scholar] [CrossRef]
- Garrett, D.; Seok, Y.-J.; Peterkofsky, A.; Clore, G.; Groneborn, A. Identification by NMR of the Binding Surface for the Histidine-Containing Phosphocarrier Protein HPr on the N-Terminal Domain of Enzyme I of the Escherichia coli Phosphotransferase System. Biochemistry 1997, 36, 4393–4398. [Google Scholar] [CrossRef]
- Baker, K.A.; Hilty, C.; Peti, W.; Prince, A.; Pfaffinger, P.J.; Wider, G.; Wüthrich, K.; Choe, S. NMR-derived dynamic aspects of N-type inactivation of a Kv channel suggest a transient interaction with the T1 domain. Biochemistry 2006, 45, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Marousis, K.D.; Tsika, A.C.; Birkou, M.; Matsoukas, M.T.; Spyroulias, G.A. Lead identification through the synergistic action of biomolecular NMR and in silico methodologies. In Methods in Molecular Biology; Humana: New York, NY, USA, 2018; Volume 1824, pp. 299–316. [Google Scholar] [CrossRef]



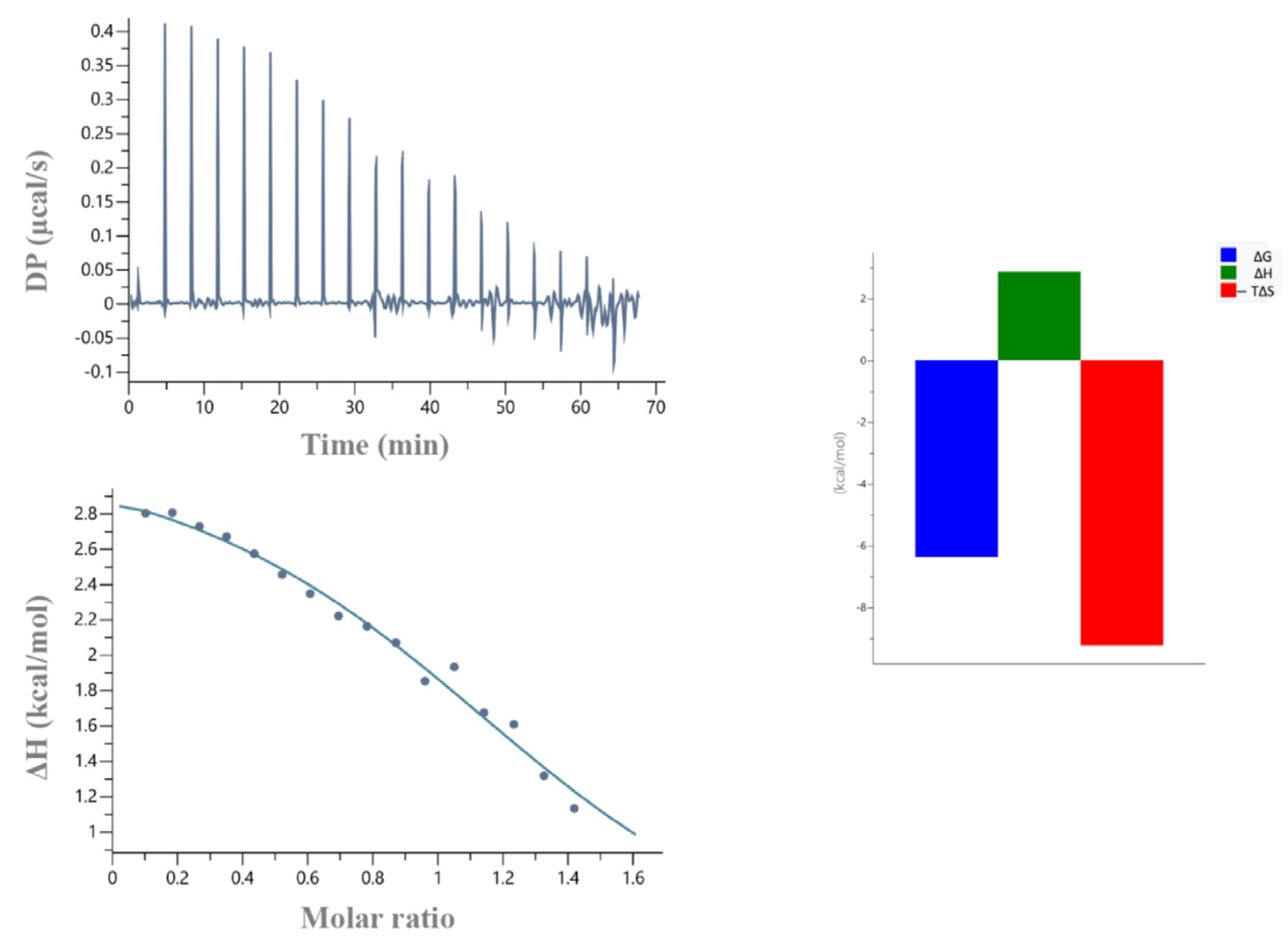

| Protein | Kd (μΜ) | N | ΔH (Kcal/mol) | −TΔS (Kcal/mol) | ΔG (Kcal/mol) |
|---|---|---|---|---|---|
| Arkadia 68 aa | 32 | 1 | 3.1 | −9.3 | −6.14 |
| Arkadia 119 aa | 2.7 | 0.6 * | −4.7 | −2.9 | −7.61 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Birkou, M.; Delegkou, G.N.; Marousis, K.D.; Fragkaki, N.; Toro, T.; Episkopou, V.; Spyroulias, G.A. Unveiling the Essential Role of Arkadia’s Non-RING Elements in the Ubiquitination Process. Int. J. Mol. Sci. 2022, 23, 10585. https://doi.org/10.3390/ijms231810585
Birkou M, Delegkou GN, Marousis KD, Fragkaki N, Toro T, Episkopou V, Spyroulias GA. Unveiling the Essential Role of Arkadia’s Non-RING Elements in the Ubiquitination Process. International Journal of Molecular Sciences. 2022; 23(18):10585. https://doi.org/10.3390/ijms231810585
Chicago/Turabian StyleBirkou, Maria, Georgia N. Delegkou, Konstantinos D. Marousis, Nefeli Fragkaki, Tamara Toro, Vasso Episkopou, and Georgios A. Spyroulias. 2022. "Unveiling the Essential Role of Arkadia’s Non-RING Elements in the Ubiquitination Process" International Journal of Molecular Sciences 23, no. 18: 10585. https://doi.org/10.3390/ijms231810585







