Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients
Abstract
:1. Introduction
2. Results
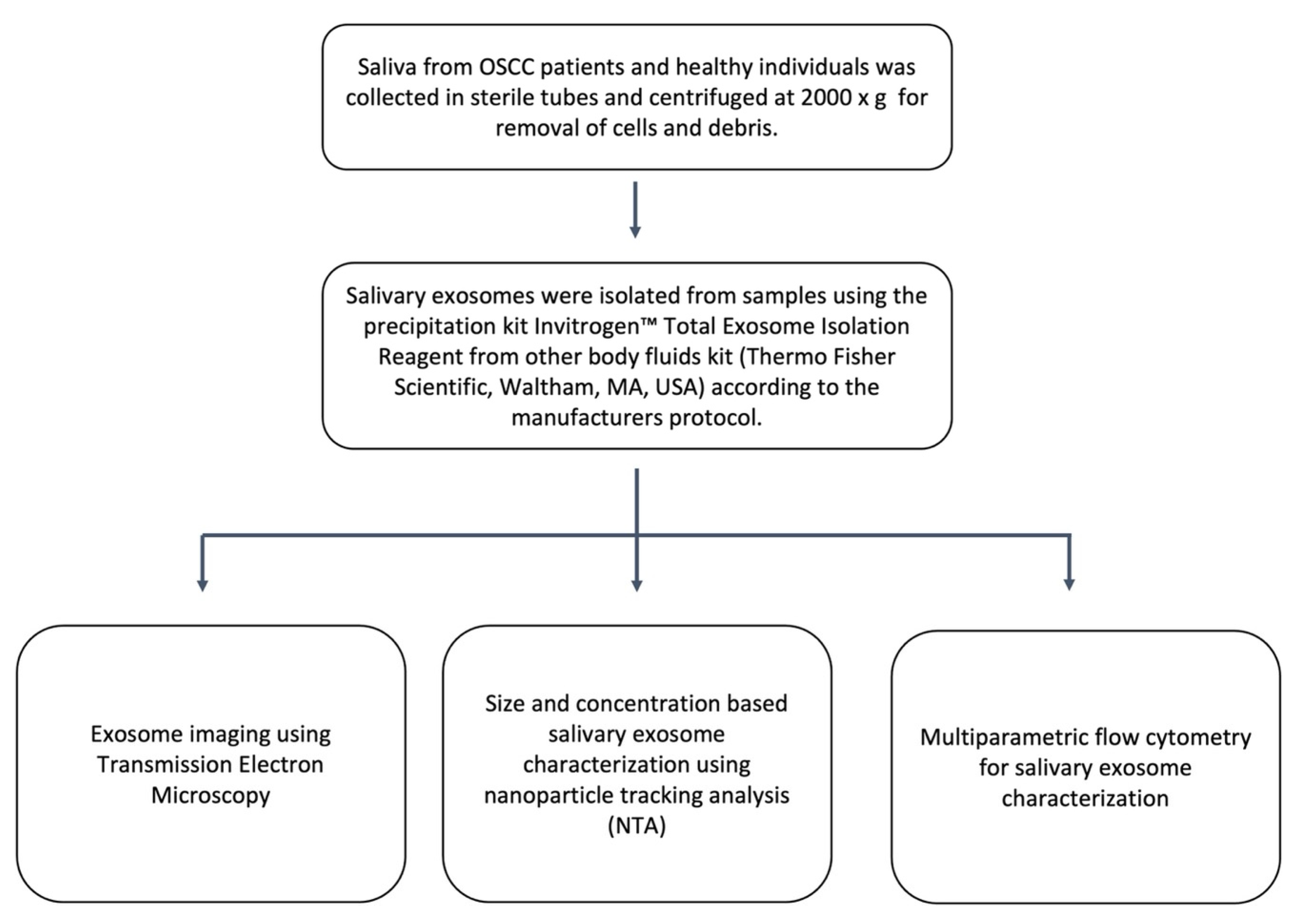
2.1. Identification and Characterisation of Salivary Exosomes Derived from Oral Cancer Patients
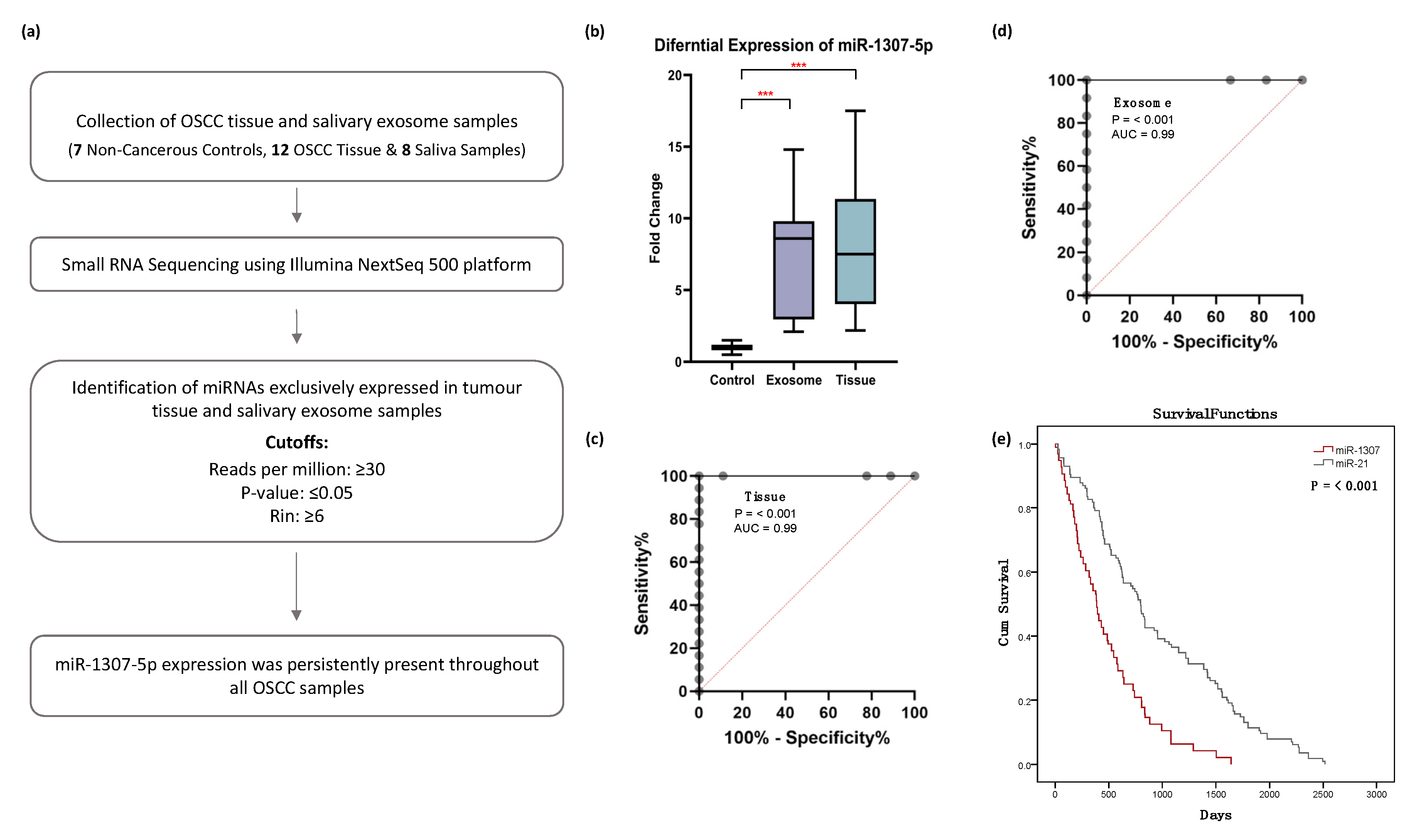
2.2. Small RNA Sequencing Analysis Identified miR-1307-5p, a miRNA Which Is Exclusively Expressed in Tissues and Salivary Exosomes of OSCC Patients
2.3. miR-1307-5p as a Potential Prognosticator in OSCC Patients
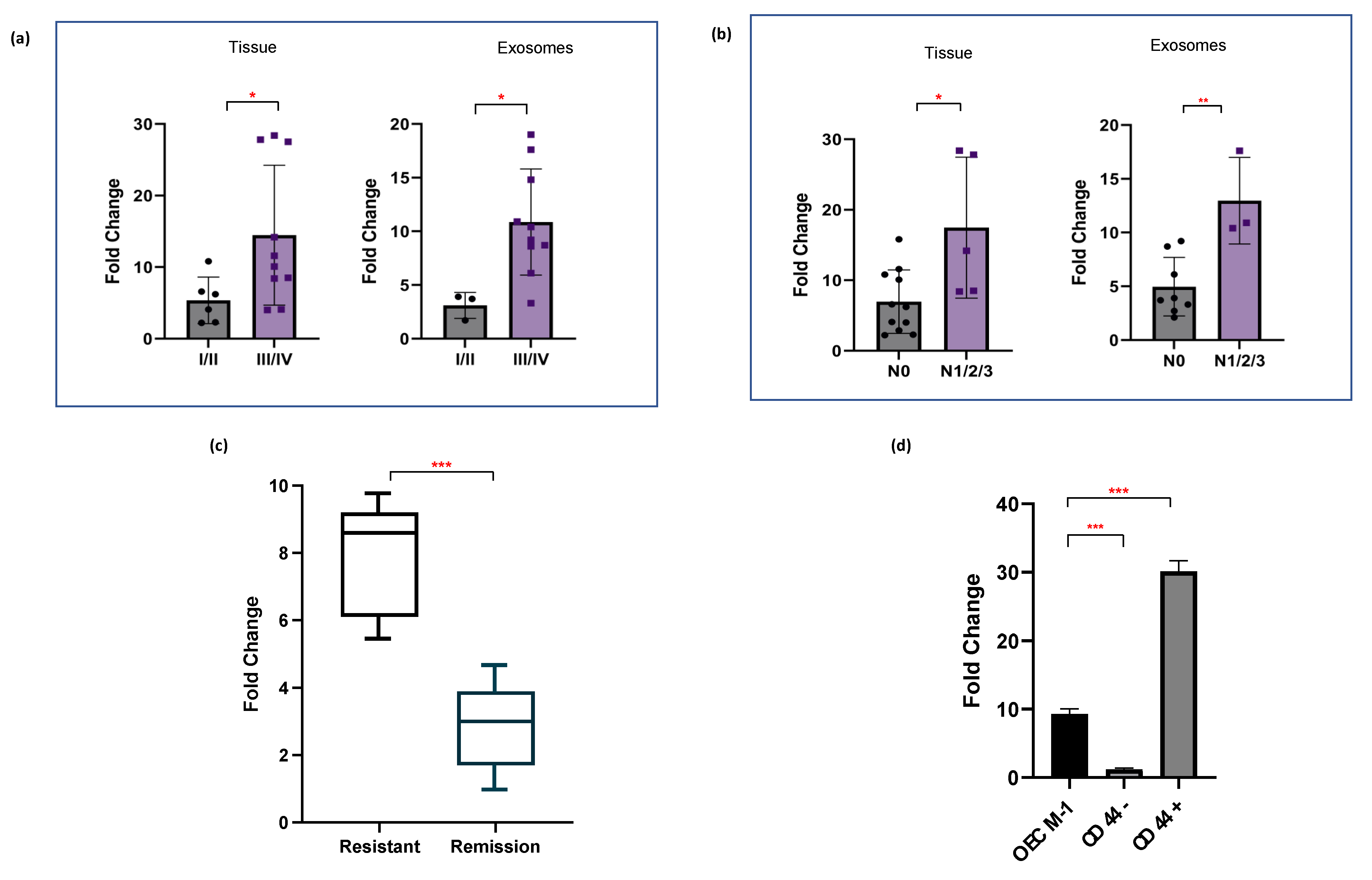
2.4. miR-1307-5p Demonstrates Clinical Association with Disease Progression, Aggressiveness, and Therapeutic Refractoriness
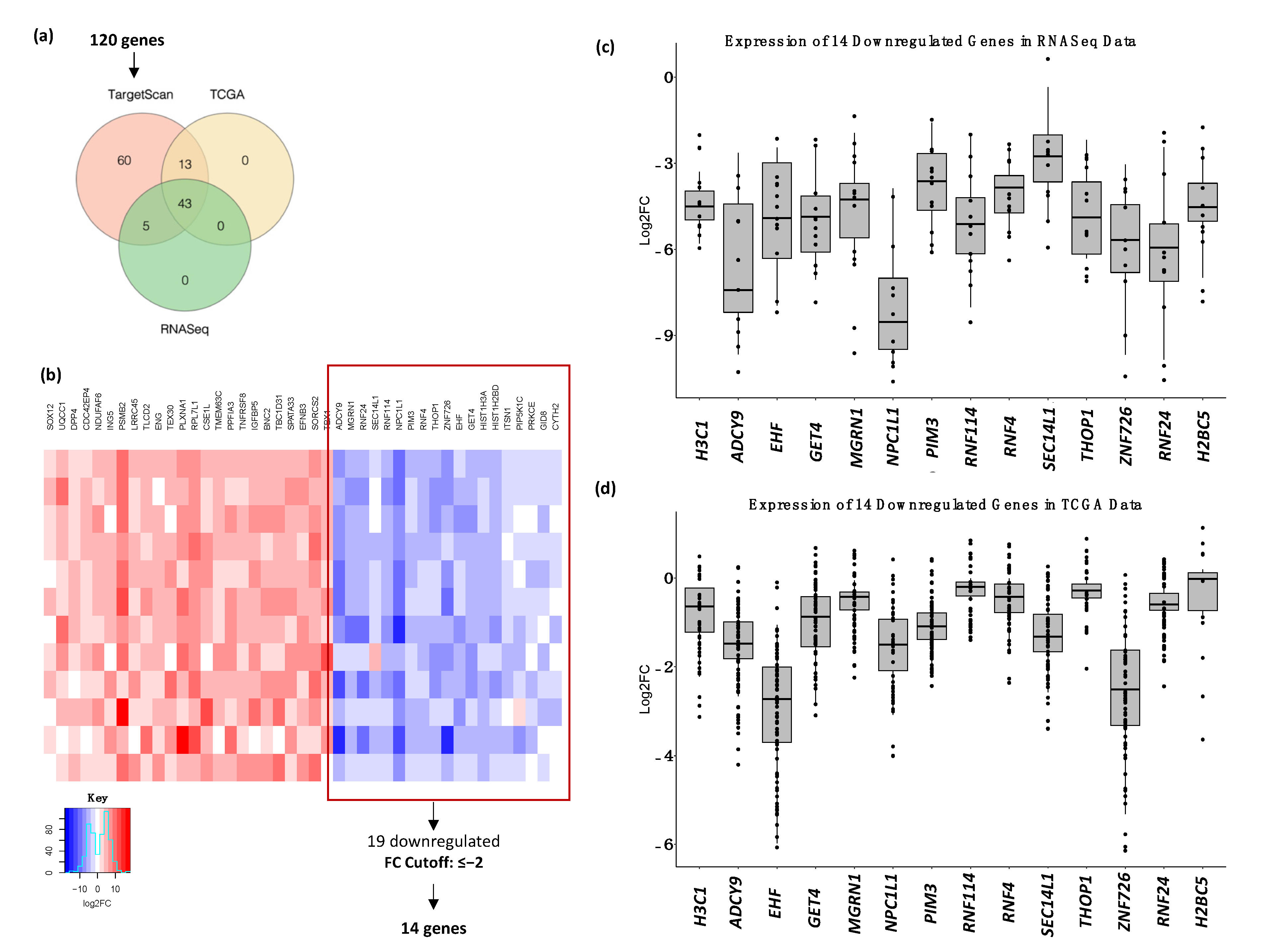
2.5. Identification of Differentially Expressed Genes in OSCC Patients Using RNA Sequencing Analysis
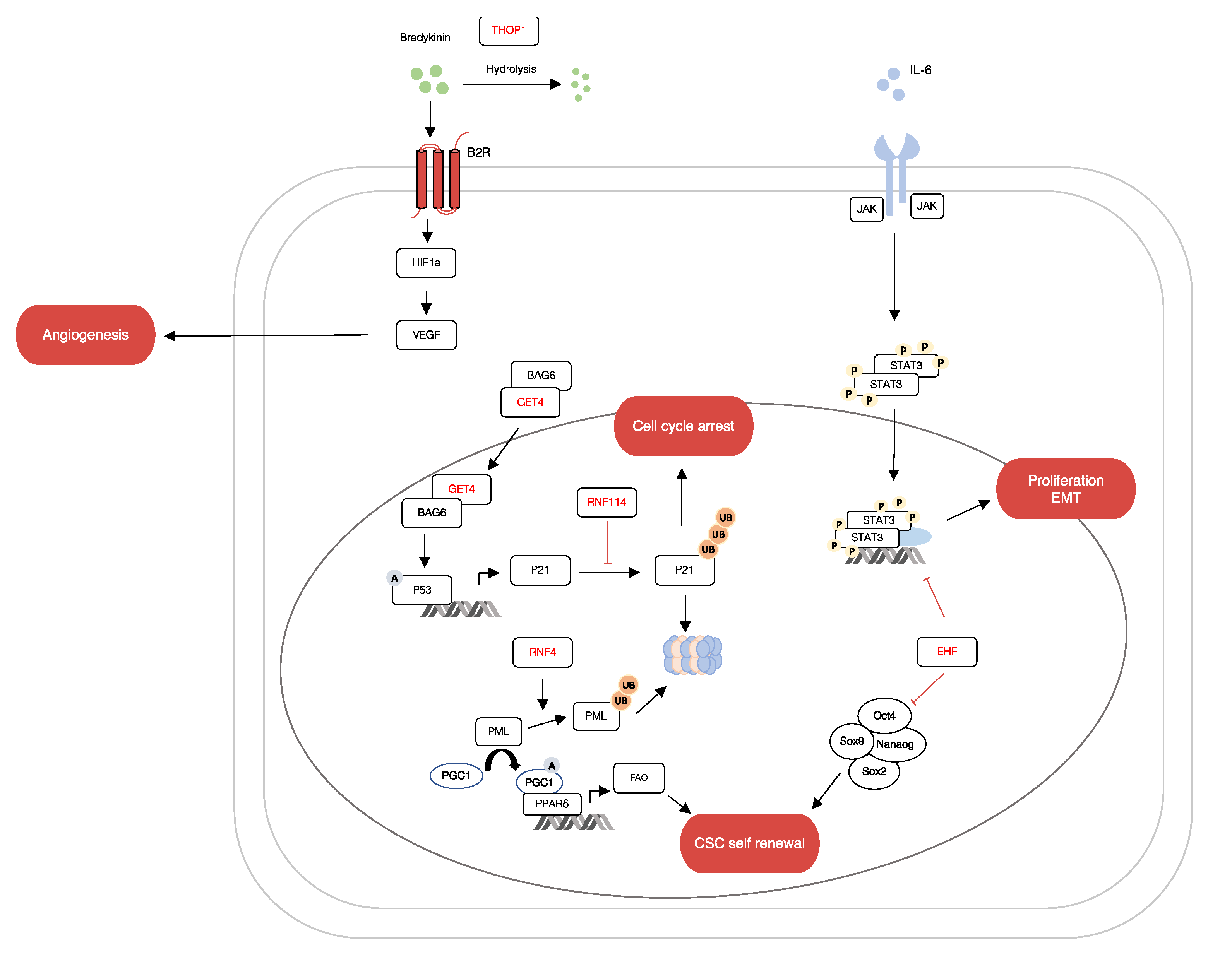
2.6. Target Gene Prediction and Functional Analysis of miR-1307-5p
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Sampling Details
4.2. Exosome Isolation
4.3. Exosome Characterisation
4.4. RNA Extraction
4.5. miRNA Sequencing
4.6. Transcriptome Sequencing
4.7. miRNA Target Prediction
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Unique miRNAs in OSCC Salivary Exosome Samples | Unique miRNAs in OSCC Tissue Samples |
|---|---|
| hsa-miR-125a-5p | hsa-miR-10b-5p |
| hsa-miR-1307-5p | hsa-miR-128-1-5p |
| hsa-miR-330-5p | hsa-miR-130a-5p |
| hsa-miR-138-5p | |
| hsa-miR-139-5p | |
| hsa-miR-146b-5p | |
| hsa-miR-214-5p | |
| hsa-miR-365b-5p | |
| hsa-miR-382-5p | |
| hsa-miR-409-5p | |
| hsa-miR-671-5p | |
| hsa-miR-1307-5p |

References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Shetty, K.S.R.; Kurle, V.; Greeshma, P.; Ganga, V.B.; Murthy, S.P.; Thammaiah, S.K.; Prasad, P.K.; Chavan, P.; Halkud, R.; Krishnappa, R. Salvage Surgery in Recurrent Oral Squamous Cell Carcinoma. Front. Oral Health 2021, 2, 815606. [Google Scholar] [CrossRef] [PubMed]
- Soung, Y.H.; Ford, S.; Zhang, V.; Chung, J. Exosomes in Cancer Diagnostics. Cancers 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Emerging roles of exosomes during epithelial–mesenchymal transition and cancer progression. In Seminars in Cell & Developmental Biology; Greening, D.W.; Gopal, S.K.; Mathias, R.A.; Liu, L.; Sheng, J.; Zhu, H.-J.; Simpson, R.J. (Eds.) Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Salehi, M.; Sharifi, M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J. Cell. Physiol. 2018, 233, 6370–6380. [Google Scholar] [CrossRef]
- Langevin, S.; Kuhnell, D.; Parry, T.; Biesiada, J.; Huang, S.; Wise-Draper, T.; Kasper, S. Comprehensive microRNA-sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget 2017, 8, 82459–82474. [Google Scholar] [CrossRef]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2019, 121, 109553. [Google Scholar] [CrossRef]
- Farag, A.F.; Sabry, D.; Hassabou, N.F.; Alaa EL-Din, Y. MicroRNA-134/MicroRNA-200a Derived Salivary Exosomes are Novel Diagnostic Biomarkers of Oral Squamous Cell Carcinoma. Egypt. Dent. J. 2021, 67, 367–377. [Google Scholar] [CrossRef]
- Dioguardi, M.; Spirito, F.; Sovereto, D.; Alovisi, M.; Troiano, G.; Aiuto, R.; Garcovich, D.; Crincoli, V.; Laino, L.; Cazzolla, A.P.; et al. MicroRNA-21 Expression as a Prognostic Biomarker in Oral Cancer: Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2022, 19, 3396. [Google Scholar] [CrossRef]
- Narasimhan, N.S.; Narasimhan, N.M. The Emerging Role of MicroRNA21 in Oral Cancer. Biomed. Pharmacol. J. 2018, 11, 1961–1966. [Google Scholar] [CrossRef]
- Dong, B.; Li, S.; Zhu, S.; Yi, M.; Luo, S.; Wu, K. MiRNA-mediated EMT and CSCs in cancer chemoresistance. Exp. Hematol. Oncol. 2021, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Valinezhad, O.A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genom. 2014, 970607. [Google Scholar] [CrossRef]
- Lee, E.; Ito, K.; Zhao, Y.; Schadt, E.E.; Irie, H.Y.; Zhu, J. Inferred miRNA activity identifies miRNA-mediated regulatory networks underlying multiple cancers. Bioinformatics 2015, 32, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, L.; Ludwig, S.; Vahl, J.M.; Brunner, C.; Hoffmann, T.K.; Theodoraki, M.-N. The emerging role of exosomes in diagnosis, prognosis, and therapy in head and neck cancer. Int. J. Mol. Sci. 2020, 21, 4072. [Google Scholar] [CrossRef]
- Patel, A.; Patel, S.; Patel, P.; Tanavde, V. Saliva Based Liquid Biopsies in Head and Neck Cancer: How Far Are We from the Clinic? Front. Oncol. 2022, 12, 828434. [Google Scholar] [CrossRef]
- Teng, Y.; Gao, L.; Loveless, R.; Rodrigo, J.P.; Strojan, P.; Willems, S.M.; Nathan, C.-A.; Mäkitie, A.A.; Saba, N.F.; Ferlito, A. The Hidden Link of Exosomes to Head and Neck Cancer. Cancers 2021, 13, 5802. [Google Scholar] [CrossRef]
- Kaur, S.; Elkahloun, A.G.; Arakelyan, A.; Young, L.; Myers, T.G.; Otaizo-Carrasquero, F.; Wu, W.; Margolis, L.; Roberts, D.D. CD63, MHC class 1, and CD47 identify subsets of extracellular vesicles containing distinct populations of noncoding RNAs. Sci. Rep. 2018, 8, 2577. [Google Scholar] [CrossRef]
- Du, Q.; Ye, X.; Lu, S.-R.; Li, H.; Liu, H.-Y.; Zhai, Q.; Yu, B. Exosomal miR-30a and miR-222 derived from colon cancer mesenchymal stem cells promote the tumorigenicity of colon cancer through targeting MIA3. J. Gastrointest. Oncol. 2021, 12, 52–68. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, M.; Wu, J.; Jie, Z.; Chang, S.; Shuang, T. The clinicopathological significance of miR-1307 in chemotherapy resistant epithelial ovarian cancer. J. Ovarian Res. 2015, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.Y.; Sun, L.; Guo, Z.R.; Li, J.C.; Bai, T.T.; Cai, X.X.; Li, W.H.; Zhu, Y.F. Upregulated expression of serum exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J. Ovarian Res. 2019, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Albino, D.; Longoni, N.; Curti, L.; Mello-Grand, M.; Pinton, S.; Civenni, G.; Carbone, G.M. ESE3/EHF Controls Epithelial Cell Differentiation and Its Loss Leads to Prostate Tumors with Mesenchymal and Stem-like Features. Cancer Res. 2012, 72, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Endo, K.; Sakamoto, K.; Kayamori, K.; Ehata, S.; Ichikawa, J.; Ando, T.; Nakamura, R.; Kimura, Y.; Yoshizawa, K.; et al. EHF suppresses cancer progression by inhibiting ETS1-mediated ZEB expression. Oncogenesis 2021, 10, 1–15. [Google Scholar] [CrossRef]
- Oyelakin, A.; Nayak, K.B.; Glathar, A.R.; Gluck, C.; Wrynn, T.; Tugores, A.; Romano, R.-A.; Sinha, S. EHF is a novel regulator of cellular redox metabolism and predicts patient prognosis in HNSCC. NAR Cancer 2022, 4, zcac017. [Google Scholar] [CrossRef]
- Senft, D.; Qi, J.; Ronai, Z. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Cancer 2017, 18, 69–88. [Google Scholar] [CrossRef]
- Tatham, M.H.; Geoffroy, M.-C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Palvimo, J.J.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef]
- Ito, K.; Carracedo, A.; Weiss, D.; Arai, F.; Ala, U.; Avigan, D.E.; Schafer, Z.T.; Evans, R.M.; Suda, T.; Lee, C.-H.; et al. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 2012, 18, 1350–1358. [Google Scholar] [CrossRef]
- Wang, X.; Wang, G.; Shi, Y.; Sun, L.; Gorczynski, R.; Li, Y.-J.; Xu, Z.; Spaner, E.D. PPAR-delta promotes survival of breast cancer cells in harsh metabolic conditions. Oncogenesis 2016, 5, e232. [Google Scholar] [CrossRef]
- Huang, W.-C.; Jang, T.-H.; Tung, S.-L.; Yen, T.-C.; Chan, S.-H.; Wang, L.-H. A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing β5-integrin/c-met signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, V.; Campos, M.; Melo, R.L.; Ferro, E.S.; Camargo, A.C.M.; Juliano, M.A.; Juliano, L. Substrate Specificity Characterization of Recombinant Metallo Oligo-Peptidases Thimet Oligopeptidase and Neurolysin. Biochemistry 2001, 40, 4417–4425. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Gewehr, M.C.F.; Navon, A. Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions. Biomolecules 2020, 10, 1229. [Google Scholar] [CrossRef] [PubMed]
- Koike, K.; Masuda, T.; Sato, K.; Motomura, Y.; Takahashi, J.; Shimizu, D.; Mimori, K. Clinical significance of GET4 expression in colorectal cancer. Cancer Res. 2019, 79 (Suppl. 13), 5238. [Google Scholar] [CrossRef]
- Koike, K.; Masuda, T.; Sato, K.; Fujii, A.; Wakiyama, H.; Tobo, T.; Nakagawa, T. GET4 is a novel driver gene in colorectal cancer that regulates the localization of BAG6, a nucleocytoplasmic shuttling protein. Cancer Sci. 2022, 113, 156. [Google Scholar] [CrossRef]
- Davidovich, E.; Aframian, D.J.; Shapira, J.; Peretz, B. A comparison of the sialochemistry, oral pH, and oral health status of down syndrome children to healthy children. Int. J. Paediatr. Dent. 2010, 20, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, E.N.; de Souza Fonseca, M.A.; Dezem, F.S.; Lester, J.; Karlan, B.Y.; Noushmehr, H.; Lin, X.; Lawrenson, K. Optimizing exosomal RNA isolation for RNA-Seq analyses of archival sera specimens. PLoS ONE 2018, 13, e0196913. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. In Babraham Bioinformatics; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]


| Gene | Log2FC | |
|---|---|---|
| TCGA | RNASeq | |
| Downregulated | ||
| NPC1L1 | −6.24 | −7.32 |
| ZNF726 | −3.05 | −5.67 |
| HIST1H3A | −4.89 | −4.65 |
| ADCY9 | −4.23 | −3.98 |
| EHF | −2.51 | −3.06 |
| RNF4 | −2.52 | −2.89 |
| HIST1H2BD | −2.95 | −2.74 |
| RNF24 | −2.67 | −2.45 |
| GET4 | −2.41 | −2.41 |
| PIM3 | −2.77 | −2.34 |
| GID8 | −1.68 | −2.27 |
| THOP1 | −2.27 | −2.19 |
| RNF114 | −2.65 | −2.02 |
| MGRN1 | −2.29 | −1.95 |
| SEC14L1 | −2.34 | −1.82 |
| CYTH2 | −1.18 | −1.72 |
| PIP5K1C | −1.98 | −1.50 |
| ITSN1 | −2.36 | −1.29 |
| PRKCE | −1.93 | −0.88 |
| Upregulated | ||
| SOX12 | 1.56 | 2.19 |
| UQCC1 | 2.17 | 2.46 |
| ENG | 3.54 | 2.79 |
| NDUFAF6 | 2.95 | 3.30 |
| TLCD2 | 3.50 | 3.31 |
| ING5 | 3.03 | 3.37 |
| LRRC45 | 3.22 | 3.40 |
| DPP4 | 2.60 | 3.63 |
| CDC42EP4 | 2.79 | 3.63 |
| PSMB2 | 3.19 | 3.90 |
| CSE1L | 3.94 | 3.97 |
| TEX30 | 3.56 | 4.05 |
| RPL7L1 | 3.86 | 4.06 |
| TMEM63C | 4.18 | 4.11 |
| PPFIA3 | 4.83 | 4.72 |
| PLXNA1 | 3.86 | 4.73 |
| IGFBP5 | 7.18 | 5.33 |
| TNFRSF8 | 6.07 | 6.78 |
| SPATA33 | 7.92 | 8.27 |
| EFNB3 | 8.48 | 8.50 |
| BNC2 | 7.29 | 8.56 |
| TBC1D31 | 7.77 | 8.98 |
| SORCS2 | 8.78 | 9.54 |
| Characteristic | Value = N% |
|---|---|
| Gender | |
| Male | 21 (100%) |
| Sample Type | |
| Tissue | 19 |
| Saliva | 12 |
| Age group at diagnosis (years) | |
| <50 | 11 (52.4%) |
| >50 | 10 (47.6%) |
| Age at diagnosis (years, M ± SD) | 48 ± 83 |
| Stage | |
| I/II | 8 (39.1%) |
| III/IV | 13 (61.9%) |
| Lymphovascular Invasion | |
| N0 | 15 (71.4%) |
| N1 | 0 |
| N2 | 2 (9.5%) |
| N3 | 4 (19.1%) |
| Relapse | |
| Yes | 3 (14.3%) |
| No | 18 (85.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, A.; Patel, S.; Patel, P.; Mandlik, D.; Patel, K.; Tanavde, V. Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients. Int. J. Mol. Sci. 2022, 23, 10639. https://doi.org/10.3390/ijms231810639
Patel A, Patel S, Patel P, Mandlik D, Patel K, Tanavde V. Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients. International Journal of Molecular Sciences. 2022; 23(18):10639. https://doi.org/10.3390/ijms231810639
Chicago/Turabian StylePatel, Aditi, Shanaya Patel, Parina Patel, Dushyant Mandlik, Kaustubh Patel, and Vivek Tanavde. 2022. "Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients" International Journal of Molecular Sciences 23, no. 18: 10639. https://doi.org/10.3390/ijms231810639
APA StylePatel, A., Patel, S., Patel, P., Mandlik, D., Patel, K., & Tanavde, V. (2022). Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients. International Journal of Molecular Sciences, 23(18), 10639. https://doi.org/10.3390/ijms231810639







