Nanotechnology Meets Oncology: A Perspective on the Role of the Personalized Nanoparticle-Protein Corona in the Development of Technologies for Pancreatic Cancer Detection
Abstract
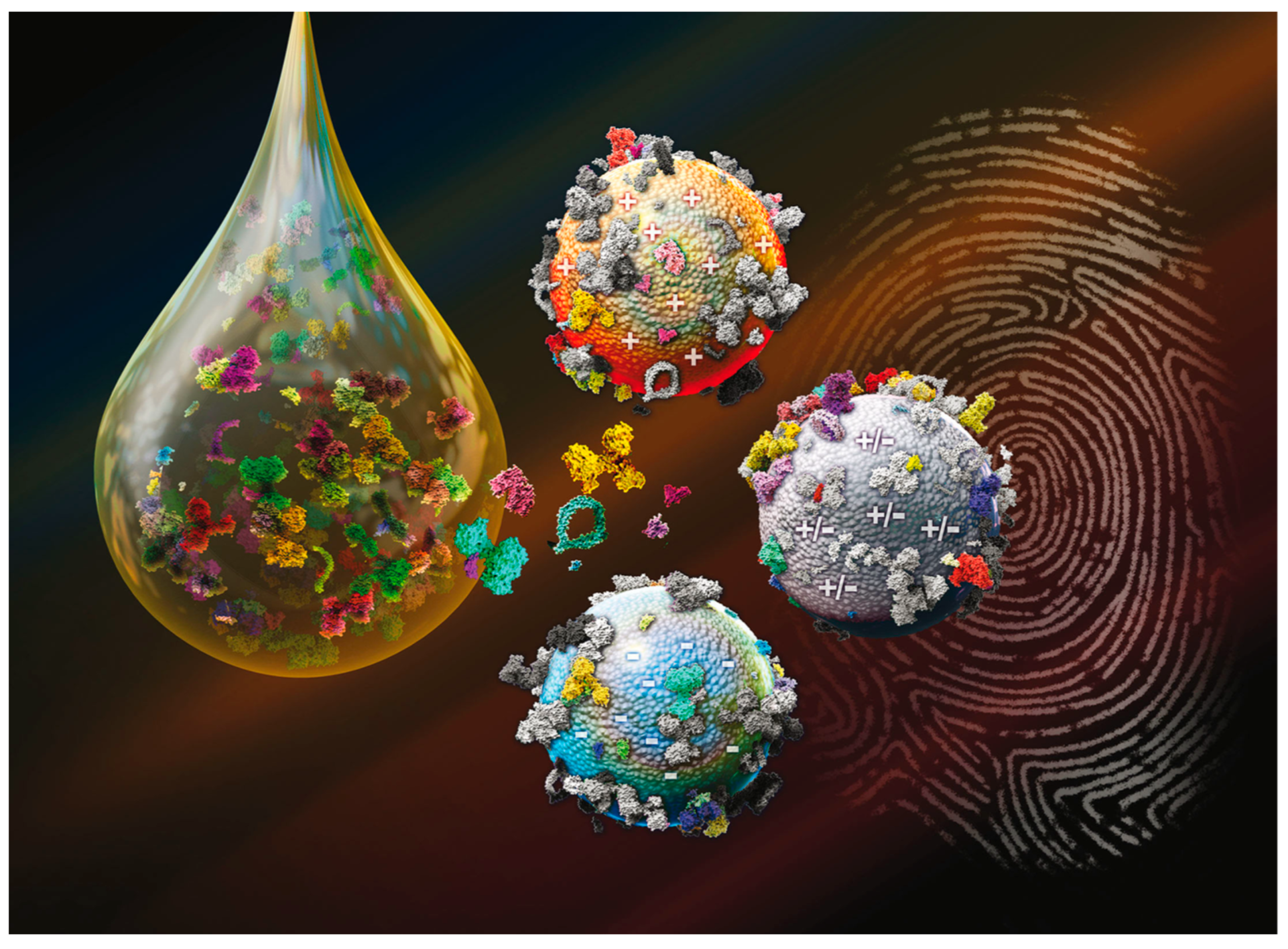
:1. Introduction
2. Personalized Protein Corona: The Missing Piece for Early-Stage Cancer Detection
3. Nanoparticle-Enabled Blood Tests
4. NEB Tests Based on Direct Characterization of the Personalized Protein Corona
5. NEB Tests Based on the Indirect Characterization of the Personalized Protein Corona
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Alghamdi, M.A.; Fallica, A.N.; Virzì, N.; Kesharwani, P.; Pittalà, V.; Greish, K. The Promise of Nanotechnology in Personalized Medicine. J. Pers. Med. 2022, 12, 673. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G. Liposome–protein corona in a physiological environment: Challenges and opportunities for targeted delivery of nanomedicines. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef]
- Papi, M.; Caracciolo, G. Principal component analysis of personalized biomolecular corona data for early disease detection. Nano Today 2018, 21, 14–17. [Google Scholar] [CrossRef]
- Ge, C.; Tian, J.; Zhao, Y.; Chen, C.; Zhou, R.; Chai, Z. Towards understanding of nanoparticle–protein corona. Arch. Toxicol. 2015, 89, 519–539. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.C.; Lin, S.; Parak, W.J.; Davis, T.P.; Caruso, F. A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. [Google Scholar] [CrossRef]
- Carrillo-Carrion, C.; Carril, M.; Parak, W.J. Techniques for the experimental investigation of the protein corona. Curr. Opin. Biotechnol. 2017, 46, 106–113. [Google Scholar] [CrossRef]
- Coglitore, D.; Janot, J.-M.; Balme, S. Protein at liquid solid interfaces: Toward a new paradigm to change the approach to design hybrid protein/solid-state materials. Adv. Colloid Interface Sci. 2019, 270, 278–292. [Google Scholar] [CrossRef]
- Docter, D.; Strieth, S.; Westmeier, D.; Hayden, O.; Gao, M.; Knauer, S.K.; Stauber, R.H. No king without a crown–impact of the nanomaterial-protein corona on nanobiomedicine. Nanomedicine 2015, 10, 503–519. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical− chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Laurent, S.; Aghaie, A.; Rezaee, F.; Mahmoudi, M. Personalized protein coronas: A “key” factor at the nanobiointerface. Biomater. Sci. 2014, 2, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Caputo, D.; Pozzi, D.; Colapicchioni, V.; Coppola, R. Size and charge of nanoparticles following incubation with human plasma of healthy and pancreatic cancer patients. Colloids Surf. B Biointerfaces 2014, 123, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Colapicchioni, V.; Tilio, M.; Digiacomo, L.; Gambini, V.; Palchetti, S.; Marchini, C.; Pozzi, D.; Occhipinti, S.; Amici, A.; Caracciolo, G. Personalized liposome–protein corona in the blood of breast, gastric and pancreatic cancer patients. Int. J. Biochem. Cell Biol. 2016, 75, 180–187. [Google Scholar] [CrossRef]
- Poruk, K.E.; Gay, D.Z.; Brown, K.; Mulvihill, J.D.; Boucher, K.M.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and prognostic updates. Curr. Mol. Med. 2013, 13, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, A.L.; Caracciolo, G.; Cavaliere, C.; Colapicchioni, V.; Piovesana, S.; Pozzi, D.; Laganà, A. Analytical Methods for Characterizing the Nanoparticle–Protein Corona. Chromatographia 2014, 77, 755–769. [Google Scholar] [CrossRef]
- Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2021014243 (accessed on 28 January 2021).
- Digiacomo, L.; Quagliarini, E.; La Vaccara, V.; Coppola, A.; Coppola, R.; Caputo, D.; Amenitsch, H.; Sartori, B.; Caracciolo, G.; Pozzi, D. Detection of Pancreatic Ductal Adenocarcinoma by Ex Vivo Magnetic Levitation of Plasma Protein-Coated Nanoparticles. Cancers 2021, 13, 5155. [Google Scholar] [CrossRef]
- Quagliarini, E.; Digiacomo, L.; Caputo, D.; Coppola, A.; Amenitsch, H.; Caracciolo, G.; Pozzi, D. Magnetic Levitation of Personalized Nanoparticle-Protein Corona as an Effective Tool for Cancer Detection. Nanomaterials 2022, 12, 1397. [Google Scholar] [CrossRef]
- Lammers, T.; Kiessling, F.; Hennink, W.E.; Storm, G. Drug targeting to tumors: Principles, pitfalls and (pre-) clinical progress. J. Control. Release 2012, 161, 175–187. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhang, F.; Xu, L.; Yue, L.; Kon, O.L.; Zhu, Y.; Guo, T. High-throughput proteomics and AI for cancer biomarker discovery. Adv. Drug Deliv. Rev. 2021, 176, 113844. [Google Scholar] [CrossRef]
- Merriel, S.W.; Pocock, L.; Gilbert, E.; Creavin, S.; Walter, F.M.; Spencer, A.; Hamilton, W. Systematic review and meta-analysis of the diagnostic accuracy of prostate-specific antigen (PSA) for the detection of prostate cancer in symptomatic patients. BMC Med. 2022, 20, 54. [Google Scholar] [CrossRef]
- Kamitani, Y.; Nonaka, K.; Isomoto, H. Current Status and Future Perspectives of Artificial Intelligence in Colonoscopy. J. Clin. Med. 2022, 11, 2923. [Google Scholar] [CrossRef] [PubMed]
- Gastounioti, A.; Desai, S.; Ahluwalia, V.S.; Conant, E.F.; Kontos, D. Artificial intelligence in mammographic phenotyping of breast cancer risk: A narrative review. Breast Cancer Res. 2022, 24, 14. [Google Scholar] [CrossRef] [PubMed]
- Bonney, A.; Malouf, R.; Marchal, C.; Manners, D.; Fong, K.M.; Marshall, H.M.; Irving, L.B.; Manser, R. Impact of low-dose computed tomography (LDCT) screening on lung cancer-related mortality. Cochrane Database Syst. Rev. 2022, 8, CD013829. [Google Scholar]
- Al-Shaheri, F.N.; Alhamdani, M.S.S.; Bauer, A.S.; Giese, N.; Büchler, M.W.; Hackert, T.; Hoheisel, J.D. Blood biomarkers for differential diagnosis and early detection of pancreatic cancer. Cancer Treat. Rev. 2021, 96, 102193. [Google Scholar] [CrossRef] [PubMed]
- Oberländer, J.; Champanhac, C.; da Costa Marques, R.; Landfester, K.; Mailänder, V. Temperature, concentration, and surface modification influence the cellular uptake and the protein corona of polystyrene nanoparticles. Acta Biomater. 2022, 148, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, S.; Pozzi, D.; Capriotti, A.L.; Barbera, G.; Chiozzi, R.Z.; Digiacomo, L.; Peruzzi, G.; Caracciolo, G.; Laganà, A. Influence of dynamic flow environment on nanoparticle-protein corona: From protein patterns to uptake in cancer cells. Colloids Surf. B Biointerfaces 2017, 153, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Hadjidemetriou, M.; Al-ahmady, Z.; Buggio, M.; Swift, J.; Kostarelos, K. A novel scavenging tool for cancer biomarker discovery based on the blood-circulating nanoparticle protein corona. Biomaterials 2019, 188, 118–129. [Google Scholar] [CrossRef]
- Caracciolo, G.; Safavi-Sohi, R.; Malekzadeh, R.; Poustchi, H.; Vasighi, M.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Hajipour, M.; Di Domenico, M. Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horiz. 2019, 4, 1063–1076. [Google Scholar] [CrossRef]
- Digiacomo, L.; Giulimondi, F.; Pozzi, D.; Coppola, A.; La Vaccara, V.; Caputo, D.; Caracciolo, G. A proteomic study on the personalized protein corona of liposomes. Relevance for early diagnosis of pancreatic DUCTAL adenocarcinoma and biomarker detection. J. Nanotheranostics 2021, 2, 82–93. [Google Scholar] [CrossRef]
- Di Santo, R.; Digiacomo, L.; Quagliarini, E.; Capriotti, A.L.; Laganà, A.; Zenezini Chiozzi, R.; Caputo, D.; Cascone, C.; Coppola, R.; Pozzi, D. Personalized graphene oxide-protein corona in the human plasma of pancreatic cancer patients. Front. Bioeng. Biotechnol. 2020, 8, 491. [Google Scholar] [CrossRef]
- Feng, L.; Wu, L.; Qu, X. New horizons for diagnostics and therapeutic applications of graphene and graphene oxide. Adv. Mater. 2013, 25, 168–186. [Google Scholar] [CrossRef] [PubMed]
- Koomen, J.M.; Shih, L.N.; Coombes, K.R.; Li, D.; Xiao, L.-c.; Fidler, I.J.; Abbruzzese, J.L.; Kobayashi, R. Plasma protein profiling for diagnosis of pancreatic cancer reveals the presence of host response proteins. Clin. Cancer Res. 2005, 11, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Bollig, A.; Wu, J.; Liao, D.J. Gene expression profiles in primary pancreatic tumors and metastatic lesions of Ela-c-myc transgenic mice. Mol. Cancer 2008, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Arvizo, R.R.; Giri, K.; Moyano, D.; Miranda, O.R.; Madden, B.; McCormick, D.J.; Bhattacharya, R.; Rotello, V.M.; Kocher, J.-P.; Mukherjee, P. Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles. PLoS ONE 2012, 7, e33650. [Google Scholar] [CrossRef]
- Digiacomo, L.; Palchetti, S.; Giulimondi, F.; Pozzi, D.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Caracciolo, G. The biomolecular corona of gold nanoparticles in a controlled microfluidic environment. Lab Chip 2019, 19, 2557–2567. [Google Scholar] [CrossRef]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef]
- Barrán-Berdón, A.L.; Pozzi, D.; Caracciolo, G.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Riccioli, A.; Palchetti, S.; Laganà, A. Time evolution of nanoparticle–protein corona in human plasma: Relevance for targeted drug delivery. Langmuir 2013, 29, 6485–6494. [Google Scholar] [CrossRef]
- Li, M.; Zhang, X.; Li, S.; Shao, X.; Chen, H.; Lv, L.; Huang, X. Probing protein dissociation from gold nanoparticles and the influence of temperature from the protein corona formation mechanism. RSC Adv. 2021, 11, 18198–18204. [Google Scholar] [CrossRef]
- Papi, M.; Palmieri, V.; Digiacomo, L.; Giulimondi, F.; Palchetti, S.; Ciasca, G.; Perini, G.; Caputo, D.; Cartillone, M.C.; Cascone, C. Converting the personalized biomolecular corona of graphene oxide nanoflakes into a high-throughput diagnostic test for early cancer detection. Nanoscale 2019, 11, 15339–15346. [Google Scholar] [CrossRef] [PubMed]
- Caputo, D.; Papi, M.; Coppola, R.; Palchetti, S.; Digiacomo, L.; Caracciolo, G.; Pozzi, D. A protein corona-enabled blood test for early cancer detection. Nanoscale 2017, 9, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Digiacomo, L.; Caputo, D.; Coppola, R.; Cascone, C.; Giulimondi, F.; Palchetti, S.; Pozzi, D.; Caracciolo, G. Efficient pancreatic cancer detection through personalized protein corona of gold nanoparticles. Biointerphases 2021, 16, 011010. [Google Scholar] [CrossRef] [PubMed]
- Caputo, D.; Digiacomo, L.; Cascone, C.; Pozzi, D.; Palchetti, S.; Di Santo, R.; Quagliarini, E.; Coppola, R.; Mahmoudi, M.; Caracciolo, G. Synergistic Analysis of Protein Corona and Haemoglobin Levels Detects Pancreatic Cancer. Cancers 2020, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.; Suguitan, M.; Chawla, A. Identification of high-risk germline variants for the development of pancreatic cancer: Common characteristics and potential guidance to screening guidelines. Pancreatology, 2022; ahead of print. [Google Scholar] [CrossRef]
- Caputo, D.; Cartillone, M.; Cascone, C.; Pozzi, D.; Digiacomo, L.; Palchetti, S.; Caracciolo, G.; Coppola, R. Improving the accuracy of pancreatic cancer clinical staging by exploitation of nanoparticle-blood interactions: A pilot study. Pancreatology 2018, 18, 661–665. [Google Scholar] [CrossRef]
- Schlitter, A.M.; Jesinghaus, M.; Jäger, C.; Konukiewitz, B.; Muckenhuber, A.; Demir, I.E.; Bahra, M.; Denkert, C.; Friess, H.; Kloeppel, G. pT but not pN stage of the 8th TNM classification significantly improves prognostication in pancreatic ductal adenocarcinoma. Eur. J. Cancer 2017, 84, 121–129. [Google Scholar] [CrossRef]
- Di Domenico, M.; Pozzi, D.; Palchetti, S.; Digiacomo, L.; Iorio, R.; Astarita, C.; Fiorelli, A.; Pierdiluca, M.; Santini, M.; Barbarino, M.; et al. Nanoparticle-biomolecular corona: A new approach for the early detection of non-small-cell lung cancer. J. Cell. Physiol. 2019, 234, 9378–9386. [Google Scholar] [CrossRef]
- Papi, M.; Palmieri, V.; Palchetti, S.; Pozzi, D.; Digiacomo, L.; Guadagno, E.; del Basso De Caro, M.; Di Domenico, M.; Ricci, S.; Pani, R. Exploitation of nanoparticle-protein interactions for early disease detection. Appl. Phys. Lett. 2019, 114, 163702. [Google Scholar] [CrossRef]
- Di Santo, R.; Quagliarini, E.; Digiacomo, L.; Pozzi, D.; Di Carlo, A.; Caputo, D.; Cerrato, A.; Montone, C.M.; Mahmoudi, M.; Caracciolo, G. Protein corona profile of graphene oxide allows detection of glioblastoma multiforme using a simple one-dimensional gel electrophoresis technique: A proof-of-concept study. Biomater. Sci. 2021, 9, 4671–4678. [Google Scholar] [CrossRef]
- Caputo, D.; Pozzi, D.; Farolfi, T.; Passa, R.; Coppola, R.; Caracciolo, G. Nanotechnology and pancreatic cancer management: State of the art and further perspectives. World J. Gastrointest. Oncol. 2021, 13, 231. [Google Scholar] [CrossRef]
- Digiacomo, L.; Giulimondi, F.; Capriotti, A.L.; Piovesana, S.; Montone, C.M.; Chiozzi, R.Z.; Laganà, A.; Mahmoudi, M.; Pozzi, D.; Caracciolo, G. Optimal centrifugal isolating of liposome–protein complexes from human plasma. Nanoscale Adv. 2021, 3, 3824–3834. [Google Scholar] [CrossRef]
- Ashkarran, A.A.; Dararatana, N.; Crespy, D.; Caracciolo, G.; Mahmoudi, M. Mapping the heterogeneity of protein corona by ex vivo magnetic levitation. Nanoscale 2020, 12, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Digiacomo, L.; Quagliarini, E.; Marmiroli, B.; Sartori, B.; Perini, G.; Papi, M.; Capriotti, A.L.; Montone, C.M.; Cerrato, A.; Caracciolo, G. Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle–Protein Complexes. Nanomaterials 2022, 12, 2376. [Google Scholar] [CrossRef] [PubMed]
- Caputo, D.; Caracciolo, G. Nanoparticle-enabled blood tests for early detection of pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 470, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Chetwynd, A.J.; Wheeler, K.E.; Lynch, I. Best practice in reporting corona studies: Minimum information about Nanomaterial Biocorona Experiments (MINBE). Nano Today 2019, 28, 100758. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caputo, D.; Quagliarini, E.; Pozzi, D.; Caracciolo, G. Nanotechnology Meets Oncology: A Perspective on the Role of the Personalized Nanoparticle-Protein Corona in the Development of Technologies for Pancreatic Cancer Detection. Int. J. Mol. Sci. 2022, 23, 10591. https://doi.org/10.3390/ijms231810591
Caputo D, Quagliarini E, Pozzi D, Caracciolo G. Nanotechnology Meets Oncology: A Perspective on the Role of the Personalized Nanoparticle-Protein Corona in the Development of Technologies for Pancreatic Cancer Detection. International Journal of Molecular Sciences. 2022; 23(18):10591. https://doi.org/10.3390/ijms231810591
Chicago/Turabian StyleCaputo, Damiano, Erica Quagliarini, Daniela Pozzi, and Giulio Caracciolo. 2022. "Nanotechnology Meets Oncology: A Perspective on the Role of the Personalized Nanoparticle-Protein Corona in the Development of Technologies for Pancreatic Cancer Detection" International Journal of Molecular Sciences 23, no. 18: 10591. https://doi.org/10.3390/ijms231810591
APA StyleCaputo, D., Quagliarini, E., Pozzi, D., & Caracciolo, G. (2022). Nanotechnology Meets Oncology: A Perspective on the Role of the Personalized Nanoparticle-Protein Corona in the Development of Technologies for Pancreatic Cancer Detection. International Journal of Molecular Sciences, 23(18), 10591. https://doi.org/10.3390/ijms231810591









