Transcriptome Analyses Reveal Essential Roles of Alternative Splicing Regulation in Heat-Stressed Holstein Cows
Abstract
:1. Introduction
2. Results
2.1. Heat Stress Affects the Productive Performance and Physiological Indicators of Holstein Cows
2.2. Heat Stress Induces Changes in Blood Biochemical Parameters of Holstein Cows
2.3. Descriptive Statistics of the RNA Sequencing Data
2.4. Overview of Alternative Splicing Events
2.5. Alternative Splicing Events Differentially Expressed between the Two Groups
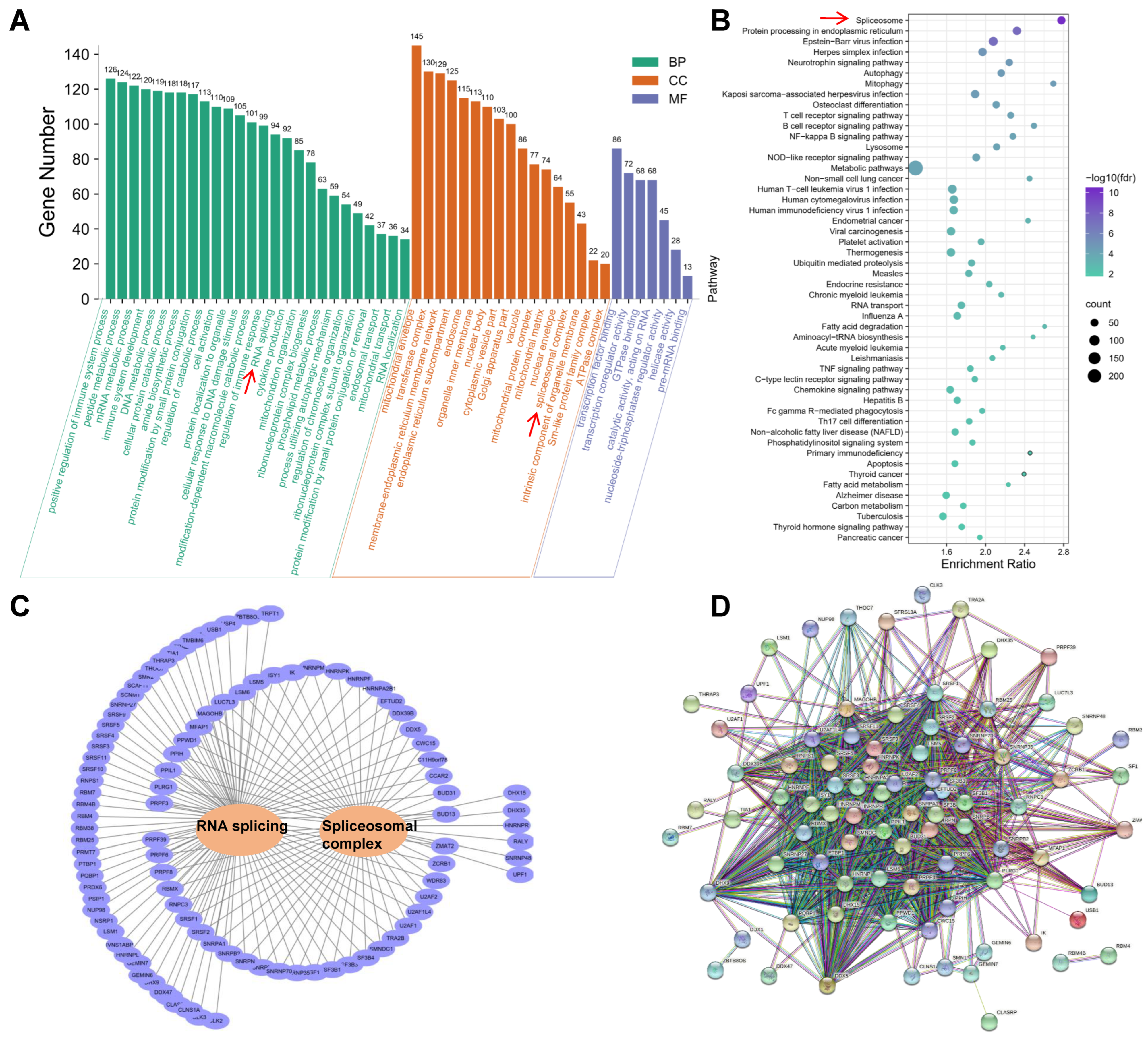
2.6. Functional Annotation of Genes with Differentially Alternatively Spliced Events
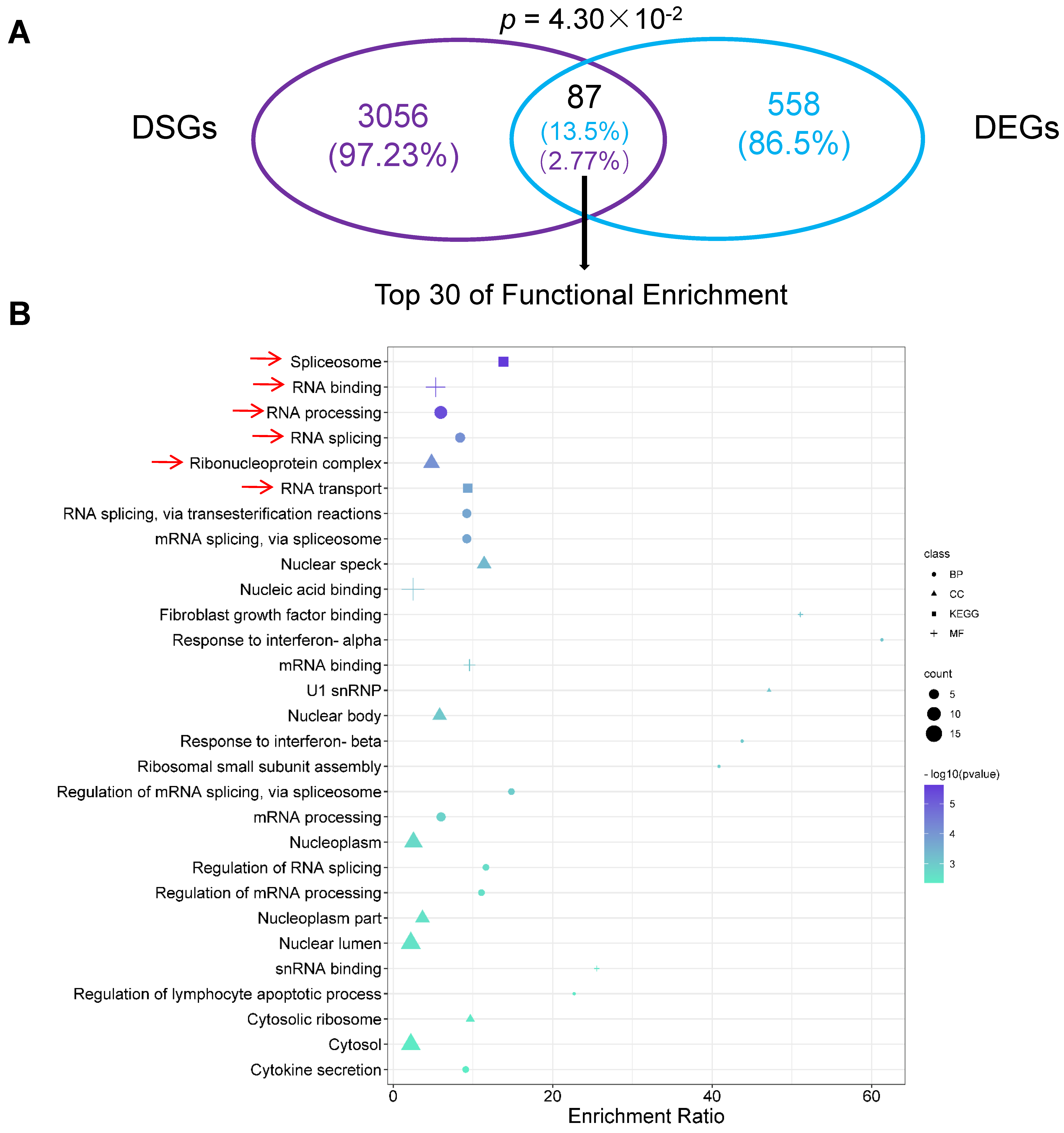
2.7. Differentially Alternatively Spliced Events in Differentially Expressed Genes
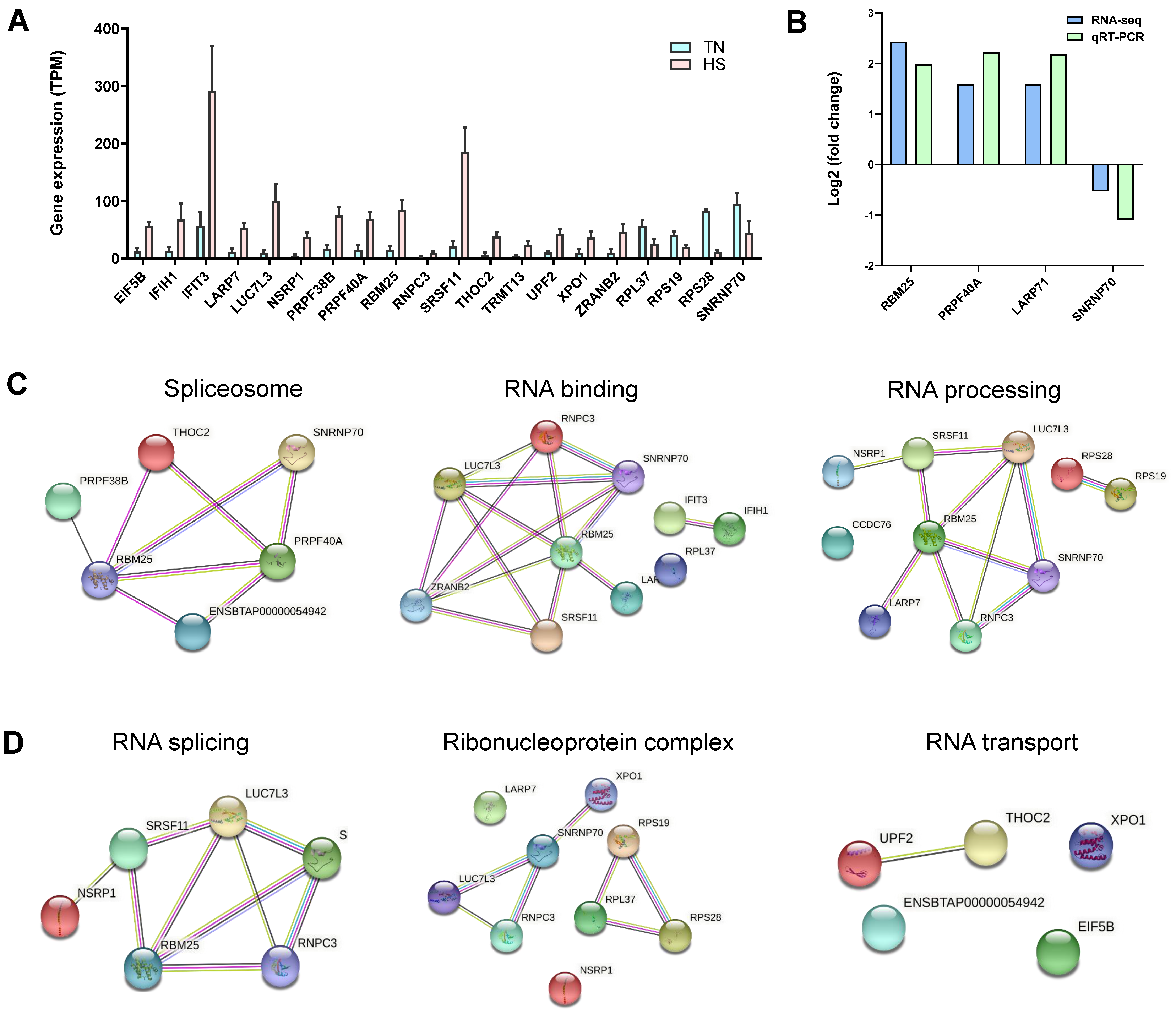
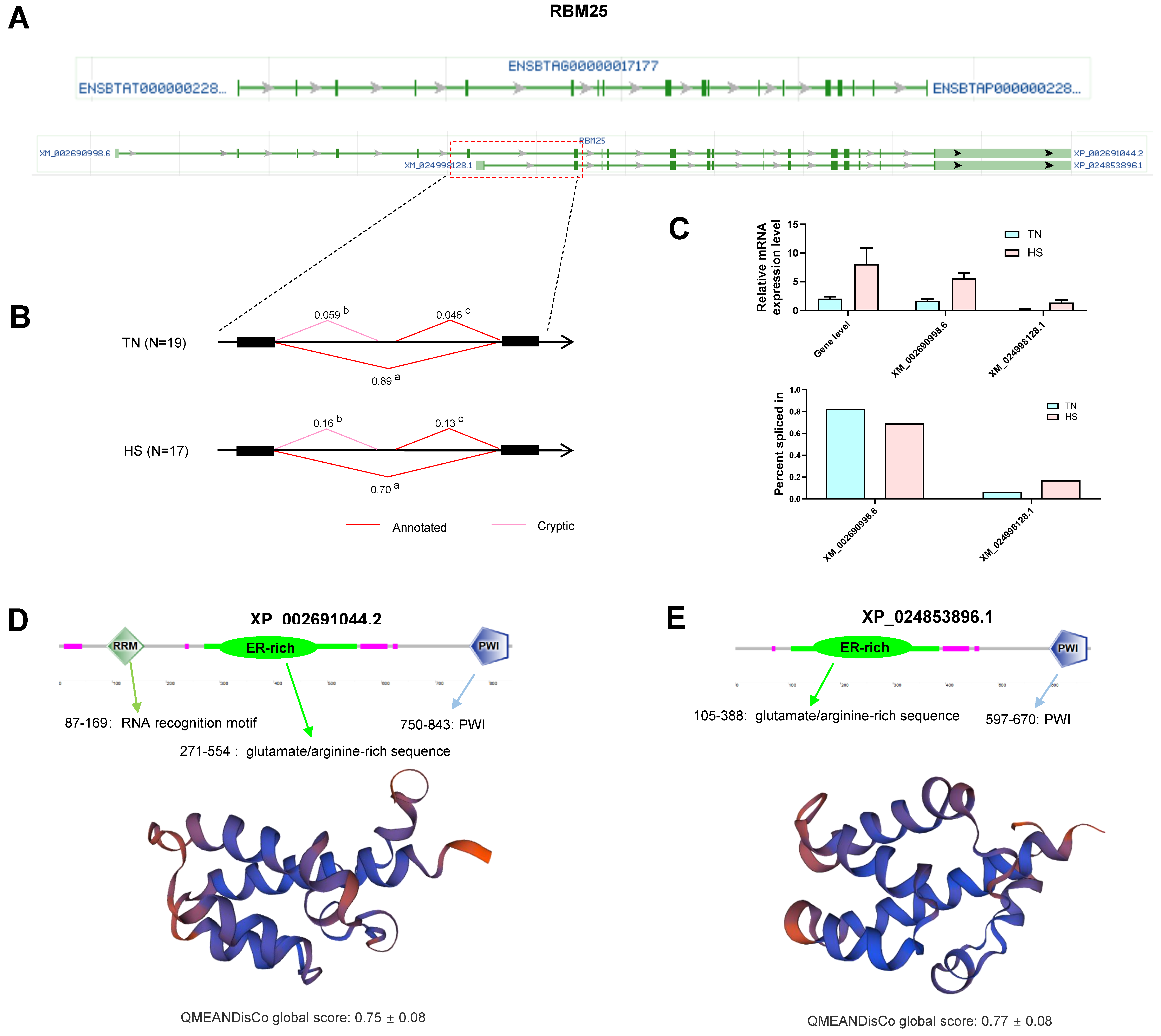
2.8. Gene Analysis for Differentially Alternative Spliced Events
3. Discussion
3.1. Effects of Heat Stress on Physiological Indicators and Milk Production
3.2. Effects of Heat Stress on Blood Biochemical Parameters
3.3. Effects of Heat Stress on Alternative Splicing
4. Materials and Methods
4.1. Sample Collection
4.2. Phenotypic Measurements
4.3. Determination of Biochemical Parameters
4.4. Library Preparation and RNA Sequencing
4.5. Quality Control and Reads Alignment
4.6. Differentially Expressed Gene Analyses
4.7. Alternative Splicing Analysis
4.8. Functional Annotation of Differentially Alternative Splicing Genes
4.9. Identification of Genes That Were Both Differentially Expressed and Differentially Alternatively Spliced
4.10. Validation of Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tao, S.; Orellana Rivas, R.M.; Marins, T.N.; Chen, Y.-C.; Gao, J.; Bernard, J.K. Impact of heat stress on lactational performance of dairy cows. Theriogenology 2020, 150, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Menta, P.R.; Machado, V.S.; Piñeiro, J.M.; Thatcher, W.W.; Santos, J.E.P.; Vieira-Neto, A. Heat stress during the transition period is associated with impaired production, reproduction, and survival in dairy cows. J. Dairy Sci. 2022, 105, 4474–4489. [Google Scholar] [CrossRef] [PubMed]
- Dahl, G.E.; Tao, S.; Laporta, J. Heat Stress Impacts Immune Status in Cows Across the Life Cycle. Front. Vet. Sci. 2020, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Brody, S. Climatic Physiology of Cattle1. J. Dairy Sci. 1956, 39, 715–725. [Google Scholar] [CrossRef]
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal 2021, 15, 100292. [Google Scholar] [CrossRef]
- Sammad, A.; Wang, Y.J.; Umer, S.; Lirong, H.; Khan, I.; Khan, A.; Ahmad, B.; Wang, Y. Nutritional Physiology and Biochemistry of Dairy Cattle under the Influence of Heat Stress: Consequences and Opportunities. Animals 2020, 10, 793. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I.; Hoogenboom, G. Genetic component of heat stress in dairy cattle, development of heat index function. J. Dairy Sci. 2000, 83, 2120–2125. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Bowman, P.J.; Haile-Mariam, M.; Pryce, J.E.; Hayes, B.J. Genomic selection for tolerance to heat stress in Australian dairy cattle. J. Dairy Sci. 2016, 99, 2849–2862. [Google Scholar] [CrossRef]
- Garner, J.B.; Chamberlain, A.J.; Vander Jagt, C.; Nguyen, T.T.T.; Mason, B.A.; Marett, L.C.; Leury, B.J.; Wales, W.J.; Hayes, B.J. Gene expression of the heat stress response in bovine peripheral white blood cells and milk somatic cells in vivo. Sci. Rep. 2020, 10, 19181. [Google Scholar] [CrossRef]
- Yue, S.; Wang, Z.; Wang, L.; Peng, Q.; Xue, B. Transcriptome Functional Analysis of Mammary Gland of Cows in Heat Stress and Thermoneutral Condition. Animals 2020, 10, 1015. [Google Scholar] [CrossRef]
- Koch, F.; Thom, U.; Albrecht, E.; Weikard, R.; Nolte, W.; Kuhla, B.; Kuehn, C. Heat stress directly impairs gut integrity and recruits distinct immune cell populations into the bovine intestine. Proc. Natl. Acad. Sci. USA 2019, 116, 10333–10338. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. Why genes in pieces? Nature 1978, 271, 501. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef]
- Chen, W.; Moore, M.J. Spliceosomes. Curr. Biol. 2015, 25, R181–R183. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef]
- Takechi, H.; Hosokawa, N.; Hirayoshi, K.; Nagata, K. Alternative 5′ splice site selection induced by heat shock. Mol. Cell. Biol. 1994, 14, 567–575. [Google Scholar]
- Fujikake, N.; Nagai, Y.; Popiel, H.A.; Kano, H.; Yamaguchi, M.; Toda, T. Alternative splicing regulates the transcriptional activity of Drosophila heat shock transcription factor in response to heat/cold stress. FEBS Lett. 2005, 579, 3842–3848. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, C.; Zhu, D.; He, H.; Wei, Z.; Yuan, Q.; Li, X.; Gao, X.; Zhang, B.; Gao, H.; et al. Identifying candidate genes and patterns of heat-stress response in rice using a genome-wide association study and transcriptome analyses. Crop J. 2022. [Google Scholar] [CrossRef]
- Sun, J.; Liu, Z.; Quan, J.; Li, L.; Zhao, G.; Lu, J. RNA-seq Analysis Reveals Alternative Splicing Under Heat Stress in Rainbow Trout (Oncorhynchus mykiss). Mar. Biotechnol. 2022, 24, 5–17. [Google Scholar] [CrossRef]
- Huang, S.; Dou, J.; Li, Z.; Hu, L.; Yu, Y.; Wang, Y. Analysis of Genomic Alternative Splicing Patterns in Rat under Heat Stress Based on RNA-Seq Data. Genes 2022, 13, 358. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.; Hoffmann, G.; Ammon, C.; Amon, B.; Heuwieser, W.; Halachmi, I.; Banhazi, T.; Amon, T. Influence of Barn Climate, Body Postures and Milk Yield on the Respiration Rate of Dairy Cows. Ann. Anim. Sci. 2019, 19, 469–481. [Google Scholar] [CrossRef]
- Sammad, A.; Umer, S.; Shi, R.; Zhu, H.; Zhao, X.; Wang, Y. Dairy cow reproduction under the influence of heat stress. J. Anim. Physiol. Anim. Nutr. 2020, 104, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Burfeind, O.; Suthar, V.S.; Heuwieser, W. Effect of heat stress on body temperature in healthy early postpartum dairy cows. Theriogenology 2012, 78, 2031–2038. [Google Scholar] [CrossRef]
- Godyń, D.; Nowicki, J.; Herbut, P. Effects of Environmental Enrichment on Pig Welfare—A Review. Animals 2019, 9, 383. [Google Scholar] [CrossRef]
- Dikmen, S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Genome-wide association mapping for identification of quantitative trait loci for rectal temperature during heat stress in Holstein cattle. PLoS ONE 2013, 8, e69202. [Google Scholar]
- Brito, L.F.; Oliveira, H.R.; McConn, B.R.; Schinckel, A.P.; Arrazola, A.; Marchant-Forde, J.N.; Johnson, J.S. Large-Scale Phenotyping of Livestock Welfare in Commercial Production Systems: A New Frontier in Animal Breeding. Front. Genet. 2020, 11, 793. [Google Scholar] [CrossRef]
- Mader, T.L.; Davis, M.S.; Brown-Brandl, T. Environmental factors influencing heat stress in feedlot cattle1,2. Anim. Sci. J. 2006, 84, 712–719. [Google Scholar] [CrossRef]
- Gaughan, J.B.; Mader, T.L.; Holt, S.M.; Sullivan, M.L.; Hahn, G.L. Assessing the heat tolerance of 17 beef cattle genotypes. Int. J. Biometeorol. 2010, 54, 617–627. [Google Scholar] [CrossRef]
- Gorniak, T.; Meyer, U.; Südekum, K.-H.; Dänicke, S. Impact of mild heat stress on dry matter intake, milk yield and milk composition in mid-lactation Holstein dairy cows in a temperate climate. Arch. Anim. Nutr. 2014, 68, 358–369. [Google Scholar] [CrossRef]
- West, J.W. Effects of heat-stress on production in dairy cattle. J. Dairy Sci. 2003, 86, 2131–2144. [Google Scholar] [CrossRef]
- Kim, W.-S.; Peng, D.-Q.; Jo, Y.-H.; Nejad, J.G.; Lee, H.-G. Responses of beef calves to long-term heat stress exposure by evaluating growth performance, physiological, blood and behavioral parameters. J. Therm. Biol. 2021, 100, 103033. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yi, H.; Wu, Q.; Jiang, Z.; Wang, L. Effects of acute heat stress on intestinal microbiota in grow-finishing pigs, and associations with feed intake and serum profile. J. Appl. Microbiol. 2020, 128, 840–852. [Google Scholar] [CrossRef] [PubMed]
- McMorris, T. Developing the catecholamines hypothesis for the acute exercise-cognition interaction in humans: Lessons from animal studies. Physiol. Behav. 2016, 165, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Cao, H.; Zhu, Y.; Wang, T.; Hu, G.; Kornmatitsuk, B.; Luo, J. Improving effects of dietary rumen protected gamma-aminobutyric acid additive on apparent nutrient digestibility, growth performance and health status in heat-stressed beef cattle. Anim. Sci. J. 2018, 89, 1280–1286. [Google Scholar] [CrossRef]
- Habashy, W.S.; Milfort, M.C.; Rekaya, R.; Aggrey, S.E. Cellular antioxidant enzyme activity and biomarkers for oxidative stress are affected by heat stress. Int. J. Biometeorol. 2019, 63, 1569–1584. [Google Scholar] [CrossRef]
- Fang, H.; Kang, L.; Abbas, Z.; Hu, L.; Chen, Y.; Tan, X.; Wang, Y.; Xu, Q. Identification of key Genes and Pathways Associated with Thermal Stress in Peripheral Blood Mononuclear Cells of Holstein Dairy Cattle. Front. Genet. 2021, 12, 662080. [Google Scholar] [CrossRef]
- Eisenhardt, S.U.; Thiele, J.R.; Bannasch, H.; Stark, G.B.; Peter, K. C-reactive protein: How conformational changes influence inflammatory properties. Cell Cycle 2009, 8, 3885–3892. [Google Scholar] [CrossRef]
- Sonna, L.A.; Fujita, J.; Gaffin, S.L.; Lilly, C.M. Invited Review: Effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. 2002, 92, 1725–1742. [Google Scholar]
- Sammad, A.; Luo, H.; Hu, L.; Zhu, H.; Wang, Y. Transcriptome Reveals Granulosa Cells Coping through Redox, Inflammatory and Metabolic Mechanisms under Acute Heat Stress. Cells 2022, 11, 1443. [Google Scholar] [CrossRef]
- Tan, S.; Wang, W.; Tian, C.; Niu, D.; Zhou, T.; Jin, Y.; Yang, Y.; Gao, D.; Dunham, R.; Liu, Z. Heat stress induced alternative splicing in catfish as determined by transcriptome analysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 29, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Bond, U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988, 7, 3509–3518. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of Alternative Pre-messenger RNA Splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Feng, Y.; Yang, P.; Cui, Y.; Liu, J.; Yang, C.; Gu, X. Transcriptome analysis reveals that constant heat stress modifies the metabolism and structure of the porcine longissimus dorsi skeletal muscle. Mol. Genet. Genom. 2016, 291, 2101–2115. [Google Scholar] [CrossRef]
- Liu, S.; Yue, T.; Ahmad, M.J.; Hu, X.; Zhang, X.; Deng, T.; Hu, Y.; He, C.; Zhou, Y.; Yang, L. Transcriptome Analysis Reveals Potential Regulatory Genes Related to Heat Tolerance in Holstein Dairy Cattle. Genes 2020, 11, 68. [Google Scholar] [CrossRef]
- Keil, L.C.; Summy-Long, J.; Severs, W.B. Release of Vasopressin by Angiotensin II1. Endocrinology 1975, 96, 1063–1065. [Google Scholar] [CrossRef] [PubMed]
- Mulrow, P.J. Angiotensin II and aldosterone regulation. Regul. Pept. 1999, 80, 27–32. [Google Scholar] [CrossRef]
- Shaji, S.; Sejian, V.; Bagath, M.; Manjunathareddy, G.B.; Kurien, E.K.; Varma, G.; Bhatta, R. Summer season related heat and nutritional stresses on the adaptive capability of goats based on blood biochemical response and hepatic HSP70 gene expression. Biol. Rhy. Res. 2017, 48, 65–83. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Servi, L.; Hernando, C.E.; Saura-Sanchez, M.; Yanovsky, M.J.; Petrillo, E.; Botto, J.F. Alternative Splicing Regulation During Light-Induced Germination of Arabidopsis thaliana Seeds. Front. Plant Sci. 2019, 10, 1076. [Google Scholar] [CrossRef]
- Carlson, S.M.; Soulette, C.M.; Yang, Z.; Elias, J.E.; Brooks, A.N.; Gozani, O. RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J. Biol. Chem. 2017, 292, 13381–13390. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Z.; Yuan, B.; Li, X. RBM25 Mediates Abiotic Responses in Plants. Front. Plant Sci. 2017, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Ou, A.C.; Cho, A.; Benz, E.J., Jr.; Huang, S.C. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell Biol. 2008, 28, 5924–5936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, Y.; Xia, K.; Yang, X.; Zhang, S.; Wang, L.; Ren, S.; Zhou, H.; Liu, Y.; Tang, F. Alternative splicing acts as an independent prognosticator in ovarian carcinoma. Sci. Rep. 2021, 11, 10413. [Google Scholar] [CrossRef] [PubMed]
- Daniels, N.J.; Hershberger, C.E.; Gu, X.; Schueger, C.; DiPasquale, W.M.; Brick, J.; Saunthararajah, Y.; Maciejewski, J.P.; Padgett, R.A. Functional analyses of human LUC7-like proteins involved in splicing regulation and myeloid neoplasms. Cell Rep. 2021, 35, 108989. [Google Scholar] [CrossRef] [PubMed]
- McDowell, R.E.; Hooven, N.W.; Camoens, J.K. Effect of Climate on Performance of Holsteins in First Lactation. J. Dairy Sci. 1976, 59, 965–971. [Google Scholar] [CrossRef]
- Luo, H.; Brito, L.F.; Li, X.; Su, G.; Dou, J.; Xu, W.; Yan, X.; Zhang, H.; Guo, G.; Liu, L.; et al. Genetic parameters for rectal temperature, respiration rate, and drooling score in Holstein cattle and their relationships with various fertility, production, body conformation, and health traits. J. Dairy Sci. 2021, 104, 4390–4403. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- DeLuca, D.S.; Levin, J.Z.; Sivachenko, A.; Fennell, T.; Nazaire, M.-D.; Williams, C.; Reich, M.; Winckler, W.; Getz, G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012, 28, 1530–1532. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.I.; Knowles, D.A.; Humphrey, J.; Barbeira, A.N.; Dickinson, S.P.; Im, H.K.; Pritchard, J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018, 50, 151–158. [Google Scholar] [CrossRef] [PubMed]



| Variable | Thermal Neutrality (TN, N = 19) 1 | Heat Stress (HS, N = 17) 1 | p-Value 2 |
|---|---|---|---|
| Milk production | |||
| 7AMY (kg/day) | 40.60 ± 8.77 | 36.95 ± 4.96 | 3.41 × 10−2 |
| MY (kg) | 41.33 ± 7.67 | 36.02 ± 5.63 | 2.67 × 10−2 |
| Rectal Temperature (°C) | |||
| 0700 h | 38.53 ± 0.19 | 38.64 ± 0.30 | 1.61 × 10−1 |
| 1400 h | 38.68 ± 0.14 | 39.00 ± 0.59 | 4.78 × 10−2 |
| 2100 h | 38.52 ± 0.19 | 39.00 ± 0.63 | 2.68 × 10−2 |
| Respiration Rate (breaths/min) | |||
| 0700 h | 35.16 ± 3.74 | 79.64 ± 18.15 | 2.41 × 10−8 |
| 1400 h | 40.52 ± 6.32 | 104.00 ± 20.30 | 3.06 × 10−10 |
| 2100 h | 35.26 ± 3.74 | 85.41 ± 25.27 | 2.13 × 10−10 |
| Drooling Score | |||
| 0700 h | 1.00 ± 0.00 | 1.24 ± 0.43 | 4.13 × 10−2 |
| 1400 h | 1.00 ± 0.00 | 1.82 ± 0.78 | 6.82 × 10−4 |
| 2100 h | 1.00 ± 0.00 | 1.41 ± 0.69 | 2.99 × 10−2 |
| Event | Gene | Description | TN (PSI 1) | HS (PSI 1) | HS-TN (ΔPSI) | p-Adjusted 2 |
|---|---|---|---|---|---|---|
| chr18:clu_38374_NA | TYROBP | Transmembrane immune signaling adaptor TYROBP | 0.39 | 0.28 | −0.11 | 1.34 × 10−86 |
| chr16:clu_44038_NA | AGTRAP | Angiotensin II receptor associated protein | 0.25 | 0.13 | −0.12 | 8.68 × 10−79 |
| chr13:clu_28601_NA | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 0.35 | 0.04 | −0.30 | 3.06 × 10−78 |
| chr12:clu_40742_NA | ENSBTAG00000053845 | Ras GTPase-activating protein 3-like | 0.67 | 0.36 | −0.32 | 1.17 × 10−70 |
| chrX:clu_27138_NA | BCAP31 | B cell receptor associated protein 31 | 0.76 | 0.41 | −0.35 | 5.54 × 10−69 |
| chr19:clu_25591_NA | JPT1; NT5C | Jupiter microtubule associated homolog 1; 5′,3′-nucleotidase, cytosolic | 0.32 | 0.19 | −0.13 | 1.54 × 10−59 |
| chr8:clu_30411_NA | CCDC107 | Coiled-coil domain containing 107 | 0.46 | 0.06 | −0.40 | 5.80 × 10−59 |
| chr15:clu_11260_NA | ILK | Integrin linked kinase | 0.69 | 0.49 | −0.20 | 4.93 × 10−58 |
| chr28:clu_4496_NA | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | 0.32 | 0.16 | −0.16 | 6.49 × 10−56 |
| chr11:clu_41811_NA | VAMP5 | Vesicle associated membrane protein 5 | 0.90 | 0.56 | −0.35 | 7.50 × 10−56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Sammad, A.; Zhang, C.; Brito, L.F.; Xu, Q.; Wang, Y. Transcriptome Analyses Reveal Essential Roles of Alternative Splicing Regulation in Heat-Stressed Holstein Cows. Int. J. Mol. Sci. 2022, 23, 10664. https://doi.org/10.3390/ijms231810664
Hu L, Sammad A, Zhang C, Brito LF, Xu Q, Wang Y. Transcriptome Analyses Reveal Essential Roles of Alternative Splicing Regulation in Heat-Stressed Holstein Cows. International Journal of Molecular Sciences. 2022; 23(18):10664. https://doi.org/10.3390/ijms231810664
Chicago/Turabian StyleHu, Lirong, Abdul Sammad, Congcong Zhang, Luiz F. Brito, Qing Xu, and Yachun Wang. 2022. "Transcriptome Analyses Reveal Essential Roles of Alternative Splicing Regulation in Heat-Stressed Holstein Cows" International Journal of Molecular Sciences 23, no. 18: 10664. https://doi.org/10.3390/ijms231810664
APA StyleHu, L., Sammad, A., Zhang, C., Brito, L. F., Xu, Q., & Wang, Y. (2022). Transcriptome Analyses Reveal Essential Roles of Alternative Splicing Regulation in Heat-Stressed Holstein Cows. International Journal of Molecular Sciences, 23(18), 10664. https://doi.org/10.3390/ijms231810664








