The Effect of Tropical Temperatures on the Quality of RNA Extracted from Stabilized Whole-Blood Samples
Abstract
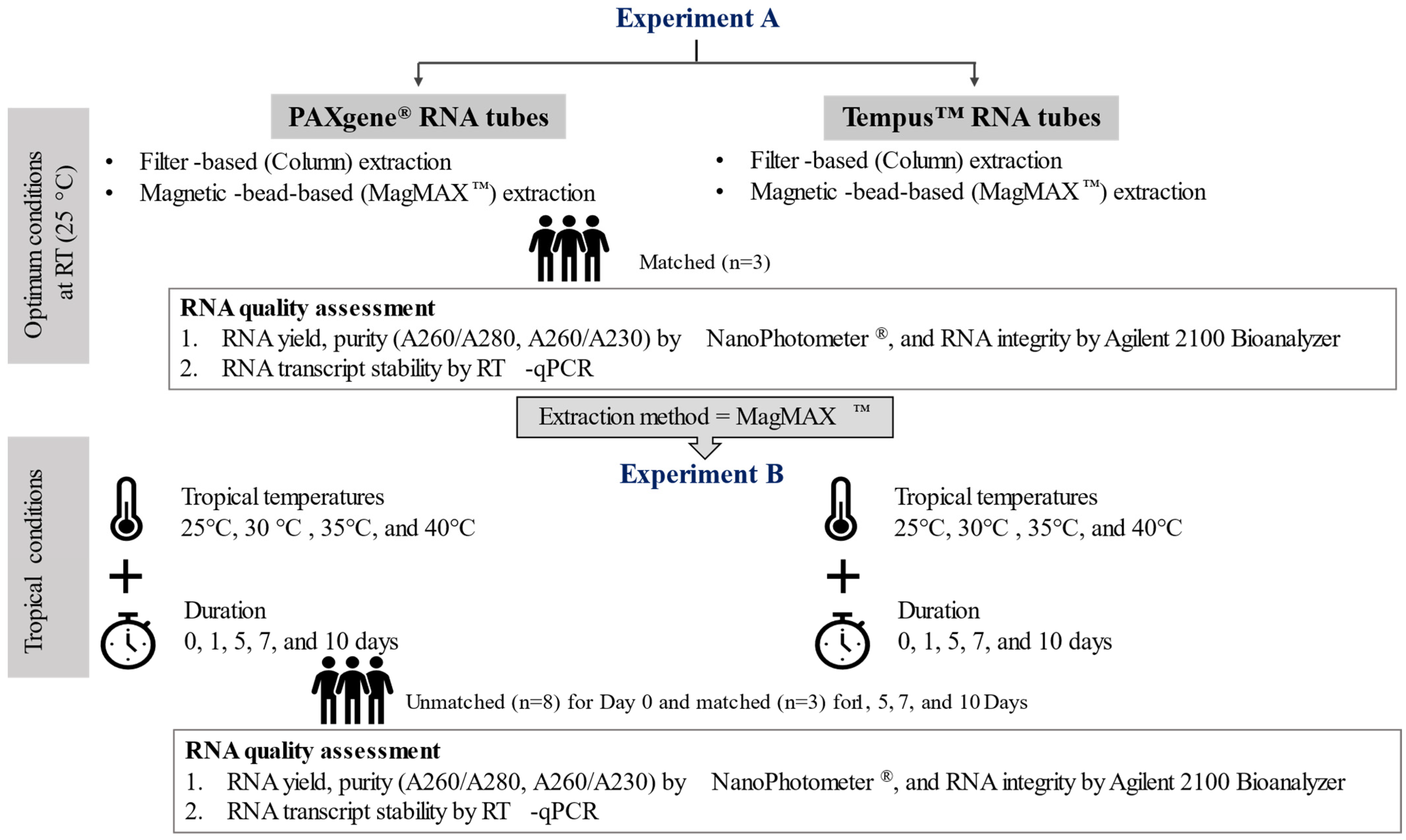
:1. Introduction
2. Results
2.1. Magnetic-Bead and Spin-Column-Based RNA Purification Systems Extracted Equivalent Whole-Blood RNA Quantity, Quality, and Purity
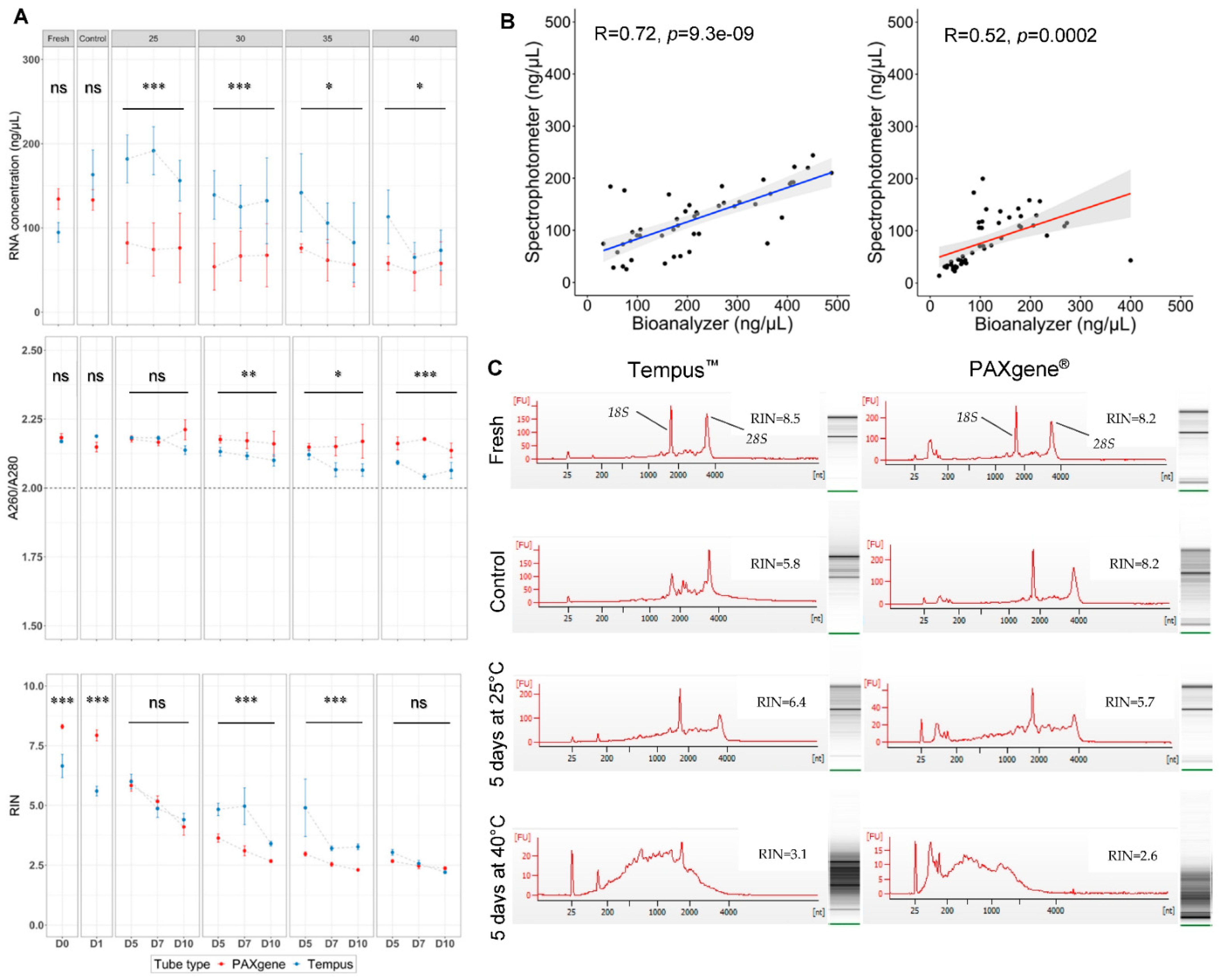
2.2. A Higher Quantity of RNA Was Obtained Using Tempus™ Blood RNA Tubes in Suboptimal Tropical Conditions
2.3. A Higher Quality of RNA Was Obtained Using PAXgene® Tubes in Optimal Laboratory Conditions
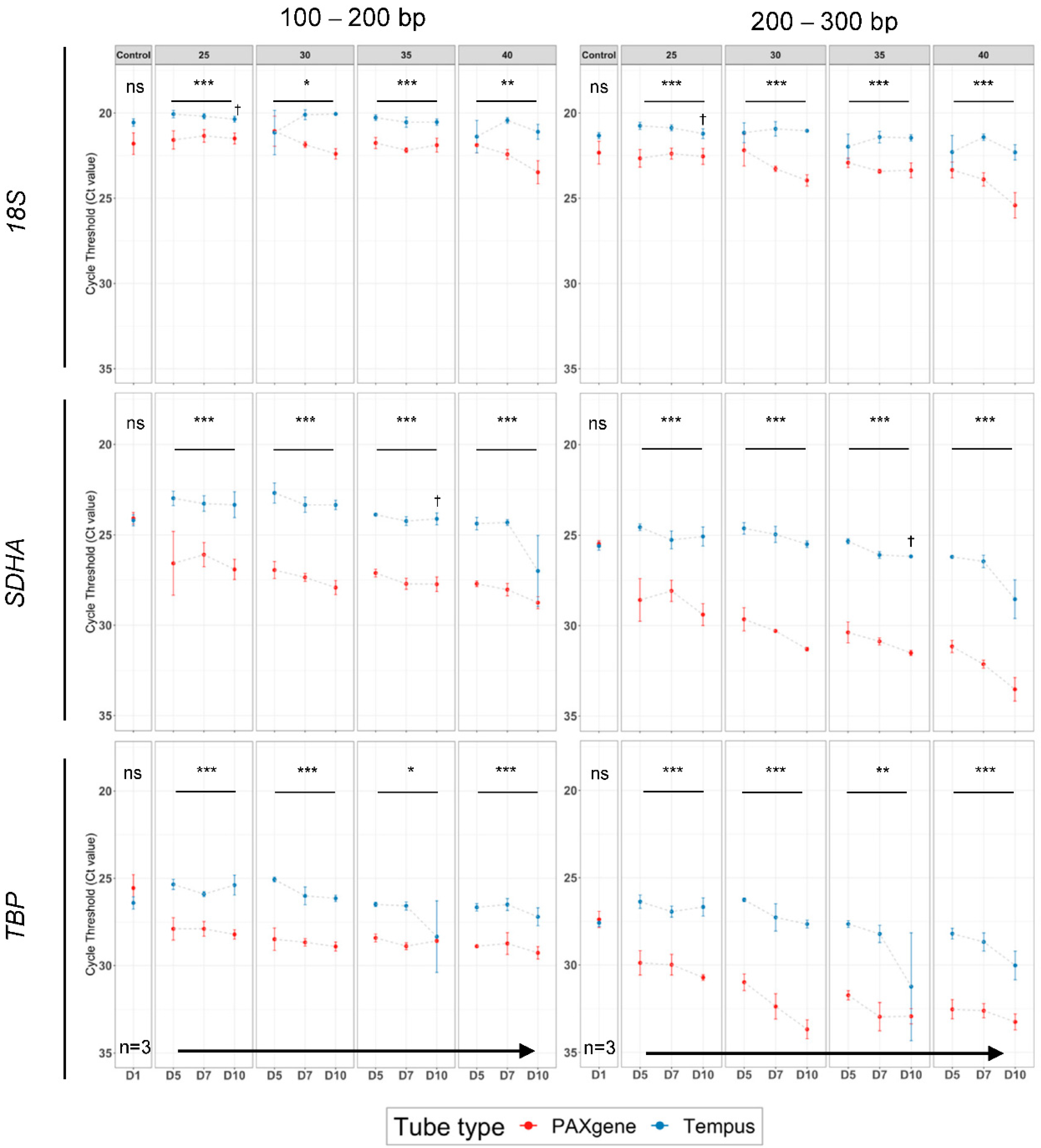
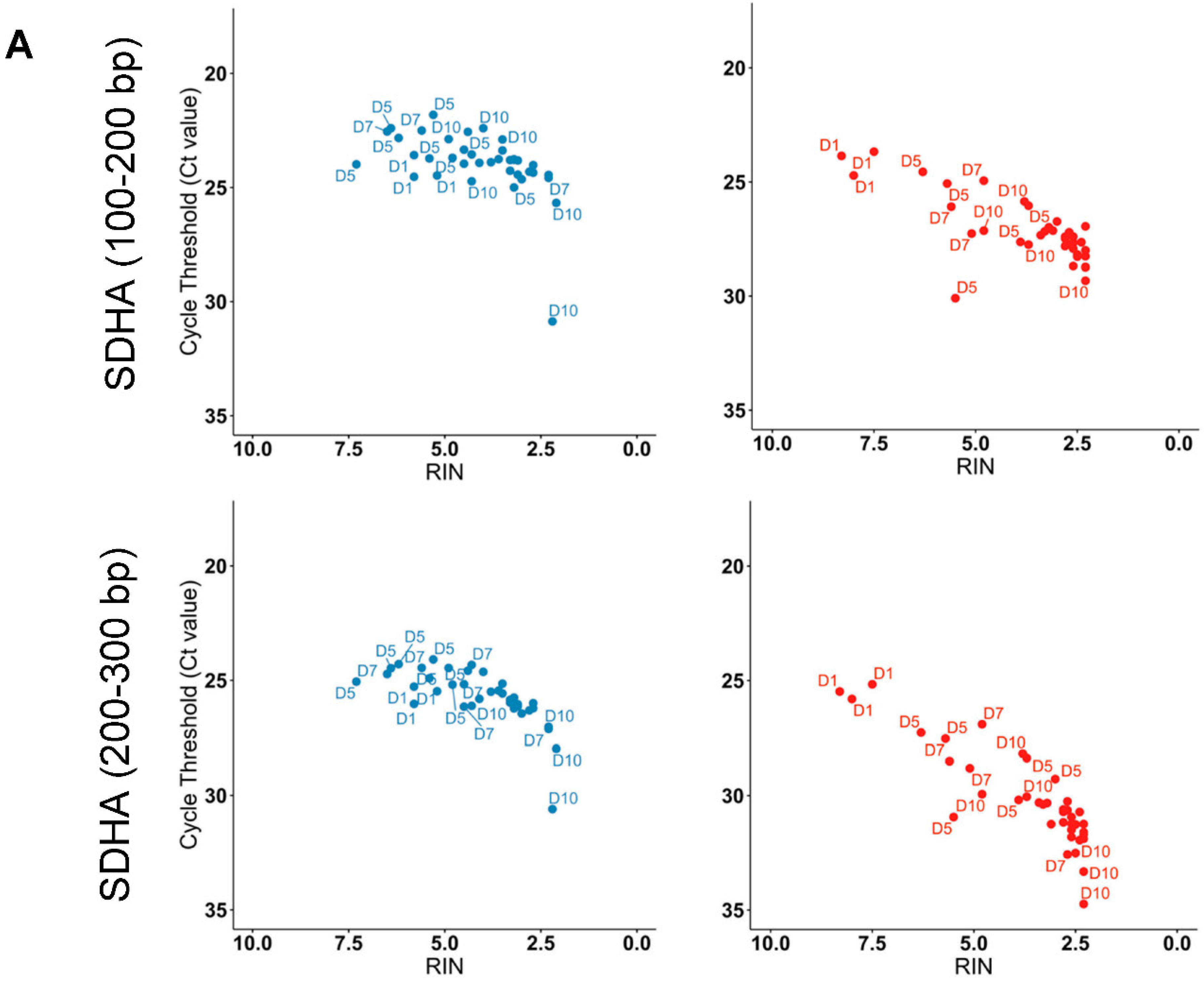
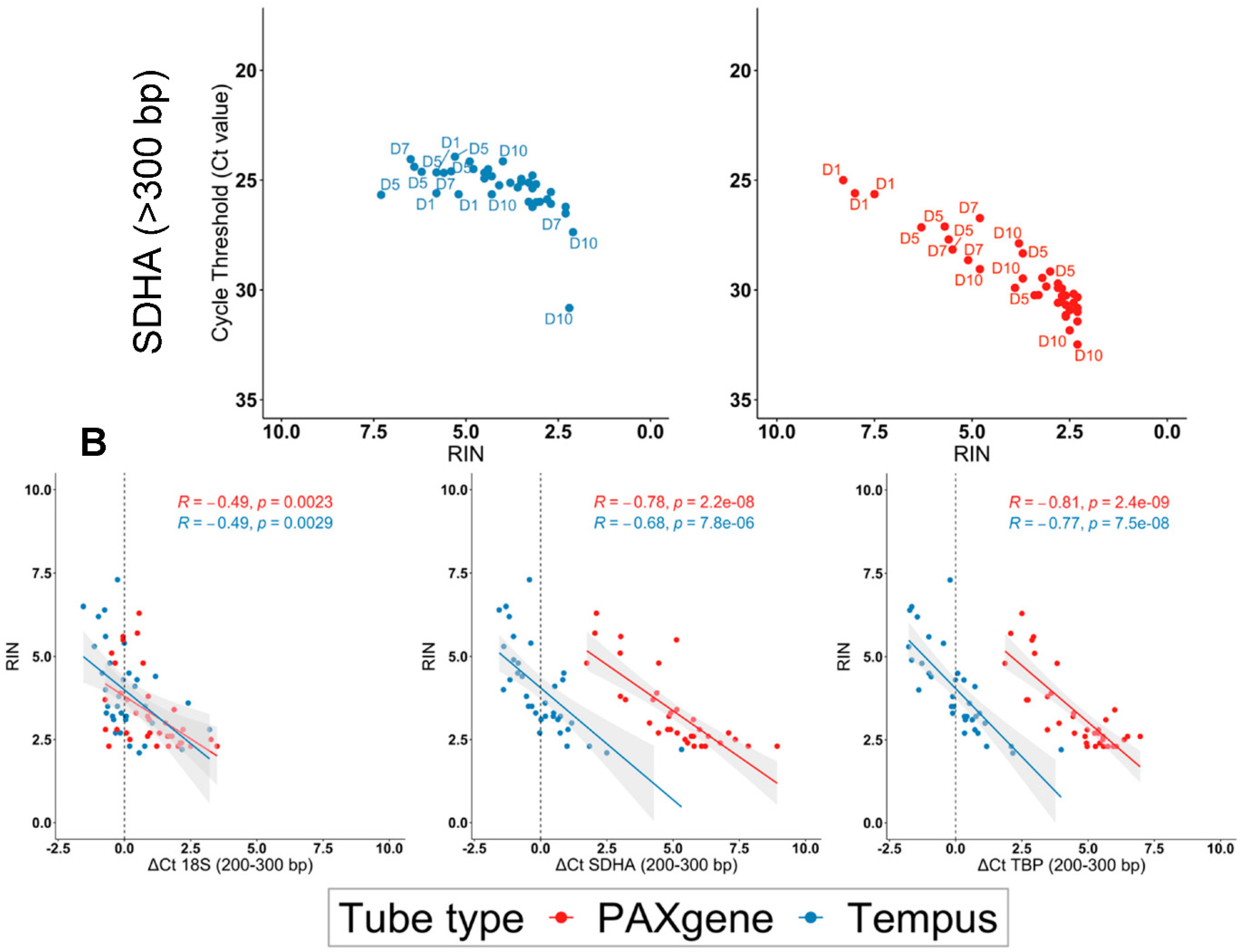
2.4. The Highest Quality RNA Was Obtained Using Tempus™ Blood RNA Tubes in Suboptimal Tropical Conditions
2.5. Tempus™ Tubes Maintain mRNA Integrity across Suboptimal Tropical Conditions
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Experimental Design
4.3. RNA Extraction
4.3.1. Column-Based RNA Purification
4.3.2. Magnetic-Bead-Based RNA Purification (MagMAX™)
4.4. RNA Yield, Purity and Integrity
4.5. Reverse Transcription Quantitative PCR (RT-qPCR)
4.5.1. Reverse Transcription
4.5.2. Quantitative PCR (qPCR)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohr, S.; Liew, C.-C. The peripheral-blood transcriptome: New insights into disease and risk assessment. Trends Mol. Med. 2007, 13, 422–432. [Google Scholar] [CrossRef]
- Zaas, A.K.; Chen, M.; Varkey, J.; Veldman, T.; Hero, A.O., 3rd; Lucas, J.; Huang, Y.; Turner, R.; Gilbert, A.; Lambkin-Williams, R.; et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009, 6, 207–217. [Google Scholar] [CrossRef]
- Holcomb, Z.E.; Tsalik, E.L.; Woods, C.W.; McClain, M.T. Host-Based Peripheral Blood Gene Expression Analysis for Diagnosis of Infectious Diseases. J. Clin. Microbiol. 2017, 55, 360–368. [Google Scholar] [CrossRef]
- Gliddon, H.D.; Herberg, J.A.; Levin, M.; Kaforou, M. Genome-wide host RNA signatures of infectious diseases: Discovery and clinical translation. Immunology 2018, 153, 171–178. [Google Scholar] [CrossRef]
- Gautam, A.; Donohue, D.; Hoke, A.; Miller, S.A.; Srinivasan, S.; Sowe, B.; Detwiler, L.; Lynch, J.; Levangie, M.; Hammamieh, R.; et al. Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods. PLoS ONE 2019, 14, e0225137. [Google Scholar] [CrossRef]
- Browne, D.J.; Brady, J.L.; Waardenberg, A.J.; Loiseau, C.; Doolan, D.L. An Analytically and Diagnostically Sensitive RNA Extraction and RT-qPCR Protocol for Peripheral Blood Mononuclear Cells. Front. Immunol. 2020, 11, 402. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Duale, N.; Lipkin, W.I.; Briese, T.; Aarem, J.; Rønningen, K.S.; Aas, K.K.; Magnus, P.; Harbak, K.; Susser, E.; Brunborg, G. Long-term storage of blood RNA collected in RNA stabilizing Tempus tubes in a large biobank—Evaluation of RNA quality and stability. BMC Res. Notes 2014, 7, 633. [Google Scholar] [CrossRef]
- Huang, L.-H.; Lin, P.-H.; Tsai, K.-W.; Wang, L.-J.; Huang, Y.-H.; Kuo, H.-C.; Li, S.-C. The effects of storage temperature and duration of blood samples on DNA and RNA qualities. PLoS ONE 2017, 12, e0184692. [Google Scholar] [CrossRef]
- Malentacchi, F.; Pizzamiglio, S.; Wyrich, R.; Verderio, P.; Ciniselli, C.; Pazzagli, M.; Gelmini, S. Effects of Transport and Storage Conditions on Gene Expression in Blood Samples. Biopreserv. Biobank. 2016, 14, 122–128. [Google Scholar] [CrossRef]
- Malentacchi, F.; Pazzagli, M.; Simi, L.; Orlando, C.; Wyrich, R.; Günther, K.; Verderio, P.; Pizzamiglio, S.; Ciniselli, C.M.; Zhang, H.; et al. SPIDIA-RNA: Second External Quality Assessment for the Pre-Analytical Phase of Blood Samples Used for RNA Based Analyses. PLoS ONE 2014, 9, e112293. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Williams, T.L.; Fernando, M.R. A novel blood collection device stabilizes cell-free RNA in blood during sample shipping and storage. BMC Res. Notes 2013, 6, 380. [Google Scholar] [CrossRef]
- Donohue, D.E.; Gautam, A.; Miller, S.A.; Srinivasan, S.; Abu-Amara, D.; Campbell, R.; Marmar, C.R.; Hammamieh, R.; Jett, M. Gene expression profiling of whole blood: A comparative assessment of RNA-stabilizing collection methods. PLoS ONE 2019, 14, e0223065. [Google Scholar] [CrossRef]
- Opitz, L.; Salinas-Riester, G.; Grade, M.; Jung, K.; Jo, P.; Emons, G.; Ghadimi, B.M.; Beißbarth, T.; Gaedcke, J. Impact of RNA degradation on gene expression profiling. BMC Med. Genom. 2010, 3, 36. [Google Scholar] [CrossRef]
- Langelaan, M.L.P.; Dylus, J.; Bock, E.; Jongen, B.; Mertens, A.A.M.; Raijmakers, M.T.M. Improved pre-analytical process for RNA isolation from whole blood samples. Ned. Tijdschr. Klin. Chemie Lab. 2014, 39, 164–165. [Google Scholar]
- Feddersen, S.; Bastholt, L.; Pedersen, S.M. Stabilization of circulating thyroglobulin mRNA transcripts in patients treated for differentiated thyroid carcinoma. Ann. Clin. Biochem. 2017, 54, 558–566. [Google Scholar] [CrossRef]
- Duale, N.; Brunborg, G.; Ronningen, K.S.; Briese, T.; Aarem, J.; Aas, K.K.; Magnus, P.; Stoltenberg, C.; Susser, E.; Lipkin, I.W. Human blood RNA stabilization in samples collected and transported for a large biobank. BMC Res. Notes 2012, 5, 510. [Google Scholar] [CrossRef]
- Aarem, J.; Brunborg, G.; Aas, K.K.; Harbak, K.; Taipale, M.M.; Magnus, P.; Knudsen, G.P.; Duale, N. Comparison of blood RNA isolation methods from samples stabilized in Tempus tubes and stored at a large human biobank. BMC Res. Notes 2016, 9, 430. [Google Scholar] [CrossRef]
- Yip, L.; Fuhlbrigge, R.; Atkinson, M.A.; Fathman, C.G. Impact of blood collection and processing on peripheral blood gene expression profiling in type 1 diabetes. BMC Genom. 2017, 18, 636. [Google Scholar] [CrossRef]
- Weber, D.G.; Casjens, S.; Rozynek, P.; Lehnert, M.; Zilch-Schöneweis, S.; Bryk, O.; Taeger, D.; Gomolka, M.; Kreuzer, M.; Otten, H.; et al. Assessment of mRNA and microRNA Stabilization in Peripheral Human Blood for Multicenter Studies and Biobanks. Biomark. Insights 2010, 5, 95–102. [Google Scholar] [CrossRef]
- Menke, A.; Rex-Haffner, M.; Klengel, T.; Binder, E.B.; Mehta, D. Peripheral blood gene expression: It all boils down to the RNA collection tubes. BMC Res. Notes 2012, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Eikmans, M.; Rekers, N.V.; Anholts, J.D.H.; Heidt, S.; Claas, F.H.J. Blood cell mRNAs and microRNAs: Optimized protocols for extraction and preservation. Blood 2013, 121, e81–e89. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Al-Soud, W.A.; Rådström, P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001, 39, 485–493. [Google Scholar] [CrossRef]
- Zhang, H.; Korenková, V.; Sjöback, R.; Švec, D.; Björkman, J.; Kruhøffer, M.; Verderio, P.; Pizzamiglio, S.; Ciniselli, C.M.; Wyrich, R.; et al. Biomarkers for Monitoring Pre-Analytical Quality Variation of mRNA in Blood Samples. PLoS ONE 2014, 9, e111644. [Google Scholar] [CrossRef]
- PAXgene Blood RNA System. Technical Note [Internet]. 2018. Available online: https://www.preanalytix.com/storage/download/_ProductResources_/TechnicalNotes/PROM-7266-002_BD-7969_TN_Blood_RNA_System_RNA_stability_over_11_years_storage_1118_WW_WEB.pdf (accessed on 24 March 2021).
- Applied BiosystemsTM TempusTM Blood RNA Tube [Internet]. Available online: https://www.thermofisher.com/order/catalog/product/4342792#/4342792 (accessed on 24 March 2021).
- Mlcochova, H.; Hezova, R.; Stanik, M.; Slaby, O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 41.e1–41.e9. [Google Scholar] [CrossRef]
- Sheng, Q.; Vickers, K.; Zhao, S.; Wang, J.; Samuels, D.C.; Koues, O.; Shyr, Y.; Guo, Y. Multi-perspective quality control of Illumina RNA sequencing data analysis. Brief. Funct. Genom. 2017, 16, 194–204. [Google Scholar] [CrossRef]
- Puchta, M.; Boczkowska, M.; Groszyk, J. Low RIN Value for RNA-Seq Library Construction from Long-Term Stored Seeds: A Case Study of Barley Seeds. Genes 2020, 11, 1190. [Google Scholar] [CrossRef]
- Shen, Y.; Li, R.; Tian, F.; Chen, Z.; Lu, N.; Bai, Y.; Ge, Q.; Lu, Z. Impact of RNA integrity and blood sample storage conditions on the gene expression analysis. Onco. Targets. Ther. 2018, 11, 3573–3581. [Google Scholar] [CrossRef]
- Yang, J.; Sui, X.; Wen, C.; Pan, C.; Zhu, Y.; Zhang, J.; Zhang, L. A hemocompatible cryoprotectant inspired by freezing-tolerant plants. Colloids Surf. B Biointerfaces 2019, 176, 106–114. [Google Scholar] [CrossRef]
- Koh, E.J.; Yu, S.Y.; Kim, S.H.; Kim, S.J.; Lee, E.I.; Hwang, S.Y. Understanding Confounding Effects of Blood Handling Strategies on RNA Quality and Transcriptomic Alteration Using RNA Sequencing. Biochip J. 2021, 15, 187–194. [Google Scholar] [CrossRef]
- Asare, A.L.; Kolchinsky, S.A.; Gao, Z.; Wang, R.; Raddassi, K.; Bourcier, K.; Seyfert-Margolis, V. Differential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolation. BMC Genom. 2008, 9, 474. [Google Scholar] [CrossRef]
- Häntzsch, M.; Tolios, A.; Beutner, F.; Nagel, D.; Thiery, J.; Teupser, D.; Holdt, L.M. Comparison of whole blood RNA preservation tubes and novel generation RNA extraction kits for analysis of mRNA and miRNA profiles. PLoS ONE 2014, 9, e113298. [Google Scholar] [CrossRef] [PubMed]
- Matheson, L.A.; Duong, T.T.; Rosenberg, A.M.; Yeung, R.S.M. Assessment of sample collection and storage methods for multicenter immunologic research in children. J. Immunol. Methods 2008, 339, 82–89. [Google Scholar] [CrossRef]
- Nikula, T.; Mykkänen, J.; Simell, O.; Lahesmaa, R. Genome-wide comparison of two RNA-stabilizing reagents for transcriptional profiling of peripheral blood. Transl. Res. 2013, 161, 181–188. [Google Scholar] [CrossRef]
- Roy, D.; Tomo, S.; Modi, A.; Purohit, P.; Sharma, P. Optimising total RNA quality and quantity by phenol-chloroform extraction method from human visceral adipose tissue: A standardisation study. MethodsX 2020, 7, 101113. [Google Scholar] [CrossRef]
- Carrillo-Ávila, J.A.; de la Puente, R.; Catalina, P.; Rejón, J.D.; Espín-Vallejo, L.; Valdivieso, V.; Aguilar-Quesada, R. Evaluation of RNA purification methods by using different blood stabilization tubes: Identification of key features for epidemiological studies. BMC Res. Notes 2020, 13, 77. [Google Scholar] [CrossRef]
- Sidova, M.; Tomankova, S.; Abaffy, P.; Kubista, M.; Sindelka, R. Effects of post-mortem and physical degradation on RNA integrity and quality. Biomol. Detect. Quantif. 2015, 5, 3–9. [Google Scholar] [CrossRef]
- Browne, D.J.; Kelly, A.M.; Brady, J.L.; Doolan, D.L. A high-throughput screening (HTS) RT-qPCR assay for quantifying surrogate markers of immunity from PBMCs. Front. Immunol. 2022, 13, 962220. [Google Scholar] [CrossRef]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38, 792–799. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
| Model 1 = RNA Concentration (ng/μL) | ||||
| Explanatory variable | Estimate | Std. Error | t value | p-value |
| (Intercept) | 66.230 | 78.853 | 0.84 | 0.404 |
| Tube type: Tempus™ | 254.894 | 47.339 | 5.384 | 1.15 × 10−6 *** |
| Days | 5.049 | 9.997 | 0.505 | 0.615 |
| Temperature | −0.106 | 2.358 | −0.045 | 0.964 |
| Tube-type Tempus™: Days | −5.859 | 3.342 | −1.753 | 0.084 |
| Tube-type Tempus™: Temperature | −4.650 | 1.228 | −3.786 | 0.0003 * |
| Days: Temperature | −0.170 | 0.299 | −0.57 | 0.571 |
| Adjusted R-squared: 0.764 (p value: <2.2 × 10−16) | ||||
| Model 2 = A260/A280 | ||||
| Explanatory variable | Estimate | Std. Error | t value | p-value |
| (Intercept) | 2.181 | 0.102 | 21.466 | <2 × 10−16 *** |
| Tube type: Tempus™ | 0.161 | 0.061 | 2.638 | 0.011 * |
| Days | 0.008 | 0.013 | 0.606 | 0.547 |
| Temperature | 0.000 | 0.003 | −0.085 | 0.933 |
| Tube-type Tempus™: Days | −0.008 | 0.004 | −1.951 | 0.055 |
| Tube-type Tempus™: Temperature | −0.005 | 0.002 | −3.075 | 0.003 ** |
| Days: Temperature | 0.000 | 0.000 | −0.567 | 0.572 |
| Adjusted R-squared:0.565 (p-value: 1.458 × 10−10) | ||||
| Model 3 = log2(RIN) | ||||
| Explanatory variable | Estimate | Std. Error | t value | p-value |
| (Intercept) | 4.905 | 0.572 | 8.581 | 3.45 × 10−12 *** |
| Tube type: Tempus™ | 0.338 | 0.343 | 0.985 | 0.328 |
| Days | −0.148 | 0.072 | −2.047 | 0.045 * |
| Temperature | −0.082 | 0.017 | −4.777 | 1.10 × 10−5 *** |
| Tube-type Tempus™: Days | −0.020 | 0.024 | −0.836 | 0.407 |
| Tube-type Tempus™: Temperature | 0.002 | 0.009 | 0.236 | 0.814 |
| Days: Temperature | 0.002 | 0.002 | 1.057 | 0.295 |
| Adjusted R-squared: 0.797 (p value: <2.2 × 10−16) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarathkumara, Y.D.; Browne, D.J.; Kelly, A.M.; Pattinson, D.J.; Rush, C.M.; Warner, J.; Proietti, C.; Doolan, D.L. The Effect of Tropical Temperatures on the Quality of RNA Extracted from Stabilized Whole-Blood Samples. Int. J. Mol. Sci. 2022, 23, 10609. https://doi.org/10.3390/ijms231810609
Sarathkumara YD, Browne DJ, Kelly AM, Pattinson DJ, Rush CM, Warner J, Proietti C, Doolan DL. The Effect of Tropical Temperatures on the Quality of RNA Extracted from Stabilized Whole-Blood Samples. International Journal of Molecular Sciences. 2022; 23(18):10609. https://doi.org/10.3390/ijms231810609
Chicago/Turabian StyleSarathkumara, Yomani D., Daniel J. Browne, Ashton M. Kelly, David J. Pattinson, Catherine M. Rush, Jeffrey Warner, Carla Proietti, and Denise L. Doolan. 2022. "The Effect of Tropical Temperatures on the Quality of RNA Extracted from Stabilized Whole-Blood Samples" International Journal of Molecular Sciences 23, no. 18: 10609. https://doi.org/10.3390/ijms231810609
APA StyleSarathkumara, Y. D., Browne, D. J., Kelly, A. M., Pattinson, D. J., Rush, C. M., Warner, J., Proietti, C., & Doolan, D. L. (2022). The Effect of Tropical Temperatures on the Quality of RNA Extracted from Stabilized Whole-Blood Samples. International Journal of Molecular Sciences, 23(18), 10609. https://doi.org/10.3390/ijms231810609






