Bioactive Compounds from Marine Sponges and Algae: Effects on Cancer Cell Metabolome and Chemical Structures
Abstract
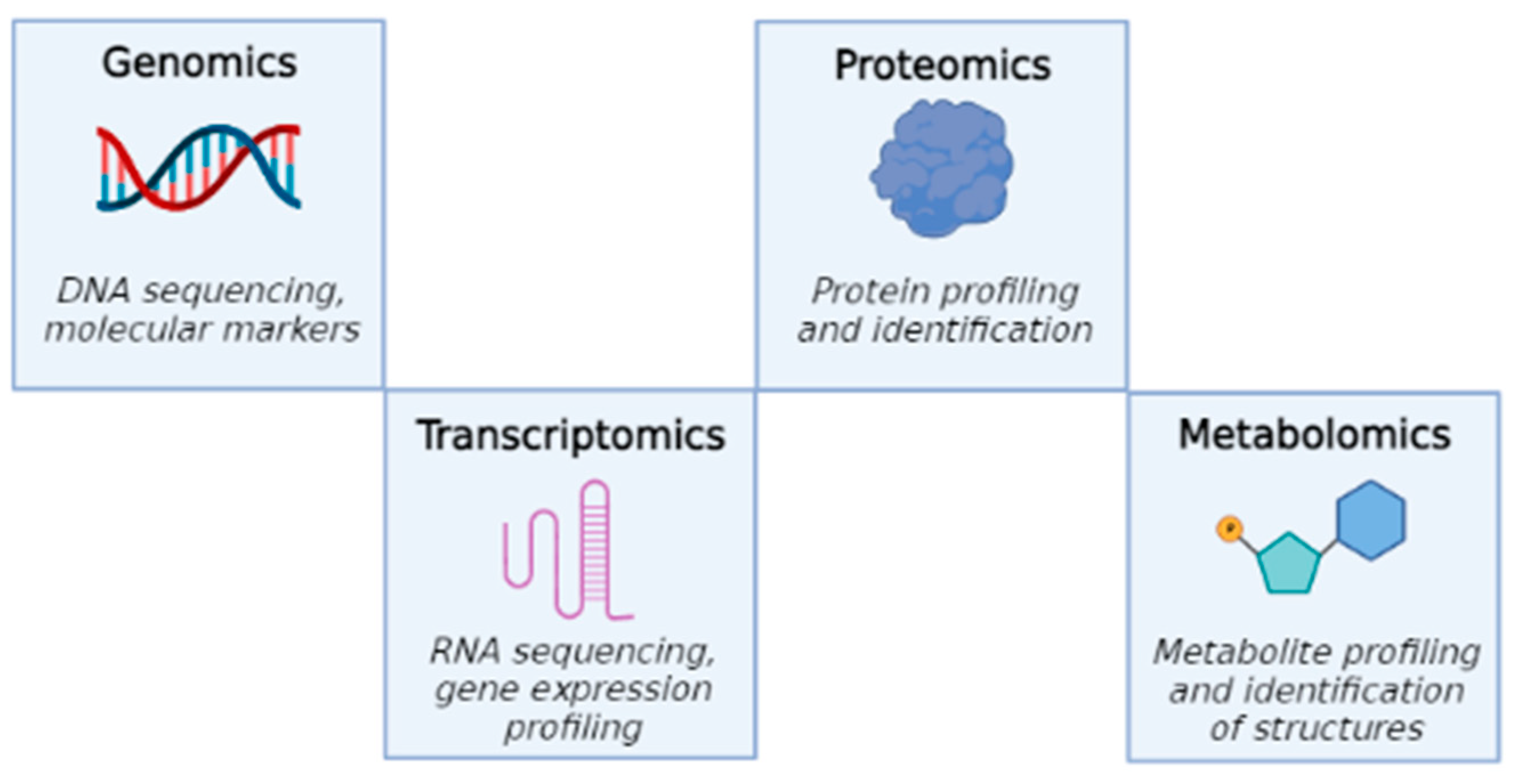
:1. Introduction: A New “Omics” Technology: Metabolomics
2. Main Metabolomics Methodologies
2.1. Sample and Extraction Techniques
2.2. Separation Techniques
2.3. Detection Techniques
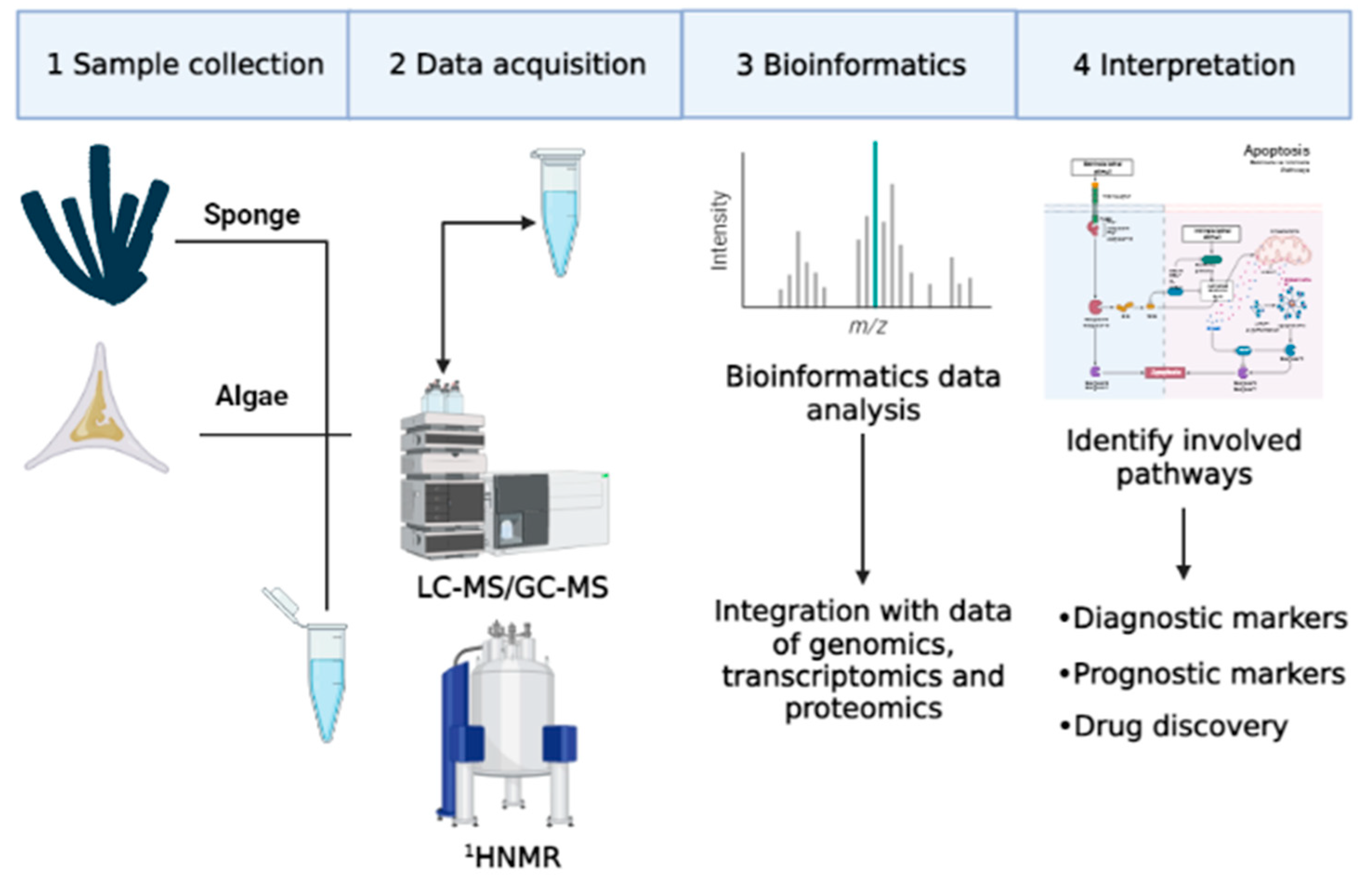
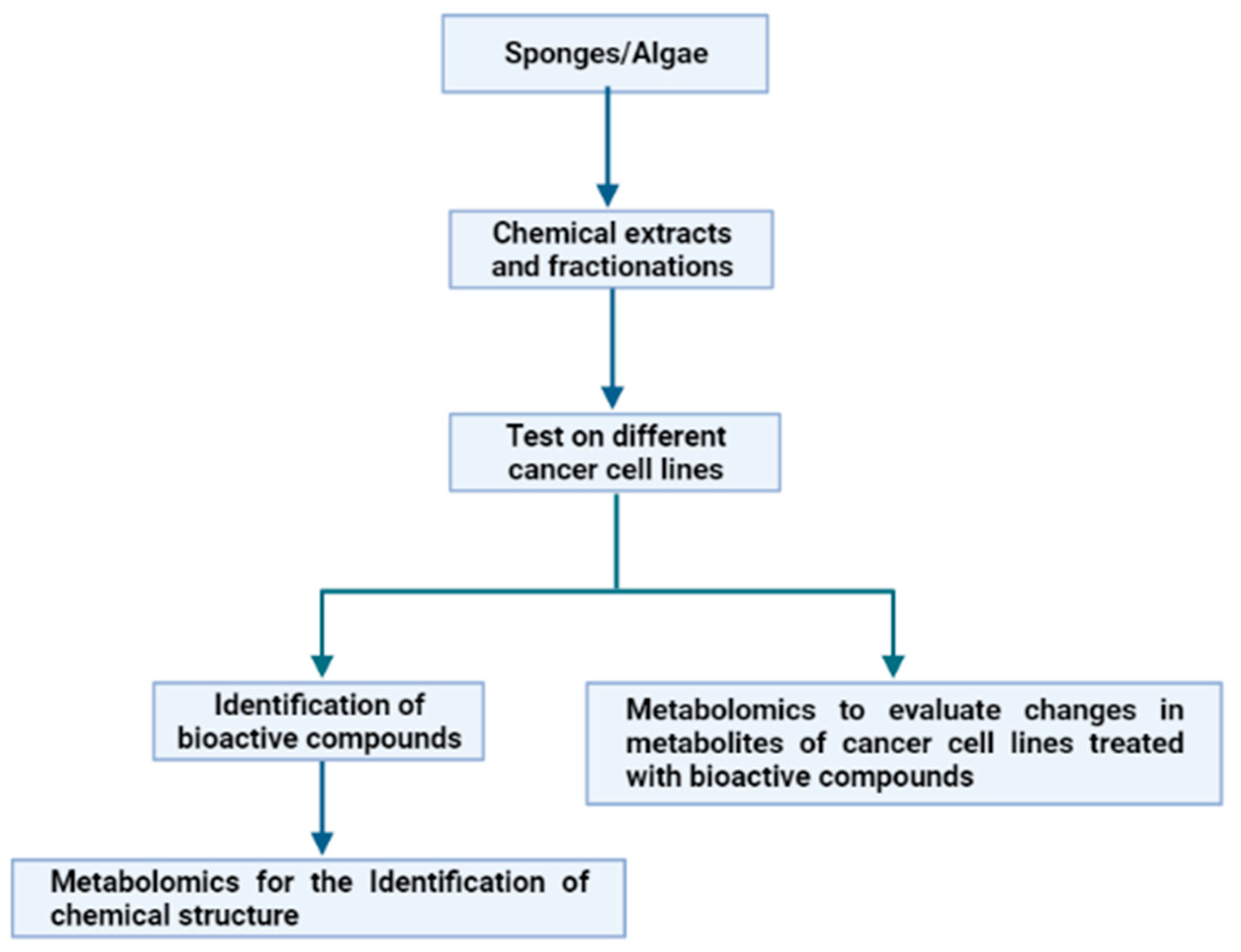
3. Metabolomics Applied to Marine Organisms in Cancer Studies
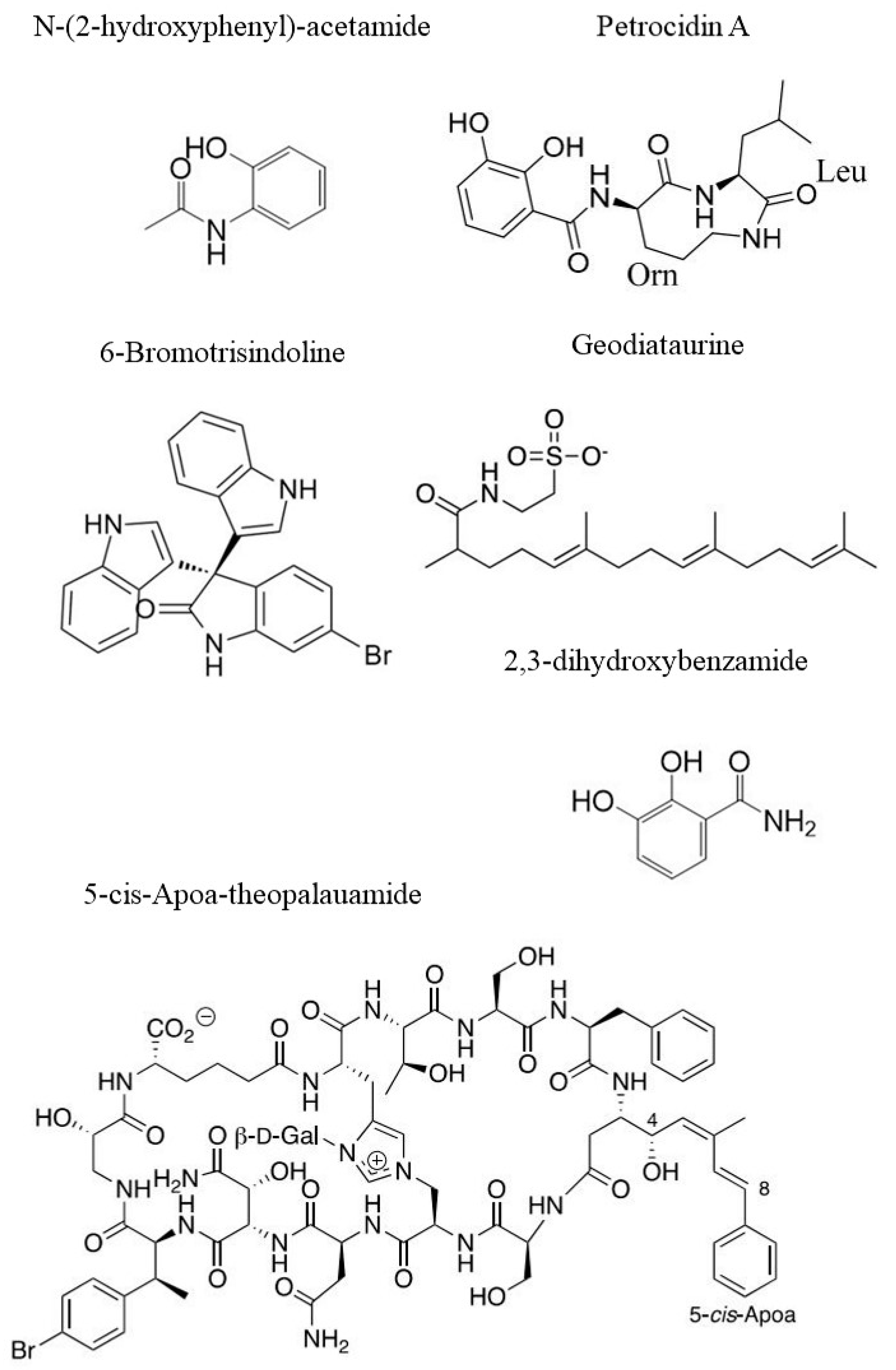
3.1. Structure Elucidation of Bioactive Molecules from Sponges
3.2. Structure Elucidation of Bioactive Molecules from Algae
3.3. Metabolite Changes in Treated Cancer Cell Lines
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, I.D.; Theodoridis, G.; Virgiliou, C. A perspective on the standards describing mass spectrometry-based metabolic phenotyping (metabolomics/metabonomics) studies in publications. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1164, 122515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst. 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Rochfort, S. Metabolomics reviewed: A new “omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Applications of metabolomics in drug discovery and development. Drugs R D 2008, 9, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics: Applications to food science and nutrition research. Trends Food Sci. Technol. 2008, 19, 482–493. [Google Scholar] [CrossRef]
- Wang, X.; Sun, H.; Zhang, A.; Sun, W.; Wang, P.; Wang, Z. Potential role of metabolomics apporoaches in the area of traditional Chinese medicine: As pillars of the bridge between Chinese and Western medicine. J. Pharm. Biomed. Anal. 2011, 55, 859–868. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, Z.; Sun, W.; Wang, P.; Wang, X. Metabolomics: Towards understanding traditional Chinese medicine. Planta Med. 2010, 76, 2026–2035. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Goulitquer, S.; Potin, P.; Tonon, T. Mass spectrometry-based metabolomics to elucidate functions in marine organisms and ecosystems. Mar. Drugs 2012, 10, 849–880. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based plant metabolomics: Where do we stand, where do we go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.C.; Helton, E.D.; Goldzieher, J.W. Chromatographiic profiling and identification of ethynyl and non-ethynyl compounds. Steroids 1975, 25, 229–244. [Google Scholar] [CrossRef]
- Krstulovic, A.M.; Matzura, C.T.; Bertani-Dziedzic, L.; Cerqueira, S.; Gitlow, S.E. Endogenous levels of free and conjugated urinary 3-methoxy-4-hydroxyphenylethyleneglycol in control subjects and patients with pheochromatography with electrochemical detection. Clin. Chim. Acta 1980, 103, 109–116. [Google Scholar] [CrossRef]
- Muskiet, F.A.J.; Stratingh, M.C.; Stob, G.J.; Wolthers, B.G. Simultaneous determination of the four major catecholamine metabolites and estimation of a serotonin metabolite in urine by capillary gas chromatography of their tert-butyldimethylsilyl derivatives. Clin Chem 1981, 27, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.; Hoult, J.R.S.; Waddell, K.A.; Blair, I.A.; Dollery, C.T. Total profiling by GC/NICIMS of the major cyclo-oxygenase products from antigen and leukotriene-challenged guinea-pig lung. Biochem. Pharmacol. 1984, 33, 395–400. [Google Scholar] [CrossRef]
- Weichert, H.; Kolbe, A.; Kraus, A.; Wasternack, C.; Feussner, I. Metabolic profiling of oxylipins in germinating cucumber seedlings-lipoxygenase-dependent degradation of triacylglycerols and biosynthesis of volatile aldehydes. Planta 2002, 215, 612–619. [Google Scholar] [CrossRef]
- Ruocco, N.; Costantini, S.; Zupo, V.; Lauritano, C.; Caramiello, D.; Ianora, A.; Budillon, A.; Romano, G.; Nuzzo, G.; D’Ippolito, G.; et al. Toxigenic effects of two benthic diatoms upon grazing activity of the sea urchin: Morphological, metabolomic and de novo transcriptomic analysis. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Miralto, A.; Barone, G.; Romano, G.; Poulet, S.A.; Ianora, A.; Russo, G.L.; Buttino, I.; Mazzarella, G.; Laablr, M.; Cabrini, M.; et al. The insidious effect of diatoms on copepod reproduction. Nature 1999, 402, 173–176. [Google Scholar] [CrossRef]
- Ruocco, N.; Varrella, S.; Romano, G.; Ianora, A.; Bentley, M.G.; Somma, D.; Leonardi, A.; Mellone, S.; Zuppa, A.; Costantini, M. Diatom-derived oxylipins induce cell death in sea urchin embryos activating caspase-8 and caspase 3/7. Aquat. Toxicol. 2016, 176, 128–140. [Google Scholar] [CrossRef]
- Sansone, C.; Braca, A.; Ercolesi, E.; Romano, G.; Palumbo, A.; Casotti, R.; Francone, M.; Ianora, A. Diatom-derived polyunsaturated aldehydes activate cell death in human cancer cell lines but not normal cells. PLoS ONE 2014, 9, e0101220. [Google Scholar] [CrossRef] [Green Version]
- González-Domínguez, R.; Sayago, A.; Fernández-Recamales, Á. Metabolomics in Alzheimer’s disease: The need of complementary analytical platforms for the identification of biomarkers to unravel the underlying pathology. J. Chromatogr. B 2017, 1071, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, P.; Schultz, A.J.; Hill, H.H. Metabolic profiling of human blood by high-resolution ion mobility mass spectrometry (IM-MS). Int. J. Mass Spectrom. 2010, 298, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Eeckhaut, A. Van Analytical techniques for metabolomic studies: A review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- Bayona, L.M.; de Voogd, N.J.; Choi, Y.H. Metabolomics on the study of marine organisms. Metabolomics 2022, 18, 17. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Houssen, W.E.; Jaspars, M. Isolation of marine natural products. In Natural Products Isolation; Humana Press: Totowa, NJ, USA, 2006; pp. 353–390. ISBN 9781617796241. [Google Scholar]
- Cutignano, A.; Nuzzo, G.; Ianora, A.; Luongo, E.; Romano, G.; Gallo, C.; Sansone, C.; Aprea, S.; Mancini, F.; D’Oro, U.; et al. Development and application of a novel SPE-method for bioassay-guided fractionation of marine extracts. Mar. Drugs 2015, 13, 5736–5749. [Google Scholar] [CrossRef]
- Ivanišević, J.; Thomas, O.P.; Lejeusne, C.; Chevaldonné, P.; Pérez, T. Metabolic fingerprinting as an indicator of biodiversity: Towards understanding inter-specific relationships among Homoscleromorpha sponges. Metabolomics 2011, 7, 289–304. [Google Scholar] [CrossRef]
- Anderson, P.E.; Mahle, D.A.; Doom, T.E.; Reo, N.V.; DelRaso, N.J.; Raymer, M.L. Dynamic adaptive binning: An improved quantification technique for NMR spectroscopic data. Metabolomics 2011, 7, 179–190. [Google Scholar] [CrossRef]
- Wu, W.; Daszykowski, M.; Walczak, B.; Sweatman, B.C.; Connor, S.C.; Haselden, J.N.; Crowther, D.J.; Gill, R.W.; Lutz, M.W. Peak alignment of urine NMR spectra using fuzzy warping. J. Chem. Inf. Model. 2006, 46, 863–875. [Google Scholar] [CrossRef]
- Forshed, J.; Schuppe-Koistinen, I.; Jacobsson, S.P. Peak alignment of NMR signals by means of a genetic algorithm. Anal. Chim. Acta 2003, 487, 189–199. [Google Scholar] [CrossRef]
- Izquierdo-García, J.L.; Villa, P.; Kyriazis, A.; Del Puerto-Nevado, L.; Pérez-Rial, S.; Rodriguez, I.; Hernandez, N.; Ruiz-Cabello, J. Descriptive review of current NMR-based metabolomic data analysis packages. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.C.; Gutenbrunner, P.; Kenar, E.; et al. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Marston, A. Role of advances in chromatographic techniques in phytochemistry. Phytochemistry 2007, 68, 2786–2798. [Google Scholar] [CrossRef]
- Soni, N.R. Improve GC separations with derivatization for selective response and detection in novel matrices. J. Environ. Life Sci. 2016, 1, 14–25. [Google Scholar]
- Issaq, H.J.; Van, Q.N.; Waybright, T.J.; Muschik, G.M.; Veenstra, T.D. Analytical and statistical approaches to metabolomics research. J. Sep. Sci. 2009, 32, 2183–2199. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.F. Stationary phases for packed-column supercritical fluid chromatography. J. Chromatogr. A 2012, 1250, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Jandera, P. Stationary and mobile phases in hydrophilic interaction chromatography: A review. Anal. Chim. Acta 2011, 692, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Pan, Z.; Xi, B.; Asiago, V.; Musselman, B.; Raftery, D. Principal component directed partial least squares analysis for combining nuclear magnetic resonance and mass spectrometry data in metabolomics: Application to the detection of breast cancer. Anal. Chim. Acta 2011, 686, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef]
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 2, 140–150. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Mas, S.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef]
- Lu, X.; Zhao, X.; Bai, C.; Zhao, C.; Lu, G.; Xu, G. LC-MS-based metabonomics analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 866, 64–76. [Google Scholar] [CrossRef]
- Wu, H.; Guo, J.; Chen, S.; Liu, X.; Zhou, Y.; Zhang, X.; Xu, X. Recent developments in qualitative and quantitative analysis of phytochemical constituents and their metabolites using liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2013, 72, 267–291. [Google Scholar] [CrossRef]
- Want, E.J.; Nordström, A.; Morita, H.; Siuzdak, G. From exogenous to endogenous: The inevitable imprint of mass spectrometry in metabolomics. J. Proteome Res. 2007, 6, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Miggiels, P.; Wouters, B.; van Westen, G.J.P.; Dubbelman, A.C.; Hankemeier, T. Novel technologies for metabolomics: More for less. Trends Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Leenders, J.; Frédérich, M.; De Tullio, P. Nuclear magnetic resonance: A key metabolomics platform in the drug discovery process. Drug Discov. Today Technol. 2015, 13, 39–46. [Google Scholar] [CrossRef]
- Weckwerth, W. Metabolomics in Systems Biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Nagana Gowda, G.A.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Batool, F.; Emwas, A.H.; Gao, X.; Munawar, M.A.; Chotana, G.A. Synthesis and Suzuki Cross-Coupling Reactions of 2,6-Bis(trifluoromethyl)pyridine-4-boronic Acid Pinacol Ester. Synthesis 2017, 49, 1327–1334. [Google Scholar] [CrossRef]
- Elbaz, A.M.; Gani, A.; Hourani, N.; Emwas, A.H.; Sarathy, S.M.; Roberts, W.L. TG/DTG, FT-ICR mass spectrometry, and NMR spectroscopy study of heavy fuel oil. Energy and Fuels 2015, 29, 7825–7835. [Google Scholar] [CrossRef]
- Abdul Jameel, A.G.; Oudenhoven, V.; Van Emwas, A.H.; Sarathy, S.M. Predicting octane number using nuclear magnetic resonance spectroscopy and artificial neural networks. Energy and Fuels 2018, 32, 6309–6329. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Reo, N.V. NMR-based metabolomics. Drug Chem. Toxicol. 2002, 25, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Godwa, G.A.N.; Raftery, D. Recent advances in NMR-based metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Evilia, R.F. Quantitative NMR spectroscopy. Anal. Lett. 2001, 34, 2227–2236. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. High resolution proton magnetic resonance spectroscopy of biological fluids. Prog. Nucl. Magn. Reson. Spectrosc. 1989, 21, 449–501. [Google Scholar] [CrossRef]
- Beger, R. A review of applications of metabolomics in cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1931, 8, 519. [Google Scholar] [CrossRef]
- Serkova, N.J.; Niemann, C.U. Pattern recognition and biomarker 1H-NMR-based metabolomics. Expert Rev. Mol. Diagn. 2006, 6, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Dang, C.V. Cancer’s molecular sweet tooth and the warburg effect. Cancer Res. 2006, 66, 8927–8930. [Google Scholar] [CrossRef]
- Pedersen, P.L. Warburg, me and Hexokinase 2: Multiple discoveries of key molecular events underlying one of cancers’ most common phenotypes, the “Warburg Effect”, i.e., elevated glycolysis in the presence of oxygen. J. Bioenerg. Biomembr. 2007, 39, 211–222. [Google Scholar] [CrossRef]
- Dunn, W.B. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys. Biol. 2008, 5, 011001. [Google Scholar] [CrossRef]
- Armitage, E.G.; Barbas, C. Metabolomics in cancer biomarker discovery: Current trends and future perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Rohde, S.; Parchemin, C.; Tapissier-Bontemps, N.; Schupp, P.J. Metabolomics and marine biotechnology: Coupling metabolite profiling and organism biology for the discovery of new compounds. Front. Mar. Sci. 2020, 7, 613471. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2012, 29, 144–222. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Costantini, M.; Sansone, C.; Lauritano, C.; Ruocco, N.; Ianora, A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar. Environ. Res. 2017, 128, 58–69. [Google Scholar] [CrossRef]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef]
- Hirata, Y.; Uemura, D. Halichondrins—Antitumor polyether macrolides from a marine sponge. Pure Appl. Chem. 1986, 58, 701–710. [Google Scholar] [CrossRef]
- Menis, J.; Twelves, C. Eribulin (Halaven): A new, effective treatment for women with heavily pretreated metastatic breast cancer. Breast Cancer Targets Ther. 2011, 3, 101–111. [Google Scholar] [CrossRef]
- Minouni, V.; Ulmann, L.; Pasquet, V.; Mathieu, M.; Picot, L.; Bougaran, G.; Cadoret, J.P.; Morant-Manceau, A.; Schoefs, B. The potential of microalgae for the production of bioactive molecules of pharmaceutical interest. Curr. Pharm. Biotechnol. 2012, 13, 2733–2750. [Google Scholar] [CrossRef]
- Atkin, S.L.; Backett, S.T.; Mackenzie, G. Topical Formulations Containing Sporopollenin. US Patent 20080311213 A1, 22 July 2014. [Google Scholar]
- Balskus, E.P.; Walsh, C.T. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 2010, 329, 1653–1656. [Google Scholar] [CrossRef]
- Dionisio-Se Se, M.L. Aquatic microalgae as potential sources of UV-screening compounds. Philipp. J. Sci. 2010, 139, 5–19. [Google Scholar]
- Priyadarshani, L.; Rath, B. Commercial and industrial applications of microalgae—A review. J. Alagl Biomass Util. 2012, 3, 89–100. [Google Scholar]
- Lordan, S.; Paul Ross, R.; Stanton, C. Marine bioactives as functional food ingredients: Potential to reduce the incidence of chronic diseases. Mar. Drugs 2011, 9, 1056–1100. [Google Scholar] [CrossRef] [PubMed]
- Gierhart, D.L.; Fox, J.A. Protection against Sunburn and Skin Problems with Orally-Ingested High Dosage Zeaxanthin. US Patent 8088363 B2, 3 January 2012. [Google Scholar]
- Guerin, M.; Huntley, M.E.; Olaizola, M. Haemotococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. [Google Scholar] [CrossRef]
- Tominaga, K.; Hongo, N.; Karato, M.; Yamashita, E. Cosmetic benefits od astaxanthin on humans sujects. Acta Biochim. Pol. 2012, 59, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.L.; Purton, S.; Becker, D.K.; Collet, C. Microalgae as bioreactors. Plant Cell Rep. 2005, 24, 629–641. [Google Scholar] [CrossRef]
- Storey, M.A.; Andreassend, S.K.; Bracegirdle, J.; Brown, A.; Keyzers, R.A.; Ackerley, D.F.; Northcote, P.T.; Owen, J.G. Metagenomic exploration of the marine sponge Mycale hentscheli uncovers multiple polyketide-producing bacterial symbionts. MBio 2020, 11, e02997-19. [Google Scholar] [CrossRef]
- Esposito, R.; Ruocco, N.; Viel, T.; Federico, S.; Zupo, V.; Costantini, M. Sponges and their symbionts as a source of valuable compounds in cosmeceutical field. Mar. Drugs 2021, 19, 444. [Google Scholar] [CrossRef]
- Mioso, R.; Marante, F.J.T.; Bezerra, R.D.S.; Borges, F.V.P.; Santos, B.V.D.O.; De Laguna, I.H.B. Cytotoxic compounds derived from marine sponges. A review (2010–2012). Molecules 2017, 22, 8. [Google Scholar] [CrossRef]
- Khan, S.; Al-Fadhli, A.A.; Tilvi, S. Discovery of cytotoxic natural products from Red Sea sponges: Structure and synthesis. Eur. J. Med. Chem. 2021, 220, 113491. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, R.F.A.; Habib, E.S.; Eltahawy, N.A.; Hassanean, H.A.; Ibrahim, A.K.; Mohammed, A.F.; Fayez, S.; Hayallah, A.M.; Yamada, K.; Behery, F.A.; et al. New cytotoxic natural products from the red sea sponge Stylissacarteri. Mar. Drugs 2020, 18, 241. [Google Scholar] [CrossRef] [PubMed]
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Hassan, H.M.; Rateb, M.E.; Amin, E.; Mohammed, T.A.; El-Mesery, M.; Muhsinah, A.B.; Alsayari, A.; et al. Bioactive brominated oxindole alkaloids from the red sea sponge Callyspongia siphonella. Mar. Drugs 2019, 17, 465. [Google Scholar] [CrossRef]
- Einarsdottir, E.; Magnusdottir, M.; Astarita, G.; Köck, M.; Ögmundsdottir, H.M.; Thorsteinsdottir, M.; Rapp, H.T.; Omarsdottir, S.; Paglia, G. Metabolic profiling as a screening tool for cytotoxic compounds: Identification of 3-alkyl pyridine alkaloids from sponges collected at a shallow water hydrothermal vent site North of Iceland. Mar. Drugs 2017, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Kliman, M.; Claude, E.; Geromanos, S.; Astarita, G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015, 407, 4995–5007. [Google Scholar] [CrossRef] [PubMed]
- Musa, A.; Elmaidomy, A.H.; Sayed, A.M.; Alzarea, S.I.; Al-Sanea, M.M.; Mostafa, E.M.; Hendawy, O.M.; Abdelgawad, M.A.; Ayoussif, K.; Refaat, H.; et al. Cytotoxic potential, metabolic profiling, and liposomes of Coscinoderma sp. crude extract supported by in silico analysis. Int. J. Nanomedicine 2021, 16, 3861. [Google Scholar] [CrossRef]
- Olsen, E.K.; Søderholm, K.L.; Isaksson, J.; Andersen, J.H.; Hansen, E. Metabolomic profiling reveals the N-Acyl-Taurine Geodiataurine in extracts from the marine sponge Geodia macandrewii (Bowerbank). J. Nat. Prod. 2016, 79, 1285–1291. [Google Scholar] [CrossRef]
- Li, F.; Peifer, C.; Janussen, D.; Tasdemir, D. New discorhabdin alkaloids from the antarctic deep-sea sponge Latrunculia biformis. Mar. Drugs 2019, 17, 439. [Google Scholar] [CrossRef]
- Hasin, O.; Shoham, S.; Kashman, Y.; Ilan, M.; Carmeli, S. Theonellamides J and K and 5-cis-Apoa-theopalauamide, bicyclic glycopeptides of the Red Sea sponge Theonella swinhoei. Mar. Drugs 2022, 20, 31. [Google Scholar] [CrossRef]
- Mohanty, I.; Podell, S.; Biggs, J.S.; Garg, N.; Allen, E.E.; Agarwal, V. Multi-omic profiling of Melophlus sponges reveals diverse metabolomic and microbiome architectures that are non-overlapping with ecological neighbors. Mar. Drugs 2020, 18, 124. [Google Scholar] [CrossRef]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Federico, S.; Bertolino, M.; Zupo, V.; Costantini, M. Marine Demospongiae: A challenging treasure of bioactive compounds. Mar. Drugs 2022, 20, 244. [Google Scholar] [CrossRef]
- Wang, G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 2006, 33, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Othman, E.M.; Stopper, H.; Edrada-Ebel, R.A.; Hentschel, U.; Abdelmohsen, U.R. Isolation of Petrocidin A, a new cytotoxic cyclic dipeptide from the marine sponge-derived bacterium Streptomyces sp. SBT348. Mar. Drugs 2017, 15, 383. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Attia, E.; Hajjar, D.; Anany, M.; Desoukey, S.; Fouad, M.; Kamel, M.; Wajant, H.; Gulder, T.; Abdelmohsen, U. New cytotoxic cyclic peptide from the marine sponge-associated Nocardiopsis sp. UR67. Mar. Drugs 2018, 16, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shady, N.H.; Tawfike, A.F.; Yahia, R.; Fouad, M.A.; Brachmann, A.O.; Piel, J.; Abdelmohsen, U.R.; Kamel, M.S. Cytotoxic activity of actinomycetes Nocardia sp. and Nocardiopsis sp. associated with marine sponge Amphimedon sp. Nat. Prod. Res. 2021, 36, 2917–2922. [Google Scholar] [CrossRef]
- Hifnawy, M.S.; Hassan, H.M.; Mohammed, R.; Fouda, M.M.; Sayed, A.M.; Hamed, A.A.; AbouZid, S.F.; Rateb, M.E.; Alhadrami, H.A.; Abdelmohsen, U.R. Induction of antibacterial metabolites by co-cultivation of two Red-Sea-sponge-associated actinomycetes Micromonospora sp. UR56 and Actinokinespora sp. EG49. Mar. Drugs 2020, 18, 243. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M.; Morrisono, L.; Rindi, F.; Mathieson, A.C.; Miranda, S.V.; Parker, B.C.; Langangen, A.; John, D.M.; Barbara, I.; et al. AlgaeBase: An on-line resource for algae. Cryptogam. Algol. 2014, 35, 105–115. [Google Scholar] [CrossRef]
- Ibanez, E.; Herrero, M.; Mendiola, J.A.; Castro-Puyana, M. Extraction and characterization of bioactive compounds with health benefits from marine resources: Macro and micro algae, cyanobacteria, and invertebrates. In Marine Bioactive Compounds: Souzrces, Characterization and Application; Springer Science & Business Media: Berlin, Germany, 2012; pp. 55–98. ISBN 9781461412472. [Google Scholar]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.; Romano, G.; Ianora, A. Bioactivity screening of microalgae for antioxidant, anti-inflammatory, anticancer, anti-diabetes, and antibacterial activities. Front. Mar. Sci. 2016, 3, 68. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122. [Google Scholar] [CrossRef]
- Lahaye, M.; Robic, A. Structure and function properties of Ulvan, a polysaccharide from green seaweeds. Biomacromolecules 2007, 8, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Thanh, T.T.T.; Quach, T.M.T.; Nguyen, T.N.; Vu Luong, D.; Bui, M.L.; Tran, T.T. Van Structure and cytotoxic activity of ulvan extracted from green seaweed Ulva lactuca. Int. J. Biol. Macromol. 2016, 93, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Mofeed, J.; Deyab, M.; El, A.; Sabry, N.; Ward, F. In vitro anticancer activity of five marine seaweeds extract from Egypt against human breast and colon cancer cell lines. Res. Sq. 2021, 11, 583–589. [Google Scholar]
- Geisen, U.; Zenthoefer, M.; Peipp, M.; Kerber, J.; Plenge, J.; Managò, A.; Fuhrmann, M.; Geyer, R.; Hennig, S.; Adam, D.; et al. Molecular mechanisms by which a Fucus vesiculosus extract mediates cell cycle inhibition and cell death in pancreatic cancer cells. Mar. Drugs 2015, 13, 4470–4491. [Google Scholar] [CrossRef] [PubMed]
- Zenthoefer, M.; Geisen, U.; Hofmann-Peiker, K.; Fuhrmann, M.; Kerber, J.; Kirchhöfer, R.; Hennig, S.; Peipp, M.; Geyer, R.; Piker, L.; et al. Isolation of polyphenols with anticancer activity from the Baltic Sea brown seaweed Fucus vesiculosus using bioassay-guided fractionation. J. Appl. Phycol. 2017, 29, 2021–2037. [Google Scholar] [CrossRef]
- Martin, L.J. Fucoxanthin and its metabolite fucoxanthinol in cancer prevention and treatment. Mar. Drugs 2015, 13, 4784–4798. [Google Scholar] [CrossRef] [Green Version]
- Rokkaku, T.; Kimura, R.; Ishikawa, C.; Yasumoto, T.; Senba, M.; Kanaya, F.; Mori, N. Anticancer effects of marine carotenoids, fucoxanthin and its deacetylated product, fucoxanthinol, on osteosarcoma. Int. J. Oncol. 2013, 43, 1176–1186. [Google Scholar] [CrossRef]
- Shao, P.; Chen, X.; Sun, P. In vitro antioxidant and antitumor activities of different sulfated polysaccharides isolated from three algae. Int. J. Biol. Macromol. 2013, 62, 155–161. [Google Scholar] [CrossRef]
- Abreu, A.C.; Molina-Miras, A.; Aguilera-Saéz, L.M.; López-Rosales, L.; Cerón-Garciá, M.D.C.; Sánchez-Mirón, A.; Olmo-Garciá, L.; Carrasco-Pancorbo, A.; Garciá-Camacho, F.; Molina-Grima, E.; et al. Production of amphidinols and other bioproducts of interest by the marine microalga Amphidinium carterae unraveled by nuclear magnetic resonance metabolomics approach coupled to multivariate data analysis. J. Agric. Food Chem. 2019, 67, 9667–9682. [Google Scholar] [CrossRef]
- AtaseverArslan, B.; Yılancıoğlu, K.; KuşoğluGültekin, S.; Albayrak, İ. Chemical constituent of Isochrysis galbana microalgae extract and its cytotoxic activities on leukemic cell lines. İstanbul J. Pharm. 2022, 52, 64–68. [Google Scholar] [CrossRef]
- Karakaş, C.Y.; TekarslanŞahin, H.; İnan, B.; Özçimen, D.; Erginer, Y. In vitro cytotoxic activity of microalgal extracts loaded nano–micro particles produced via electrospraying and microemulsion methods. Biotechnol. Prog. 2019, 35, e2876. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.A.; Mohamad, H.; Ghazaly, M.M.; Laith, A.A.; Abdullah, M.A. Cytotoxic effects of Tetraselmis suecica chloroform extracts with silver nanoparticle co-application on MCF-7, 4 T1, and Vero cell lines. J. Appl. Phycol. 2020, 32, 127–143. [Google Scholar] [CrossRef]
- Hussein, H.A.; Kassim, M.N.I.; Maulidiani, M.; Abas, F.; Abdullah, M.A. Cytotoxicity and 1H-NMR metabolomics analyses of microalgal extracts for synergistic application with Tamoxifen on breast cancer cells with reduced toxicity against Vero cells. Heliyon 2022, 8, e09192. [Google Scholar] [CrossRef] [PubMed]
- Fayyad, R.J.; Ali, A.N.M.; Dwaish, A.S.; Al-Abboodi, A.K.A. Anticancer activity of Spirulina platensis methanolic extracts against L20B and MCF7 human cancer cell lines. Plant Arch. 2019, 19, 1419–1426. [Google Scholar]
- El-Baz, F.K.; Hussein, R.A.; Mahmoud, K.; Abdo, S.M. Cytotoxic activity of carotenoid rich fractions from Haematococcus pluvialis and Dunaliella salina microalgae and the identification of the phytoconstituents using LC-DAD/ESI-MS. Phyther. Res. 2018, 32, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Savio, S.; Farrotti, S.; Paris, D.; Arnaìz, E.; Díaz, I.; Bolado, S.; Muñoz, R.; Rodolfo, C.; Congestri, R. Value-added co-products from biomass of the diatoms Staurosirella pinnata and Phaeodactylum tricornutum. Algal Res. 2020, 47, 101830. [Google Scholar] [CrossRef]
- Costantini, S.; Guerriero, E.; Teta, R.; Capone, F.; Caso, A.; Sorice, A.; Romano, G.; Ianora, A.; Ruocco, N.; Budillon, A.; et al. Evaluating the effects of an organic extract from the mediterranean sponge Geodia cydonium on human breast cancer cell lines. Int. J. Mol. Sci. 2017, 18, 2112. [Google Scholar] [CrossRef] [Green Version]
- Heavisides, E.; Rouger, C.; Reichel, A.F.; Ulrich, C.; Wenzel-Storjohann, A.; Sebens, S.; Tasdemir, D. Seasonal variations in the metabolome and bioactivity profile of Fucus vesiculosus extracted by an optimised, pressurised liquid extraction protocol. Mar. Drugs 2018, 16, 503. [Google Scholar] [CrossRef]
- Ale, M.T.; Maruyama, H.; Tamauchi, H.; Mikkelsen, J.D.; Meyer, A.S. Fucoidan from Sargassum sp. and Fucus vesiculosus reduces cell viability of lung carcinoma and melanoma cells in vitro and activates natural killer cells in mice in vivo. Int. J. Biol. Macromol. 2011, 49, 331–336. [Google Scholar] [CrossRef]
- Zaragozá, M.C.; López, D.; Sáiz, M.P.; Poquet, M.; Pérez, J.; Puig-Parellada, P.; Màrmol, F.; Simonetti, P.; Gardana, C.; Lerat, Y.; et al. Toxicity and antioxidant activity in vitro and in vivo of two Fucus vesiculosus extracts. J. Agric. Food Chem. 2008, 56, 7773–7780. [Google Scholar] [CrossRef]

| Sponge | Compound/Extract | Cell Lines | Metabolomics | Reference |
|---|---|---|---|---|
| S. carteri | Stylissamide A and Stylissoside A | MCF7 and HepG2 | LC-MS | [93] |
| C. siphonella | 5-bromotrisindoline and 6-bromotrisindoline | HT29, OVCAR3 and MM.1S | LC-MS | [94] |
| H. rosea | 3-alkyl pyridine alkaloids | SKBR3 | MS | [95] |
| Coscinoderma sp. | Crude extract | HepG2, MCF7 and Caco2 | LC-MS | [97] |
| G. macandrewii | Geodiataurine | A2058 | UHPLC and MS | [98] |
| L. biformis | (−)-discorhabdin L, (−)-1-acetyl-discorhabdin L and (+)-1-octacosatrienoyl-discorhabdin L | HCT116 | 1H-NMR and MS | [99] |
| T. swinhoei | Theopalauamide, 5-cis-Apoa-theopalauamide and Theonellamide K | HCT116 | 1D and 2D NMR; MS | [100] |
| P. ficiformis | Petrocidin A and 2,3-Dihydroxybenzamide | HL60 and HT29 | LC-MS | [105] |
| Callyspongia sp. | Nocardiotide A | CT26, HeLa and MM.1S | MS | [106] |
| Amphimedon sp. | Crude extract | HepG2, MCF7 and Caco2 | MS | [107] |
| Callyspongia sp. and S. vagabunda | N-(2-hydroxyphenyl)-acetamide | HCT116, HepG2 and MCF7 | LC–MS | [108] |
| Algae | Compound/Extract | Cell Lines | Metabolomics | Reference |
|---|---|---|---|---|
| U. lactuca | Ulvan | HepG2, MCF7 and Hela | 1H-NMR-MS | [114] |
| U. fasciata, U. lactuca, A. anceps, C. mediterranea and S. filipendula | Organic extract | MCF7 and HTC116 | GC-MS | [115] |
| F. vesiculosus | Hydrophilic extract | Panc1, PancTU1, Panc89 and Colo 357 | HPLC | [116] |
| F. vesiculosus | Crude extracts | Panc89 and PancTU1 | 1H-NMR | [117] |
| C. okamuranus | Fucoxanthin and fucoxanthinol | Saos-2, MNNG/HOS, 143 B and LM8 | HPLC-MS | [119] |
| U. fasciata, G. furcata and S. henslouianum | Sulphated polysaccharides | MKN45 and DLD | RFC | [120] |
| A. carterae | Methanolic extract | A549, HT29, MDA-MB-231 and PSN-1 | 1H-NMR and MS | [121] |
| I. galbana | Crude extract | K562, MOLT-4, U937, HL60 and Burkitt′s lymphoma | 1H-NMR, GC-MS | [122] |
| C. protothecoides and N. oculata | Crude extract | A172 and HTC116 | GC-MS | [123] |
| T. suecica | Crude extract | A172 and HTC116 | GC-MS | [124] |
| N. oculata, T. suecica and Chlorella sp. | Water, ethanolic and methanolic extracts | MCF7 and 4 T1 | 1H-NMR | [125] |
| S. platensis | Methanolic extract | L20 B and MCF7 | GC-MS | [126] |
| H. pluvialis and D. salina | Carotenoid fractions and polar fractions | HepG2, MCF7, HCT116 and A549 | LC-MS | [127] |
| P. tricornutum and S. pinnata | Hydrophilic and lipophilic fractions | CHL-1 | 1H-NMR | [128] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, R.; Federico, S.; Glaviano, F.; Somma, E.; Zupo, V.; Costantini, M. Bioactive Compounds from Marine Sponges and Algae: Effects on Cancer Cell Metabolome and Chemical Structures. Int. J. Mol. Sci. 2022, 23, 10680. https://doi.org/10.3390/ijms231810680
Esposito R, Federico S, Glaviano F, Somma E, Zupo V, Costantini M. Bioactive Compounds from Marine Sponges and Algae: Effects on Cancer Cell Metabolome and Chemical Structures. International Journal of Molecular Sciences. 2022; 23(18):10680. https://doi.org/10.3390/ijms231810680
Chicago/Turabian StyleEsposito, Roberta, Serena Federico, Francesca Glaviano, Emanuele Somma, Valerio Zupo, and Maria Costantini. 2022. "Bioactive Compounds from Marine Sponges and Algae: Effects on Cancer Cell Metabolome and Chemical Structures" International Journal of Molecular Sciences 23, no. 18: 10680. https://doi.org/10.3390/ijms231810680
APA StyleEsposito, R., Federico, S., Glaviano, F., Somma, E., Zupo, V., & Costantini, M. (2022). Bioactive Compounds from Marine Sponges and Algae: Effects on Cancer Cell Metabolome and Chemical Structures. International Journal of Molecular Sciences, 23(18), 10680. https://doi.org/10.3390/ijms231810680








